Abstract
The Rv0183 gene of the Mycobacterium tuberculosis H37Rv strain, which has been implicated as a lysophospholipase, was cloned and expressed in Escherichia coli. The purified Rv0183 protein did not show any activity when lysophospholipid substrates were used, but preferentially hydrolysed monoacylglycerol substrates with a specific activity of 290 units·mg−1 at 37 °C. Rv0183 hydrolyses both long chain di- and triacylglycerols, as determined using the monomolecular film technique, although the turnover was lower than with MAG (monoacyl-glycerol). The enzyme shows an optimum activity at pH values ranging from 7.5 to 9.0 using mono-olein as substrate and is inactivated by serine esterase inhibitors such as E600, PMSF and tetrahydrolipstatin. The catalytic triad is composed of Ser110, Asp226 and His256 residues, as confirmed by the results of site-directed mutagenesis. Rv0183 shows 35% sequence identity with the human and mouse monoglyceride lipases and well below 15% with the other bacterial lipases characterized so far. Homologues of Rv0183 can be identified in other mycobacterial genomes such as Mycobacterium bovis, Mycobacterium smegmatis, and even Mycobacterium leprae, which is known to contain a low number of genes involved in the replication process within the host cells. The results of immunolocalization studies performed with polyclonal antibodies raised against the purified recombinant Rv0183 suggested that the enzyme was present only in the cell wall and culture medium of M. tuberculosis. Our results identify Rv0183 as the first exported lipolytic enzyme to be characterized in M. tuberculosis and suggest that Rv0183 may be involved in the degradation of the host cell lipids.
Keywords: exported, expression, immunolocalization, lipase, monoglyceride, mycobacterium, Rv0183
Abbreviations: β-CD, β-cyclodextrin; DAG, diacylglycerol; DiC10, 1,2 dicaprin; DGL, dog gastric lipase; DLPC, dilauroyl phosphatidylcholine; DLPE, dilauroyl phosphatidylethanolamine; DLPG, dilaroyl phosphatidyl glycerol; DPPC, dipalmitoyl phosphatidylcholine; DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); E600, diethyl p-nitrophenyl phosphate; HPL, human pancreatic lipase; IPTG, isopropyl β-D-thiogalactoside; MAG, monoacylglycerol; 2-MAG, α-2-MAG; MGL, monoglyceride lipase; NaTDC, sodium taurodeoxylcholate; ORF, open reading frame; PLA, phospholipase A; PLC, phospholipase C; rRv0183, recombinant Rv0183; TAG, triacylglycerol; TEV, tobacco etch virus; THL, tetrahydrolipstatin
INTRODUCTION
Although much research has been focused on tuberculosis during the last century, it is still the most deadly infectious disease worldwide. It has been estimated that one third of the world's population has been contaminated by Mycobacterium tuberculosis, the aetiological agent responsible for this disease. Despite the existence of effective drug treatments, 2.5 to 3 million people die of this disease every year (http://www.who.int/mediacentre/factsheets/fs104/en/print.html). The rise of tuberculosis is directly linked to the spread of AIDS and the evolution of new multi-drug-resistant strains. M. tuberculosis is a highly complex micro-organism, and a better understanding of the biochemistry and the physiological role of the enzymes involved in the various metabolic pathways taken by the process of infection could lead to the design of new therapeutic agents to fight the bacillus.
Among the various metabolic pathways involved, lipid metabolism is thought to be one of the main pathways taken by mycobacteria, and this pathway therefore needs to be studied in detail. Studies on the lipids present in mycobacteria have been conducted for the last 40 years, and considerable attention has focused on the lipids involved in the cell wall. Many unusual lipids, including phthiocerol dimycocerosates, mycolic acid, glycolipids, polyketides and glycans, have been found to be involved in the virulence and pathogenicity of M. tuberculosis [1–4].
An intracellular lipid storage process was recently described in M. tuberculosis, forming organites known as lipid bodies. These organites are composed of TAG (triacylglycerol) and other lipids, such as wax esters, polyhydroxyalkanoic acids and monomeromycolyldiacylglycerol [4–9]. It has been proposed that these lipids probably provide a carbon source, promoting the growth of the bacteria during the chronic infection phase [7,10]. It might therefore store the energy required during the long periods of bacterial dormancy [11] and serve as a storage form of fatty acids required for membrane lipid formation [4]. The storage of these lipids in vivo may therefore be crucial to the survival of the bacteria and allow them to adapt to the environment provided by the host cells.
It has been suggested by in vitro studies that the accumulation or the presence of fatty acids in the culture medium may stimulate the dormancy phase of the bacteria: the growth rate of most mycobacteria can be enhanced by adding oleic or palmitic acid to the culture medium [12–14]. Fatty acids can be generated in vivo by the hydrolysis of host cell lipids [7,10]. During the formation of granulomas, mycobacteria are mixed with phospholipids originating from dead cells (host cells and bacteria), which may be hydrolysed by PLA (phospholipase A) and PLC (phospholipase C), leading to the release of lysophospholipids, fatty acids and DAG (diacylglycerol) [15]. The resulting DAG may then be hydrolysed by a secreted or exported lipase and the micro-organism will absorb the fatty acids released. A synergy of this kind between lipases and phospholipases has been previously described in the case of another pathogen, Pseudomonas aeruginosa, which degrades lung surfactants (phospholipids) via the joint action of a PLC and a lipase [16]. Excreted PLA and PLC have been previously described in M. tuberculosis [17,18], but no extracellular lipase has been identified so far in this strain. Two intracellular lipolytic enzymes which may participate in the degradation of intracellular TAG have been identified and purified so far in mycobacteria. The first of these is a native enzyme which was purified from Mycobacterium phlei [19] and characterized. The second lipolytic enzyme was the expression product of the Rv3097c gene from the strain M. tuberculosis H37Rv in Escherichia coli [20].
Using the proteins deduced from the complete genome sequence of M. tuberculosis [21], it has now become possible to identify proteins which might be involved in the lipid metabolism by comparing their amino acid sequences with those of other known lipases. Here we report on the cloning and biochemical characterization of a lipase exported by M. tuberculosis, H37Rv (accession number Rv0183). This enzyme shows 34% and 36% amino acid sequence identity with monoglyceride lipases from human and mouse adipocytes respectively [22,23]. Contrary to what was suggested by genome annotation, which identified Rv0183 as a lysophospholipase, this enzyme does not hydrolyse lysophospholipids but hydrolyses neutral lipids, showing a clear-cut preference for MAG (monoacylglycerol). Polyclonal antibodies specific to Rv0183 were prepared and used to determine the location of the enzyme in M. tuberculosis. Based on our results, it seems possible that Rv0183 may contribute to the growth of M. tuberculosis bacilli and be involved in the hydrolysis of host cell lipids during the infection process.
MATERIALS AND METHODS
Materials
Pfx DNA polymerase, pDonR221 and pDest14 plasmids were purchased from Invitrogen. Rosetta pLysS E. coli cells were purchased from Novagen. Ni2+-agarose gel was obtained from Amersham Biosciences. All vinyl esters, TAGs, cholesterol oleate, PMSF, NaTDC (sodium taurodeoxylcholate), DOC (sodium deoxylcholate), gum arabic, Triton X-100, E600 (diethyl p-nitrophenyl phosphate), IPTG (isopropyl β-D-thiogalactoside) and goat anti-rabbit immunoglobulins conjugated to peroxidase were obtained from Sigma-Aldrich. Tripropionin was purchased from Acros Organics, and pure mono-olein and diolein were purified from a commercial low grade DL-α-mono-olein from Fluka. THL (tetrahydrolipstatin) was a gift from Hoffmann la Roche (Roche). Phthiocerol dimycocerosates, glycerol monomycolates and phosphatidylinositol dimannoside were provided by Dr. Martine Gilleron from IPBS (Institut de Pharmacologie et Biologie Structurale) UMRS 5089 (Toulouse, France).
Cloning
The DNA fragment of the Rv0183 ORF (open reading frame) was amplified by PCR from the bacmid BAC-Rv165 provided by the Pasteur Institute [21,24]. The full Rv0183 sequence proposed by Camus et al. [25] was analysed using the software program SignalP 3.0 Server, which identified a putative signal sequence. A truncated form of Rv0183 without the signal sequence (starting from the Thr46 residue) was therefore produced. The primers used, which contained the attB1, Shine–Dalgarno, Kozak, His6 and a TEV (tobacco etch virus) NIa site sequences at the 5′ end and the attB2 recombination site at the 3′ end, are presented in Table 1. The PCR product was cloned into the expression vector pDest14 following the manufacturer's instructions (Gateway, Invitrogen). The DNA sequence of the Rv0183 ORF was confirmed by performing DNA sequencing (MilleGen Biotechnologies, France). The rRv0183 (recombinant Rv0183) protein without the His6 tail and the TEV site is composed of 279 residues and has a calculated molecular mass of 30187 Da, a theoretical isoelectric point of 5.8 and a theoretical molar absorption coefficient of 0.858 M−1·cm−1 at 280 nm.
Table 1. Oligonucleotides used in this study.
The underlined bases indicate the site of the mutation.
| Forward | Reverse | |
|---|---|---|
| Cloning | 5′tcgaaggagatagaaccatgcatcaccatcaccatcac gaaaacctgtacttccagggtactaccacccggactgaacgg3′ | 5′cctacaaccgctcggtgagccag3′ |
| Mutagenesis | ||
| S110A | 5′gtgctcgggcacgccatgggcggcggc3′ | 5′gccgccgcccatggcgtgcccgagcac3′ |
| D226A | 5′cacggcaccgatgcccggctgatcccc3′ | 5′ggggatcagccgggcatcggtgccgtg3′ |
| D226N | 5′cacggcaccgataaccggctgatcccc3′ | 5′ggggatcagccggttatcggtgccgtg3′ |
| H256A | 5′cccgggctgtacgccgaggtgttcaac3′ | 5′gttgaacacctcggcgtacagcccggg3′ |
Mutagenesis
Three Rv0183 mutants in which the putative catalytic residues Ser110, Asp226 and His256 were mutated into Ala were obtained using the Quickchange® site-directed mutagenesis system (Stratagene). The Asp226 residue was also mutated to asparagine. The oligonucleotides used for PCR mutagenesis are specified in Table 1.
Expression and purification of recombinant and Rv0183 mutants
E. coli Rosetta pLysS cells were transformed with the pDest14-His-Rv0183 plasmid and grown overnight in LB medium with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C. On the following day, the culture medium was diluted 20 times with Terrific Broth medium containing antibiotics and grown at 37 °C. Protein expression was induced with 1 mM IPTG when the D600 had reached 0.6–0.8. The temperature was then decreased to 25 °C and cells were grown overnight. After 16 h, cells were harvested at 4 °C by centrifugation at 6500 g for 20 min. The pellet was resuspended in ice-cold lysis buffer (50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.25 mg/ml lysozyme; 30 ml per litre of initial culture) and stored overnight at −80 °C. The cell suspension was thawed on ice for 1 h and 10 μg/ml DNAse I and 20 mM MgSO4 (final concentration) were then added. The mixture was incubated for 30 min, then the supernatant containing the recombinant protein was separated from the cell debris by centrifugation at 17000 g for 30 min. This supernatant was loaded (3 ml/min) using a FPLC chromatography system (Amersham Biosciences) onto a Ni2+-agarose column (1 ml resin per 5 mg of recombinant protein) previously equilibrated with buffer A [10 mM Tris/HCl (pH 8.5), 150 mM NaCl] containing 10 mM imidazole. The column was then washed sequentially using 5 column volumes of 5% and 10% buffer B (buffer A containing 500 mM imidazole). rRv0183 was eluted with 100% buffer B. After analysis by SDS/PAGE [26], the eluted fractions containing the enzyme were pooled at a concentration of around 4 mg/ml and loaded on to a size-exclusion chromatography column (Superdex 200) in buffer A. The His6 tag in the N-terminal position of rRv0183 was removed by TEV digestion and purified by exclusion from a Ni2+-agarose column. Proteolytic digestion of rRv0183 by TEV was performed overnight at 25 °C using a TEV/rRv0183 ratio of 1:10. During the purification process, the amounts of rRv0183 were monitored by measuring the enzyme activity. After the purification process, the protein concentration was calculated from the A280 value using the molar absorption coefficient (0.858 M−1·cm−1). The enzyme was then concentrated to 12 mg/ml and stored at −80 °C.
pH-stat kinetic assays
Enzymatic hydrolysis of solutions and emulsions of various esters was monitored potentiometrically for at least 5 min at various temperatures using a pH-stat (TTT 80, Radiometer, Copenhagen, Denmark). Assays were performed in 15 ml of 2.5 mM Tris/HCl (pH 7.5) containing 150 mM NaCl and 3 mM NaTDC. Fatty acid release was automatically titrated with 0.1 M NaOH. The specific activity of the Rv0183 enzyme was expressed in units per mg of protein (units·mg−1), where one unit corresponds to the release of one micromole of fatty acid per minute.
Analysis of lipolysis products by TLC
In order to check whether rRv0183 was able to hydrolyse 2-MAG (α-2-MAG), this compound was generated from triolein using HPL (human pancreatic lipase). HPL is known to specifically hydrolyse the sn-1 and sn-3 ester bonds of TAG and to specifically release 2-MAG.
Assays were performed at 37 °C using HPL (140 μg) and colipase (molar ratio 1:2) in 15 ml of 1 mM Tris/HCl buffer (pH 7.5) containing 2 mM CaCl2, 3 mM NaTDC, 150 mM NaCl, and triolein (460 μmol) emulsified with gum arabic (3.1%, w/v). Aliquots of 500 μl of the reaction mixture were collected at various times: before adding HPL (T0), after 1 h of incubation with HPL alone and 10, 20 and 120 min after adding rRv0183 (780 μg). The reaction products were then extracted with 5 ml of a chloroform/methanol (2:1, v/v) mixture. The organic phase was separated from the water phase and dried over magnesium sulfate, and the neutral lipids were separated by TLC using silica gel plates from Merck and n-heptane/diethyl ether/acetic acid (55:45:1, by vol.) as the eluent. After spraying the plates with a cupric acetate-phosphoric acid solution, the neutral lipid bands were revealed by heating the plates at 180 °C for 10–15 min.
Monolayer assays
Lipase activities were determined using a zero order trough, as described by Verger and de Haas [27], using DiC10 (1,2 dicaprin), mono-olein, diolein, purified soybean oil and the phospholipids DLPE (dilauroyl phosphatidylethanolamine), DLPG (dilauroyl phosphatidylglycerol), DLPC (dilauroyl phosphatidylcholine) and DPPC (dipalmitoyl phosphatidylcholine) as substrates. The aqueous subphase was composed of 10 mM Tris/HCl (pH 8.0), 100 mM NaCl, 21 mM CaCl2 and 1 mM EDTA. In addition, 13 mM β-CD (β-cyclodextrin) was added when long-chain glycerides were used as substrates. The long chain fatty acids released upon substrate hydrolysis were solubilized by β-CD to prevent any interference with the surface pressure. The reactions were performed at 25 °C. The subphase in the reaction compartment was continuously agitated with a 2.0 cm magnetic stirrer set at 250 rev/min. The enzyme solution (final concentration 5 nM) was injected through the film over the stirrer. The reaction compartment had a surface area of 100 cm2 and a volume of 130 ml. The reservoir compartment was 21.9 cm long and 14.8 cm wide.
Effects of pH, temperature and NaTDC on enzyme stability and activity
The pH stability profile of rRv0183 was determined after incubating the enzyme for 90 min in 1 ml of buffers containing 100 mM NaCl, set at various pH values: 100 mM sodium acetate at pH 4.0 and 5.0; 100 mM Mes at pH 6.0 and 6.5; 100 mM Hepes at pH 7.0 and 7.5; 100 mM Tris/HCl at pH 8.0 and 100 mM glycine at pH 9.0. The residual activity was determined potentiometrically at 37 °C at pH 7.5 using a mono-olein substrate (see pH-stat assays) in the presence of 3 mM NaTDC (final concentration). To draw up the pH and temperature dependence profile, buffer and substrate were pre-equilibrated at selected pH levels. The NaTDC dependence was assessed potentiometrically at 37 °C in 2.5 mM Tris/HCl buffer (pH 7.5), 150 mM NaCl containing NaTDC at concentrations ranging from 0 to 6 mM.
Inhibition assay
Purified rRv0183 (0.7 nmole) in 5 mM Hepes buffer (pH 7.5) containing 500 mM NaCl was pre-incubated at 25 °C with several inhibitors (THL, E600 and PMSF) at a lipase/inhibitor molar ratio of 1:200 in all cases. Each inhibitor was previously solubilized in anhydrous ethanol (10 mM of a stock solution). The reaction was performed in a final volume of 70 μl in the absence or presence of 3 mM NaTDC. The remaining activity was measured potentiometrically as a function of time using mono-olein as substrate.
Titration of the sulfhydryl group
Reaction mixtures were prepared as follows in the presence and absence of 2% (w/v) SDS: 190 μl of protein solution (0.4 mg/ml) was mixed with 810 μl of 5 mM Hepes (pH 7.5), 150 mM NaCl containing 0.35 mM DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] at 25 °C. A 5 mM stock solution of DTNB was initially prepared in methanol. The reactions were carried out in 1 cm pathlength cells with stirring. The changes in absorbance occurring at 412 nm, corresponding to the formation of the TNB product (ϵ412=13.9 mM−1·cm−1 [28]), were monitored on a Kontron spectrophotometer (model SFM 25).
Production and purification of anti-rRv0183 polyclonal antibodies
Antiserum against rRv0183 was prepared in rabbits (Bourgogne Fawn) using 4 mg of purified antigen. Rabbits were injected subcutaneously every 2 weeks with 0.5 mg antigen emulsified with complete Freund's adjuvant. Rabbits were bled by ear vein puncture before the beginning of the immunization procedure (pre-immune sera), then 1 week after the second booster injection, and weekly injections were administered thereafter. The antibody response was assessed by performing a direct binding ELISA using rRv0183 antigen. This immunization procedure yielded an anti-Rv0183 serum with a titre of 1/40000. Polyclonal antibodies were purified from the serum by performing affinity chromatography using a column of immobilized rRv0183 prepared as follows: 11 mg of immobilized rRv0183 were coupled to 3 ml of AffiGel-10 (Bio-Rad) with a high efficiency of up to 90% and packed into an IBF column (diameter 1 cm). Serum was loaded on to the AffiGel-10-rRv0183 column and incubated overnight at 4 °C. After washing the column with 10 mM Tris/HCl (pH 7.0) and then eluting the pure immunoglobulins at an acidic pH (pH 2.4), the solution was immediately neutralized to pH 7.0 with 2 M Tris/HCl (pH 9.0). Purified antibodies (20 mg) were concentrated at 2 mg/ml and stored at −20 °C.
Western blotting analysis of Rv0183 in cellular fractions from M. tuberculosis
Samples of M. tuberculosis were provided by the “TB (tuberculosis) Vaccine Testing and Research Materials” Contract funded by the NIH (National Institutes of Health) and NIAID (National Institute of Allergy and Infectious Diseases, Colorado State University, U.S.A.). Proteins were separated on SDS/PAGE (12% gel) before transfer on to a nitrocellulose membrane [29]. The purified polyclonal antibodies directed against rRv0183 (final dilution 1:1000) were incubated for 1 h with the nitrocellulose membrane previously saturated with 3% (w/v) dry milk powder (Régilait®). After three washes, the membrane was incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG (at a dilution of 1:4000). The immunoreactive proteins were revealed after covering the membrane with ECL® (enhanced chemiluminescence) reagent (Amersham Biosciences, ref. RPN 2109), and exposing Kodak Biomax MS film to the membrane.
RESULTS
Selection of Rv0183
BLAST searches against all available microbial genomes showed that homologues of the Rv0183 gene (without its putative signal peptide sequence) are to be found in many mycobacterial species and other micro-organisms such as Burkholderia and Streptomyces sp. Sequence identity between the mycobacteria is 100% in the case of Mycobacterium bovis, 79% in that of Mycobacterium leprae and 68% in that of Mycobacterium smegmatis, corresponding to E values ranging from 0 to 10−111 (Figure 1).
Figure 1. Amino acid sequence alignment between Rv0183, without its putative signal peptide identified from the bioinformatic results, and three homologous proteins from M. bovis (Mb0189, 100%), M. leprae (ML2603, 79%) and M. smegmatis (68%), and a human (34%) and a mouse (36%) MGL.
This figure was generated by the ESPrit program (available from the Expasy web site) using the alignment function performed by Clustal W [from the EBI (European Bioinformatics Institute) web site] and the secondary element structure obtained from the PredictProtein website [50]. Conserved residues are shown with a black background with similar residues in grey. Residues potentially involved in the catalytic triad are indicated by a triangle (▲). The two lipase motifs are indicated by circles (●). The secondary structure elements (β-strands and α-helices) are indicated at the top.
To our knowledge, none of the homologues identified among the microbial genomes have been purified and characterized so far. Despite their low sequence identity with lysophospholipases (14 to 18%), these proteins have been proposed as putative lysophospholipases. However, a BLAST search against human and mouse genomes identified two hits with E values ranging from 10−35 to 10−37 corresponding to a sequence identity ranging from 34% to 36%. Both hits correspond to MGLs (monoglyceride lipases), which are involved in fat mobilization processes in adipose tissues [22,30]. In Rv0183 and all the mycobacterial homologues, two lipase motifs including the GXSXG pentapeptide containing a catalytic serine residue and a His-Gly dipeptide are conserved. In addition, the histidine and methionine residues present in the pentapeptide containing the active serine (GHSMG) are strictly conserved in Rv0183 and in human and mouse MGL. Secondary structure predictions suggest that Rv0183 may belong to the α/β hydrolase fold family and that it is composed of eight β-strands and nine α-helices (Figure 1), but no significant hits have been obtained from the PDB database with which to build a dependable model.
Cloning, expression and purification of rRv0183
Under the conditions described in the Materials and methods section, between 14 to 15 mg of pure rRv0183 was purified from the 23 mg of protein produced per litre of culture (giving a yield of 60–65%). The migration of purified rRv0183 observed upon performing SDS/PAGE corresponds to a molecular mass of approx. 30 kDa, which is in agreement with the theoretical mass deduced from the DNA sequence (Figure 2, lane 2). The His6 tag in the N-terminal position was removed by performing TEV cleavage with a yield of 70%. The difference between the molecular masses of rRv0183 with and without the His6 tag can be seen in Figure 2. Masses determined by MALDI-TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS) were 31940 Da and 30223 Da, respectively. About 10 mg of purified rRv0183 without the His tail was obtained from 1 litre of culture medium. All biochemical results (see below) were obtained with both forms of the rRv0183, and it was noted that the presence of the His6 tag did not affect the biochemical behaviour of the enzyme. Further characterization using CD and dynamic light scattering methods showed that rRv0183 is a monodisperse α/β protein in solution (results not shown).
Figure 2. SDS/PAGE of the purification and the TEV cleavage of rRv0183.
Protein samples were loaded onto a SDS/PAGE (12% gel) under reducing conditions. The gel was stained with Coomassie brilliant blue R250. Lane 1, molecular mass markers from Amersham Biosciences; lane 2, purified rRv0183 (10 μg); lane 3, purified rRv0183 after TEV cleavage and after exclusion from Ni2+-NTA agarose (10 μg).
Biochemical properties
Substrate specificity
To investigate the potential role of Rv0183, activity measurements using various substrates were performed using the pH-stat, TLC and monomolecular film techniques (Table 2 and Figures 3 and 4).
Table 2. Specific activities in the hydrolysis of lipids.
Experiments were performed with tri-, di- and mono-acylglycerides at 37 °C in 2.5 mM Tris/HCl (pH 7.5) buffer, 150 mM NaCl with stirring. Assays with vinyl esters were performed at 25 °C in the same buffer. The procedure used in the experiments on cholesterol oleate was strictly the same as that described by Ben Ali et al. [49]. Activities on phospholipids were determined on monomolecular film at 25 °C (see the Materials and methods section). 1 unit corresponds to 1 μmole of fatty acid released per min. Values are means±S.D. (n=4). * 3% gum arabic±3 mM NaTDC.
| Substrates | Specific activity (units·mg−1) |
|---|---|
| Triacylglycerides (at 37 °C) | |
| Triacetin (TC2) | 0 |
| Tripropionin (TC3) | 0 |
| Tributyrin (TC4) | 0 |
| Trioctanoin (TC8)* | 0 |
| Triolein (TC18)* | 0 |
| Diacylglycerides (at 37 °C) | |
| DiC6* | 27±S.D. |
| Dicaprin (DiC10)* | 46±S.D. |
| DiC14* | 0 |
| Diolein (DiC18)* | 0 |
| Monoacylglycerides (at 37 °C) | |
| 1-monobutyroyl-rac-glycerol (MC4) | 31±S.D. |
| 1-mono-octanoyl-rac-glycerol (MC8) | 307±S.D. |
| 1-monodecanoyl-rac-glycerol (MC10) | 334±S.D. |
| 1-monolauroyl-rac-glycerol (MC12)* | 328±S.D. |
| 1-monomyristoyl-rac-glycerol (MC14)* | 95±S.D. |
| Mono-olein (MC18)* | 290±S.D. |
| Vinyl esters (at 25 °C) | |
| Vinyl acetate (VC2) | 0 |
| Vinyl propionate (VC3) | 57±S.D. |
| Vinyl butyrate (VC4) | 45±S.D. |
| Vinyl laurate (VC12)* | 64±S.D. |
| Cholesterol oleate | 0 |
| Phospholipids (at 25 °C) | |
| DLPE | 0 |
| DLPG | 0 |
| DLPC | 0 |
| Dipalmitoyl phosphatidylcholine (DPPC) | 0 |
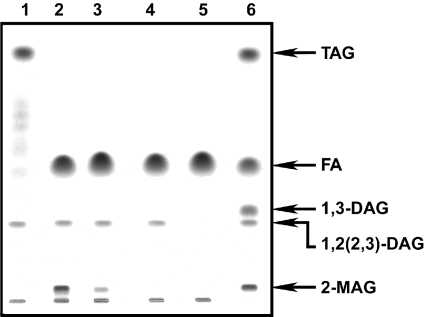
Figure 3. TLC analysis of triolein degradation by HPL and rRv0183.
The reaction was performed on triolein emulsion with HPL (140 μg) for 1 h and rRv0183 (780 μg) was then added to the mixture containing the HPL degradation products. Lipids were extracted with a chloroform/methanol solution and separated on silica gel by TLC (see Materials and methods section). Lane 1, reaction mixture before incubation with enzymes; lane 2, products released after 1 h of incubation with HPL; lanes 3, 4 and 5, products released 10, 20 and 120 min respectively after adding rRv0183; lane 6, lipid markers. FA, freeoleic acid.
Figure 4. Lipolytic activity of rRv0183 on DiC10, mono-olein, diolein and purified soybean oil monolayers as a function of the surface pressure.
Lipolytic activity was determined after injecting 15 μg of rRv0183 into the reaction compartment. Collapse point of each substrate was reached at 41, 32, 28 and 13.8 mN·m−1 in the case of DiC10, mono-olein, diolein and purified soybean oil respectively. Each experiment was performed in triplicate (error ±5%).
The rRv0183 activity measured using the pH-stat technique showed that this enzyme was not able to hydrolyse emulsified TAGs, whatever the chain length of the substrates tested (Table 2). Only DAGs with short and medium fatty acid chains were hydrolysed, and specific activities of 27 and 46 units·mg−1 were measured with DiC6 and DiC10 emulsified with gum arabic. No activity was recorded with the DAGs having the longest chain fatty acids, even in the presence of gum arabic and detergents (NaTDC, CHAPS or Tween). rRv0183 was found to be active, however, on synthetic MAGs with short, medium and long chain fatty acids [MC4, 1-monobutyroyl-rac-glycerol; MC8, 1-mono-octanoyl-rac-glycerol; MC10, 1-monodecanoyl-rac-glycerol; MC12, 1-monolauroyl-rac-glycerol; MC14, 1-monomyristoyl-rac-glycerol; MC18:1, 1-mono-oleyl-rac-glycerol (mono-olein)] (Table 2). The maximum specific activity recorded using mono-olein as substrate was obtained at 37 °C and reached 290 units·mg−1. MC16 (1-monopalmitoyl-rac-glycerol) was also tested, but under our assay conditions, MC16 was found to be insoluble due to the absence of double bonds, and no lipase activity was recorded.
Activity of rRv0183 was also investigated with 2-MAG, the physiological MAG usually produced by 1,3-regioselective lipase. Triolein was first converted into 2-MAG and free oleic acid by adding HPL prior to rRv0183 (Figure 3, lane 2). Ten min after rRv0183 was added to the reaction mixture, 2-MAG decreased rapidly, and was completely hydrolysed within 20 min (Figure 3, lanes 3 and 4). These results show that rRv0183 is able to hydrolyse both 2-MAG and 1-3-MAG. Since we also established that rRv0183 is not able to hydrolyse dioleylglycerols (Table 2), the disappearance of 1,2-dioleyl-rac-glycerol [1,2(2,3)-DAG] observed after adding rRv0183 (Figure 3, lanes 3–5) can be attributed to the HPL activity also present in the reaction mixture. Overall, the concomitant action of HPL and rR0183 results in the complete conversion of triolein into free oleic acid (Figure 3, lane 5). These results support the idea that rRv0183 is a monoglyceride lipase, and that this enzyme is responsible for completing the process of TAG lipolysis initiated by other lipases.
Using the monolayer technique, we also observed that rRv0183 shows considerable rates of activity on mono-olein and DiC10, as well as being able to hydrolyse diolein and purified soybean oil monomolecular films (Figure 4). The activities measured using a DiC10 substrate were comparable to those recorded with dog gastric lipase (DGL) under similar conditions [31]. When mono-olein was used as the substrate, the lipase activity increased with the pressure up to the collapse value (30 mN·m−1) without reaching a maximum. With DiC10, diolein and purified soybean oil substrates, the activities reached a maximum of 11, 8 and 3 mmole·cm−2·min−1·M−1 at 15, 20 and 7 mN·m−1 respectively, and decreased at higher surface pressure values. The activity recorded with diolein and purified soybean oil was found to be almost zero before the collapse of the monomolecular film. When DiC10 was used as the substrate, the hydrolytic activity decreased more slowly as a function of the surface pressure without reaching zero, even near the collapse value. This result might explain why the rRv0183 activity was recorded with emulsified dicaprin using the pH-stat technique (46 units·mg−1), whereas no activity was found to occur with diolein, triolein or purified soybean oil under these assay conditions. Although the monomolecular film technique does not make it possible to determine specific activities with each of the substrates, these results clearly indicate that rRv0183 is able to hydrolyse TAG, DAG and MAG and that these compounds can therefore be accommodated in the active site of the enzyme. However, the absence of activity observed on long chain DAG and TAG emulsions under bulk conditions may be due to the fact that the physical state of the substrate induces denaturation of the enzyme at the oil/water interface, or else it may show that rRv0183 has a poor affinity for oil/water interfaces.
The fact that rRv0183 showed high rates of activity on monoglycerides under bulk conditions probably results from rRv0183 having a better affinity for micellar substrates rather than emulsified substrates. This is an important finding for understanding the role of Rv0183 in vivo.
Since Rv0183 has been annotated in the genome as a lysophopholipase, we checked its activity using lyso-PC (lysophosphatidylcholine) as a substrate (results not shown). Despite many attempts to induce activity in this case, we did not observe any hydrolysis of this substrate. Actually, this result was not very surprising, since the homologous human and mouse monoglyceride lipases do not hydrolyse lysophospholipids [22,23,30]. In order to investigate other potential substrates, trials were performed with several phospholipids using the monomolecular film technique. rRv0183 was not found to hydrolyse cationic (dilauroylphosphatidylethanolamine, DiC12PE), anionic (dilauroylphosphatidylglycerol, DiC12PG) or zwitterionic (DPPC; dilauroylphosphatidylcholine, DiC12PC) phospholipids (Table 2). We also checked the activity of rRv0183 on lipids constituting the cell wall such as phthiocerol dimycocerosates, glycerol monomycolates and phosphatidylinositol dimannoside. No activity was detected on these various substrates (results not shown).
Chain length dependence
rRv0183 was also found to hydrolyse synthetic vinyl esters with various acyl chain lengths. To determine the acyl chain length specificity, the specific activity of rRv0183 was measured on vinyl esters and MAGs with 4 to 18 carbon atoms, and the highest values were those recorded with MAGs with between 8 and 18 carbon atoms (results not shown). Similar specific activities were recorded with vinyl laurate (VC12), vinyl butyrate (VC4) and vinyl propionate (VC3) (64, 45 and 57 units·mg−1 respectively; Table 2). These results show that the enzyme activity does not depend significantly on the chain length, and that the active site can therefore accommodate substrates with short, medium and long acyl chains.
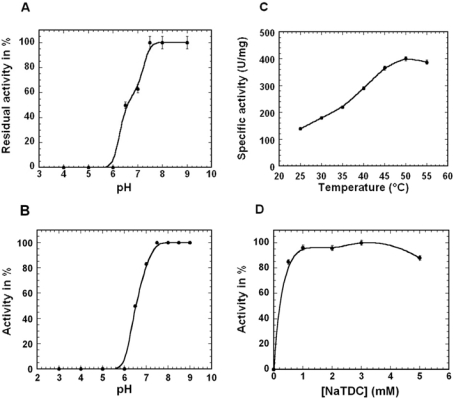
pH stability and pH dependent activity
The pH stability and the pH dependent activity of rRv0183 were investigated using a mono-olein substrate (Figures 5A and 5B). The results obtained show that rRv0183 is stable at alkaline pH values ranging from 7.5 to 9 and that 100% residual activity occurs at pH 9 after 1 h of incubation at 25°C (Figure 5A). At lower pH values, the residual activity decreased rapidly to 50% at pH 6.5, and it was completely abolished after 1 h of incubation at pH levels of 6.0 and below. The curve describing the activity of rRv0183 as a function of pH (Figure 5B) is superimposable on the pH stability curve (Figure 5A). Since the purified rRv0183 was found to be unstable at pH values below pH 6.5, it was impossible to determine the real effects of pH on the rate of enzyme turnover at lower pH levels.
Figure 5. Kinetic assays on rRv0183 using a mono-olein substrate.
(A) pH stability, (B) pH effects on rRv0183 activity, (C) temperature dependence and (D) NaTDC dependence (for details of the experimental procedure, see Materials and methods section).
Temperature dependence
The temperature dependence profile obtained using the mono-olein substrate (Figure 5C) shows that the specific activity of rRv0183 increased 3-fold in a linear fashion from 25 °C to 50 °C before decreasing slowly at higher temperatures. Although rRv0183 showed a high rate of activity at 55 °C, it cannot be classified as a thermostable enzyme, since no residual activity was detected after 1 h of incubation at 55 °C, which suggests that rRv0183 is completely denatured at this temperature. Based on these results, 37 °C was used in all the present biochemical studies. At this temperature, it was possible to test volatile vinyl substrates as well as triglycerides and the activity of the enzyme remained linear for several minutes with all the substrates tested.
NaTDC dependence
Lipase activity on MAG and DAG was enhanced by adding various detergents, such as tetradecyltrimethyl-ammonium bromide (a cationic detergent), Triton X-100 (a non-ionic detergent), CHAPS (a zwitterionic detergent), DOC (an anionic detergent) (results not shown) and NaTDC (an anionic detergent) (Figure 5D). In the absence of detergent, no activity was recorded with mono-olein, DiC6, DiC10 and VC12 substrates. When mono-olein was used as the substrate, the lipolytic activity increased with the NaTDC concentration, peaked at 1 mM NaTDC, and then remained stable up to 5 mM NaTDC (Figure 5D).
The fact that the activity profiles recorded with other detergents under the same assay conditions were similar to those obtained with NaTDC shows that the charge of the detergent does not affect the activity of rRv0183. These results suggest that the binding of rRv0183 to substrate micelles, which is a prerequisite for lipolytic activity to occur, is probably not mediated by electric charges but only by hydrophobic interactions. NaTDC, which is commonly used in lipase assays, was used here to measure the activity of rRv0183. The standard activity assay developed for use with rRv0183 was performed in 2.5 mM Tris/HCl, 150 mM NaCl, 3 mM NaTDC, pH 7.5, at 37 °C using 9.3 mM of mono-olein (50 μl) as substrate.
Absence of the disulfide bridge
The amino acid sequence of Rv0183 possesses two cysteine residues. Under native conditions, no free sulfhydryl groups were detected during the incubation of rRv0183 with the DTNB reagent. However, after adding 2% (w/v) SDS to the rRv0183 protein solution in the absence of reducing agent, two moles of sulfhydryl group were titrated per mole of enzyme. These results show that two free cysteine residues are present but that they are not involved in a disulfide bridge and not accessible to the solvent in the native Rv0183.
Identification of the catalytic triad
The high level of sequence identity observed between Rv0183 and the human and mouse MGLs, in which a catalytic triad has been identified [22,30], led us to study the possible involvement of Ser110, Asp226 and His256 residues in the catalytic triad of Rv0183 (Figure 1). The corresponding alanine mutants were expressed and purified and correct folding was checked using the CD technique. The enzymatic activity on vinyl esters and monoglycerides was found to be completely abolished upon the following mutations: Ser110 into Ala, Asp226 into Ala or Asn, and His256 into Ala.
Effects of serine esterase inhibitors on rRv0183 activity
After preincubating pure rRv0183 with E600, PMSF or THL at a molar excess of 200 in the absence of NaTDC, the lipolytic activity observed on mono-olein was inhibited (Figure 6A): after a 180 min incubation step, the inhibition rate of rRv0183 was 100% with PMSF, 80% with E600 and only 22% with THL. In the presence of 3 mM NaTDC, the inhibitory process was accelerated. The activity was completely inhibited after a 25 min incubation step with PMSF and a 103 min incubation step with E600. After a 150 min incubation step with THL, the rate of rRv0183 inhibition was 62% (Figure 6B).
Figure 6. Inhibitory effects of E600 (○), THL (◇) and PMSF (□) on rRv0183 activity as a function of time.
(A) Without NaTDC and (B) with 3 mM NaTDC. The protein/inhibitor ratio was 1:200 in all experiments. Enzyme activity was determined as described in the Materials and methods section.
These results differ from those obtained with classical lipases such as HPL, DGL and TLL (Thermocyces lanuginosa lipase), the inhibition of which requires the presence of a detergent such as NaTDC: these enzymes are known to have a lid domain covering the active site, and only in the presence of NaTDC does the lid adopt an open conformation giving inhibitors access to the active site [32,33]. The kinetic behaviour of rRv0183 in the presence of an inhibitor supports the idea that the catalytic site of this enzyme is probably not covered by a lid.
Immunolocalization of Rv0183 in cell fractions
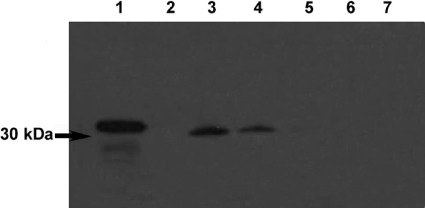
Anti-rRv0183 polyclonal antibodies were used to determine the cellular and subcellular distribution of Rv0183 in M. tuberculosis. As shown in Figure 7, polyclonal antibodies recognized the pure rRv0183 (lane 1) as well as the native Rv0183 present in the M. tuberculosis culture medium (lane 3) and the cell wall (lane 4). As can be seen from Figure 7, Rv0183 was not detected in the culture medium filtrate when passed through a 0.22 μm filter (lane 2), which suggests that Rv0183 is bound to large particles. No trace of enzyme was observed in either the membrane or the cytosol fractions (lanes 5, 6 and 7). The fact that native Rv0183 was detected in the culture medium is in agreement with results obtained in studies in which the M. tuberculosis proteome was analysed [34,35].
Figure 7. Location of Rv0183 in M. tuberculosis cellular compartments.
Aliquots (15 μg of proteins per well) of M. tuberculosis cellular compartments were subjected to immunoblotting analysis using purified polyclonal antibodies directed against rRv0183. Lane 1, rRv0183 purified protein (400 ng); lane 2, protein from culture medium obtained using a 0.22 μm filter; lane 3, protein from culture medium; lane 4, cell wall; lane 5, membrane; lane 6, Triton X-114 extracted proteins; lane 7, cytosol. M. tuberculosis cellular samples were kindly provided by the TB (tuberculosis) Vaccine Testing and Research Materials funded by NIH and NIAID (Colorado State University).
DISCUSSION
Understanding the molecular mechanisms involved in lipid metabolism in M. tuberculosis is likely to be of both fundamental and clinical importance. Although many studies have been performed on the synthesis of lipids in M. tuberculosis [11], little is known so far about their degradation [20]. No secreted or exported lipases have been identified up to now in M. tuberculosis, although lipids play an important role in this micro-organism: phospholipids in the host cells may be hydrolysed into fatty acids and glycerol and absorbed by the mycobacteria. These fatty acids are then used to synthesize the lipids (TAGs) occurring in the M. tuberculosis lipidic inclusion bodies, which are thought to be required for the survival of the bacteria during the dormant and reactivation phases [7]. During these phases, intra- and extra-cellular lipases may play an important role in lipid metabolism processes. In the M. tuberculosis genome, which was completed in a recent study, 24 genes have been annotated as putative lipases (Lip C to Z, except K and S) [20,21]. However, among these 24 targets, nine do not possess the GXSXG pentapeptide, which is the minimum moiety included in the larger lipase consensus pattern [LIV]-{KG}-[LIVFY]-[LIVMST]-G-[HYWV]-S-{YAG}-G-[GSTAC], according to the PROSITE database. Among the 15 remaining targets, only Lip Y (Rv3097c) has been found so far to be a lipolytic enzyme [20], while Lip H (Rv1399c) and Lip F (Rv3487c) were not classified as lipolytic enzymes after being biochemically characterized [36,37]. However, more specific research on the M. tuberculosis genome based on comparisons with genes coding for biochemically characterized enzymes found to be lipolytic has shown that other genes may be lipases (results not shown). One of these potential lipases, the Rv0183 gene, which codes for a putative lysophopholipase [21], was selected because of its high amino acid sequence identity (∼34%) with known mammalian monoglyceride lipases. After having cloned, expressed and biochemically characterized the rRv0183, this protein was found to be a lipolytic enzyme hydrolyzing long chain MAGs which does not show any lysophospholipase activity. This enzyme is the first exported lipase to be identified in M. tuberculosis. The optimum conditions of expression obtained in Rosetta pLysS E. coli cells at 37 °C using Terrific Broth as growth medium yielded around 10.5 mg of purified soluble recombinant protein per litre of culture medium. The optimum assay conditions were then established using the pH-stat technique, which is a useful means of performing kinetic studies. The biochemical properties of rRv0183 are very similar to those of human and mouse MGL [22,30,38,39]: amino acid sequence alignment (Figure 1) and mutagenesis experiments showed that the catalytic triad is composed of residues Ser110, Asp226 and His256. The results of inhibition studies performed in the absence and presence of NaTDC support the idea that, unlike many other lipases, rRv0183 does not possess a lid covering its active site. This is actually not very surprising, since many bacterial lipases have been found to be devoid of a lid domain [40].
The biochemical characteristics of rRv0183 determined using the monomolecular film technique clearly show that this enzyme is also potentially active on long chain TAGs and DAGs. Therefore, rRv0183 possesses an active site which can accommodate various glycerides. The absence of rRv0183 activity on TAG and DAG in the pH-stat assay probably resulted from the occurrence of poor interactions between the enzyme and the lipid/water interface, whereas the monomolecular film technique made it possible to screen a larger range of interfacial conditions.
The high levels of expression of rRv0183 obtained here will hopefully lead to further crystallization studies. Information about the rRv0183 three dimensional structure will help to explain the substrate specificity of the enzyme, which is currently difficult to interpret both at the molecular level and in terms of the putative physiological role of Rv0183.
The cellular location of Rv0183 means that the native enzyme can be exported through the cell wall and may play a role both in the cell wall and in the culture medium: Rv0183 may act outside the bacilli and be directly involved in the host lipid degradation process initiated by another partner. Many studies have shown that protein fractions originating from the culture medium are potential candidates for the production of vaccines, and therefore Rv0183 might be one of them. Since no Rv0183 protein can be detected inside the cytoplasm, Rv0183 obviously does not play the same physiological role in vivo as the first intracellular lipase identified [20]. Several hypotheses can be put forward: (i) Rv0183 may be able to hydrolyse extracellular TAG, DAG and MAG with different rates, leading to the uptake of free fatty acids and their further storage in lipid inclusion bodies when M. tuberculosis infects adipocytes [10]; (ii) Rv0183 may hydrolyse diglycerides released by PLC and then the resulting monoglycerides, thus triggering the release of glycerol and absorbable fatty acids; (iii) Rv0183 may hydrolyse the monoglycerides resulting from the action of another, as yet unknown, exported lipase. The latter scheme is known to occur in human and mouse adipocytes, where the homologous MGL catalyses the hydrolysis of monoglycerides after the action of adipose triglyceride lipase and hormono-sensitive lipase on TAGs and DAGs respectively [41,42].
The absence of Rv0183 in the culture medium passed through a 0.22 μm filter suggests that Rv0183 may aggregate in large vesicles after being exported, or that Rv0183 may be exported from the membrane by these extracellular vesicles. Vesicular trafficking from phagosomes containing bacteria has been found to occur in previous studies [43,44]. Mycobacterial lipids were found to be present in extracellular vesicles isolated from the culture medium. By the release of these vesicles, the effects of the bacteria may be exerted on more distal cells or tissues rather than being restricted to the nearest cells. In this context, the possibility can not be ruled out that Rv0183 may be involved in trafficking processes of this kind during the infection stage. Although Rv0183 was detected in the culture medium, the processes underlying its up-regulation are not yet known. Its physiological role will now have to be investigated using specific antibodies. This powerful tool combined with electron microscopy will make it possible to detect the presence of Rv0183 during the process of phagocytosis of mycobacteria by macrophages.
Although Rv0183 is not thought to be one of the main targets involved in the infection process [45,46], this does not rule out the possibility that this enzyme may contribute to the reactivation of the bacteria and help the mycobacteria to survive in the mature granulomas during the dormant phase. This hypothesis is in agreement with previous studies showing that bacteria grown in vitro had a preference for carbohydrates, whereas bacteria grown in vivo preferred fatty acids as their carbon source [47,48].
Likewise, the presence of a homologous enzyme in M. leprae (ML2603), which is known to have only the enzymes necessary for its replication in the host cells, suggests that this enzyme may play an important role in the life cycle of the micro-organism. Studies on the behaviour of Rv0183 might also provide indirect information about the lipid metabolism in M. leprae.
In conclusion, the results obtained in the present study show the existence of an exported lipolytic enzyme, which preferentially hydrolyses MAGs and is closely related to human and mouse MGLs. Based on the present biochemical findings, this exported enzyme may be involved in the hydrolysis of host cell lipids during the infection process, but its exact physiological contribution to the process of lipid metabolism still remains to be elucidated.
Acknowledgments
We thank Dr. Steward Cole and Dr. Nadine Honoré at the Pasteur Institute, Paris, France, for providing us with the BACs (bacterial artificial chromosomes) library, Marilyne Blémont, CNRS (Centre National de la Recherche Scientifique), UPR 9025, Marseille, France, and Emilie Layre, IPBS (Institut de Pharmacologie et de Biologie Structurale), UMR 5089, Toulouse, France, for their technical assistance. We are indebted to Dr. Jessica Blanc for revising the English manuscript before submission. This work was supported in part by the CNRS and the GIP ANR (National Research Agency Public Interest Group) 06-JCJC-0067–01.
References
- 1.Daffe M., Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 2.Minnikin D. London: Academic Press; 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles. [Google Scholar]
- 3.Minnikin D. E., Kremer L., Dover L. G., Besra G. S. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 2002;9:545–553. doi: 10.1016/s1074-5521(02)00142-4. [DOI] [PubMed] [Google Scholar]
- 4.Kremer L., de Chastellier C., Dobson G., Gibson K. J., Bifani P., Balor S., Gorvel J. P., Locht C., Minnikin D. E., Besra G. S. Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol. Microbiol. 2005;57:1113–1126. doi: 10.1111/j.1365-2958.2005.04717.x. [DOI] [PubMed] [Google Scholar]
- 5.Dawes E. A., Senior P. J. The role and regulation of energy reserve polymers in microorganisms. Adv. Microb. Physiol. 1973;10:135–266. doi: 10.1016/s0065-2911(08)60088-0. [DOI] [PubMed] [Google Scholar]
- 6.Koval'chuk L. P., Donets A. T., Razumovskii P. N. Lipid biosynthesis by actinomycetes cultured on different media. Mikrobiologiia. 1973;42:637–642. [PubMed] [Google Scholar]
- 7.Garton N., Christensen H., Minnikin D., Adegbola R., Barer M. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148:2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- 8.Waltermann M., Steinbuchel A. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 2005;187:3607–3619. doi: 10.1128/JB.187.11.3607-3619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez H. M., Steinbuchel A. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 2002;60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- 10.Neyrolles O., Hernandez-Pando R., Pietri-Rouxel F., Fornes P., Tailleux L., Payan J. A., Pivert E., Bordat Y., Aguilar D., Prevost M. C., et al. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS ONE. 2006;1:e43. doi: 10.1371/journal.pone.0000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel J., Deb C., Dubey V. S., Sirakova T. D., Abomoelak B., Morbidoni H. R., Kolattukudy P. E. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedgecock L. W. Nutritional characteristics of the atypical mycobacteria. J. Bacteriol. 1968;96:306–313. doi: 10.1128/jb.96.2.306-313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer W. B., Lewis C. W., Jr Effect of oleic acid on growth and cell structure of mycobacteria. J. Bacteriol. 1965;90:1438–1447. doi: 10.1128/jb.90.5.1438-1447.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy C. Utilization of palmitic acid by Mycobacterium avium. Infect. Immun. 1971;4:199–204. doi: 10.1128/iai.4.3.199-204.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höner zu Bentrup K., Russell D. G. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 2001;9:597–605. doi: 10.1016/s0966-842x(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 16.Konig B., Jaeger K., Konig W. Induction of inflammatory mediator release (12-hydroxyeicosatetraenoic acid) from human platelets by Pseudomonas aeruginosa. Int. Arch. Allergy Immunol. 1994;104:33–41. doi: 10.1159/000236706. [DOI] [PubMed] [Google Scholar]
- 17.Raynaud C., Guilhot C., Rauzier J., Bordat Y., Pelicic V., Manganelli R., Smith I., Gicquel B., Jackson M. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2002;45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 18.Stonehouse M. J., Cota-Gomez A., Parker S. K., Martin W. E., Hankin J. A., Murphy R. C., Chen W., Lim K. B., Hackett M., Vasil A. I., Vasil M. L. A novel class of microbial phosphocholine-specific phospholipases C. Mol. Microbiol. 2002;46:661–676. doi: 10.1046/j.1365-2958.2002.03194.x. [DOI] [PubMed] [Google Scholar]
- 19.Paznokas J. L., Kaplan A. Purification and properties of a triacylglycerol lipase from Mycobacterium phlei. Biochim. Biophys. Acta. 1977;487:405–421. doi: 10.1016/0005-2760(77)90212-0. [DOI] [PubMed] [Google Scholar]
- 20.Deb C., Daniel J., Sirakova T. D., Abomoelak B., Dubey V. S., Kolattukudy P. E. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson M., Contreras J. A., Hellman U., Tornqvist H., Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson M., Reue K., Xia Y. R., Lusis A. J., Langin D., Tornqvist H., Holm C. Exon-intron organization and chromosomal localization of the mouse monoglyceride lipase gene. Gene. 2001;272:11–18. doi: 10.1016/s0378-1119(01)00559-5. [DOI] [PubMed] [Google Scholar]
- 24.Philipp W. J., Poulet S., Eiglmeier K., Pascopella L., Balasubramanian V., Heym B., Bergh S., Bloom B. R., Jacobs W. R., Jr, Cole S. T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camus J., Pryor M., Medigue C., Cole S. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Verger R., de Haas G. H. Enzyme reactions in a membrane model. 1: A new technique to study enzyme reactions in monolayers. Chem. Phys. Lipids. 1973;10:127–136. doi: 10.1016/0009-3084(73)90009-1. [DOI] [PubMed] [Google Scholar]
- 28.Ellman G. L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 29.Gershoni J. M., Palade G. E. Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to a positively charged membrane filter. Anal. Biochem. 1982;124:396–405. doi: 10.1016/0003-2697(82)90056-2. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson M., Tornqvist H., Holm C. Expression, purification, and characterization of histidine-tagged mouse monoglyceride lipase from baculovirus-infected insect cells. Protein Expr. Purif. 2000;18:286–292. doi: 10.1006/prep.1999.1194. [DOI] [PubMed] [Google Scholar]
- 31.Rogalska E., Nury S., Douchet I., Verger R. Lipase stereoselectivity and regioselectivity toward three isomers of dicaprin: a kinetic study by the monomolecular film technique. Chirality. 1995;7:505–515. [Google Scholar]
- 32.Ben Ali Y., Chahinian H., Petry S., Muller G., Carriere F., Verger R., Abousalham A. Might the kinetic behavior of hormone-sensitive lipase reflect the absence of the lid domain? Biochemistry. 2004;43:9298–9306. doi: 10.1021/bi049479o. [DOI] [PubMed] [Google Scholar]
- 33.Belle V., Fournel A., Woudstra M., Ranaldi S., Prieri F., Thome V., Currault J., Verger R., Guigliarelli B., Carriere F. Probing the opening of the pancreatic lipase lid using site-directed spin labeling and EPR spectroscopy. Biochemistry. 2007;46:2205–2214. doi: 10.1021/bi0616089. [DOI] [PubMed] [Google Scholar]
- 34.Rosenkrands I., King A., Weldingh K., Moniatte M., Moertz E., Andersen P. Towards the proteome of Mycobacterium tuberculosis. Electrophoresis. 2000;21:3740–3756. doi: 10.1002/1522-2683(200011)21:17<3740::AID-ELPS3740>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Jungblut P. R., Schaible U. E., Mollenkopf H. J., Zimny-Arndt U., Raupach B., Mattow J., Halada P., Lamer S., Hagens K., Kaufmann S. H. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 36.Canaan S., Maurin D., Chahinian H., Pouilly B., Durousseau C., Frassinetti F., Scappuccini-Calvo L., Cambillau C., Bourne Y. Expression and characterization of the protein Rv1399c from Mycobacterium tuberculosis. A novel carboxyl esterase structurally related to the HSL family. Eur. J. Biochem. 2004;271:3953–3961. doi: 10.1111/j.1432-1033.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M., Wang J. D., Li Z. F., Xie J., Yang Y. P., Zhong Y., Wang H. H. Expression and characterization of the carboxyl esterase Rv3487c from Mycobacterium tuberculosis. Protein Expr. Purif. 2005;42:59–66. doi: 10.1016/j.pep.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda Y., Okamura K., Fujii S. Purification and characterization of rat liver microsomal monoacylglycerol lipase in comparison to the other esterases. Biochim. Biophys. Acta. 1977;488:128–139. [PubMed] [Google Scholar]
- 39.Somma-Delpero C., Valette A., Lepetit-Thevenin J., Nobili O., Boyer J., Verine A. Purification and properties of a monoacylglycerol lipase in human erythrocytes. Biochem. J. 1995;312:519–525. doi: 10.1042/bj3120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Pouderoyen G., Eggert T., Jaeger K. E., Dijkstra B. W. The crystal structure of Bacillus subtilis lipase: a minimal α/β hydrolase fold enzyme. J. Mol. Biol. 2001;309:215–226. doi: 10.1006/jmbi.2001.4659. [DOI] [PubMed] [Google Scholar]
- 41.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 43.Beatty W. L., Rhoades E. R., Ullrich H. J., Chatterjee D., Heuser J. E., Russell D. G. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 44.Beatty W. L., Ullrich H. J., Russell D. G. Mycobacterial surface moieties are released from infected macrophages by a constitutive exocytic event. Eur. J. Cell. Biol. 2001;80:31–40. doi: 10.1078/0171-9335-00131. [DOI] [PubMed] [Google Scholar]
- 45.Sassetti C. M., Boyd D. H., Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 46.Sassetti C. M., Rubin E. J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segal W., Bloch H. Pathogenic and immunogenic differentiation of Mycobacterium tuberculosis grown in vitro and in vivo. Am. Rev. Tuberc. 1957;75:495–500. doi: 10.1164/artpd.1957.75.3.495. [DOI] [PubMed] [Google Scholar]
- 48.Bloch H., Segal W. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben Ali Y., Carriere F., Verger R., Petry S., Muller G., Abousalham A. Continuous monitoring of cholesterol oleate hydrolysis by hormone-sensitive lipase and other cholesterol esterases. J. Lipid. Res. 2005;46:994–1000. doi: 10.1194/jlr.M400509-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]