Abstract
Previous work [Metcalfe, Ott, Patel, Singh, Mistry, Goff and Raven (2004) J. Am. Chem. Soc. 126, 16242–16248] has shown that the introduction of a methionine residue (S160M variant) close to the 2-vinyl group of the haem in ascorbate peroxidase leads to the formation of a covalent haem–methionine linkage under oxidative conditions (i.e. on reaction with H2O2). In the present study, spectroscopic, HPLC and mass spectrometric evidence is presented to show that covalent attachment of the haem to an engineered cysteine residue can also occur in the S160C variant, but, in this case, under reducing conditions analogous to those used in the formation of covalent links in cytochrome c. The data add an extra dimension to our understanding of haem to protein covalent bond formation because they show that different types of covalent attachment (one requiring an oxidative mechanism, the other a reductive pathway) are both accessible within same protein architecture.
Keywords: ascorbate peroxidase, c-type cytochrome, haem vinyl group, thioether bond, peroxidase
Abbreviations: APX, ascorbate peroxidase; DTT, dithiothreitol; MALDI, matrix-assisted laser-desorption ionization; rpAPX, recombinant pea cytosolic APX; TFA, trifluoroacetic acid; TOF, time-of-flight
INTRODUCTION
The haem group provides proteins with a remarkable range of functionalities. For example, gas binding or sensing, electron transfer and catalysis are all mediated by haem, which in many cases is non-covalently bound to the protein, e.g. in haemoglobin and b-type cytochromes. There are, however, several types of haem protein in which haem is covalently attached through a post-translational modification process. Seemingly, the most common class of protein of this type is the c-type cytochrome in which haem is attached to the polypeptide via thioether bonds formed from the vinyl groups of haem and the two thiol groups provided by a typical CXXCH motif in the polypeptide [1]. However, there are other examples, and these include the mammalian enzymes lactoperoxidase and myeloperoxidase which contain sulfonium links between the haem 2-vinyl group and a methionine residue and/or two ester links between haem methyl groups and glutamate/aspartate residues. Similarly modified haems are thought to be used more widely in other, more complex, mammalian peroxidases (e.g. thyroid peroxidases, eosinophil peroxidase) [2]. Other examples of unexpected haem–protein links have also come to light. These include the CYP4 family of cytochrome P450s, which contain ester links between the 5-methyl group and a glutamate residue [2–4], cyanobacterial haemoglobin, which contains a covalent link between the 2-vinyl group and a histidine residue [5], and the haem chaperone CcmE, which uses a different vinyl–histidine covalent link [6,7].
Neither the significance nor the mechanisms of formation of these bonds is fully understood. In some instances, e.g. the c-type cytochromes, the covalent bond is formed by the action of a post-translational apparatus, of which there are curiously three or four types, but, in other cases, e.g. myeloperoxidase, the bond is thought to result from the inherent activity of the haem centre itself. An obstacle to mechanistic studies on the formation of covalent bonds between haem and polypeptide is the production of protein without the covalent bond and yet still in a folded state that can bind haem correctly, so that the appropriate amino acid side chain is positioned sufficiently close to the correct part of the haem molecule in order for bond formation to be possible. Recently, we have made progress in two directions. First, it proved possible to prepare an apo form of a mono haem c-type cytochrome, from a thermophilic bacterium and named cytochrome c552, that had intact thiol groups in the CXXCH motif and which would adopt a near-native three-dimensional fold on non-covalent binding of haem. Incubation of the non-covalent haem–apocytochrome complex under reducing conditions resulted in the formation in vitro of material that was spectroscopically indistinguishable from the native c-type cytochrome [8]. It is believed that a close juxtaposition of the thiol groups to the vinyl groups of the haem promoted the formation of covalent bonds through attack of the thiols on the vinyl groups, which are not especially activated for either electrophilic or nucleophilic attack, of the ferrous haem. Some previous work on in vitro formation of such thioether bonds had envisaged the need for some oxidant to be provided [1,9]. Currently, there are insufficient data available to conclude that only reductive conditions will promote thioether bond formation either in vitro or in vivo where three different post-translational sets of apparatus have been identified.
The second development has been the demonstration that the introduction of a methionine residue close to the 2-vinyl group in an S160M variant of rpAPX (recombinant pea cytosolic ascorbate peroxidase) leads to the formation of a vinyl–methionine link, analogous to that found in human myeloperoxidase, by means of an oxidized Compound I intermediate [10]. These results provide the first direct evidence that the formation of a vinyl–methionine covalent linkage occurs as an H2O2-dependent (oxidative) process.
Both of these developments indicate that the examination of the in vitro reaction of S160C APX (ascorbate peroxidase) with haem under both reducing and oxidizing conditions would provide further insight into the formation of covalent bonds between haem and protein. This therefore was the aim of the present study. Apart from the mechanistic interest, understanding how such bonds can be formed may be of value in stabilizing haem proteins and in the field of de novo protein engineering, because several lines of evidence suggest that such covalent bond formation enhances stability [11,12]. Being able to tune the attachment of haem to protein can also further the possibilities of designing proteinaceous structures and enhance the ability to form functional synthetic proteins.
EXPERIMENTAL
MATERIALS AND METHODS
H2O2 solution (33%) was purchased from BDM Laboratory Supplies. All molecular biology kits and enzymes were used according to manufacturers' protocols. Oligonucleotide synthesis and DNA sequencing was carried out by the Protein and Nucleic Acid Laboratory at the University of Leicester. Water was purified from an Elga Purelab purification system, and all buffers were filtered through a 0.2-μm-pore-size filter before use.
Mutagenesis, protein expression and purification
The wild-type pea cytosolic APX template constructed previously was modified by site-directed mutagenesis using a QuikChange™ mutagenesis kit (Stratagene) according to the manufacturer's instructions. For the S160C mutation, the primers were: 5′-ATTGTTGCTCTATGTGGTGGTCACACC-3′ (forward primer) and 5′-GGTGTGACCACCACATAGAGCAACAAT-3′ (reverse primer), with the mutation shown in bold. The PCR products were subcloned into XL1-blue supercompetent Escherichia coli cells (Stratagene), and overnight cultures containing 100 μg·ml−1 ampicillin (Sigma–Aldrich) were incubated at 37 °C with vigorous shaking. The plasmid DNA was isolated using the Qiagen Miniprep plasmid system. The mutation was confirmed by sequencing across the entire rpAPX-coding gene. Plasmids containing the desired mutation were transformed into the E. coli BL21 (DE3) expression strain (Stratagene), and protein expression was carried out as reported previously [10].
Purification of the S160C variant was achieved using Ni-NTA (Ni2+-nitrilotriacetate) agarose resin (Qiagen) according to the manufacturer's instructions, but elution of the protein was achieved using low-pH buffer (pH 4.2) and not imidazole. The protein was isolated as apo-enzyme. Reconstitution with haemin was as described previously [13]. Excess haemin was removed using Fast Flow Q-Sepharose anion-exchange resin (Amersham Biosciences) (elution with 250 mM KCl). A high-molecular-mass impurity was removed with FPLC using a Superdex HR75 size-exclusion column (isocratic gradient of 150 mM potassium phosphate buffer, pH 7.0, flow rate of 0.2 ml/min). The purified enzyme showed an Asoret/A280 ratio of >2.0 and migrated as a single band on SDS/PAGE.
Electronic absorption spectroscopy
Spectra were collected using a PerkinElmer Lambda 35 or 40 spectrophotometer, linked to a PC workstation running UV-Winlab software. The temperature was controlled (±0.1 °C) using either a Julabo U3 water circulator (Lambda 40) or a PerkinElmer PT01 Peltier system (Lambda 35). The molar absorption coefficient for S160C (ϵ=8.8×104 M−1·cm−1) was determined using the pyridine haemochromagen method [14].
Dithionite reduction
A sample of S160C (∼10 μM in 0.1 M potassium phosphate, pH 7 or 5.5) was placed in a 1 ml cuvette which was sealed with a rubber septum. The solution was gently purged with N2 for 30 min, after which DTT (dithiothreitol) was added to a final concentration of 5 mM. Sodium dithionite was carefully titrated into the solution until the A290 reached approx. 1 and the protein was fully reduced. The presence of both DTT and dithionite has been used in other work with cytochrome c (see, for example, [8,14,15]) in which we have observed that DTT is needed to maintain cysteine thiol group integrity and dithionite is needed to maintain the ferrous state of the haem. The UV–visible spectrum of the solution was taken every 30 min for 18 h, at which point the reaction was deemed to be complete. After 18 h, a sample of the protein was removed and its pyridine haemochromagen spectrum was recorded.
High-performance liquid chromatography
All HPLC assays were conducted on a Varian Star™ HPLC system with an analytical Vydac C4 reverse-phase HPLC column under PC control. Solvents were: A=0.1% (w/v) TFA (trifluoroacetic acid) in water; B=0.1% (w/v) TFA in methyl cyanide. Enzyme (10 μl) was mixed with an equal volume of 2% SDS. After 1 h, the sample was made up to 110 μl total volume with water, and 100 μl was injected on to the column which had been pre-equilibrated with solvent A. Separation of the haem and protein was achieved using the following elution gradient: 30–42% solvent B for 36 min, 42% solvent A for 10 min and 42–100% solvent B for 1 min, followed by a washing sequence. UV detection was at 215 nm for protein and 404 nm for haem.
Mass spectrometry
Samples of S160C were exchanged into 0.4 M NH4HCO3, pH 8.0, and concentrated to 10 μl. The samples were mixed in a 1:1 ratio with matrix (sinapinic acid, saturated solution in 1:1 methyl cyanide/water containing 0.1% TFA) and were spotted on to a MALDI (matrix-assisted laser-desorption ionization) target plate using the drying droplet method (∼10 pM per drop). Data were collected using an Applied Biosystems Voyager Workstation within the range 10000–40000 Da. Each spectrum was a result of 100 laser shots with ten such spectra combined to give the final average mass. The spectrometer was operated in linear mode and was calibrated with horse heart myoglobin.
Samples for trypsin digestion were treated with 50:1 (w/w) (enzyme/trypsin) in NH4HCO3 buffer (0.4 M, pH 8.0, 37 °C) for 18 h. These samples (1 μl) were then mixed with 1 μl of matrix (α-cyano-4-hydroxycinnamic acid, saturated solution in 1:1 methyl cyanide/water containing 0.1% TFA). Analysis of the peptide–haem adducts by MALDI–TOF (time-of-flight) MS was carried out as follows: 1 μl of the 1:1 peptide/matrix mixture was spotted on to a MALDI target plate using the drying droplet method. The MALDI–TOF mass spectrometer was calibrated in the range 500–4500 Da with a peptide mass calibration kit (Sigma), used according to the manufacturer's instructions. Data were collected in the same mass range using an average of at least 100 laser shots. Spectra were analysed using Data Viewer software (Applied Biosystems).
RESULTS
Molecular modelling
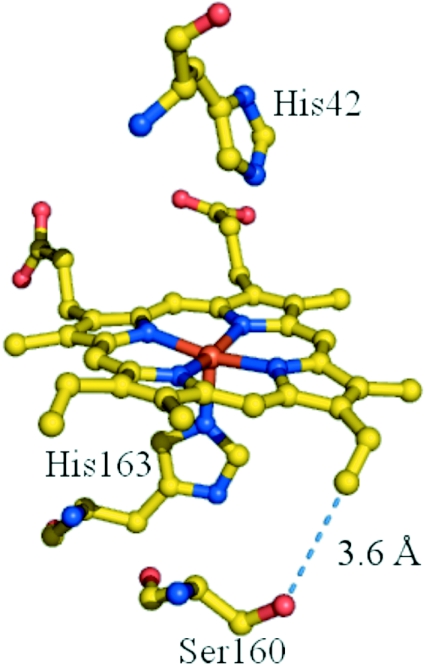
Predictions of cysteine geometry were obtained using structure-based [16] molecular modelling of the rpAPX crystal structure (Figure 1). An energy-minimized mutation of Ser160 to cysteine predicted that the sulfur of Cys160 will be placed close [≈1.8–2.5 Å (1 Å=0.1 nm)] to the 2-vinyl group, and thus appropriately positioned for covalent bond formation.
Figure 1. The active site of rpAPX, showing the position of Ser160.
The shortest distance between Ser160 and the vinyl group (broken line) is 3.6 Å. The Figure was prepared using PyMOL (DeLano Scientific; http://www.pymol.org).
Properties of ferric S160C
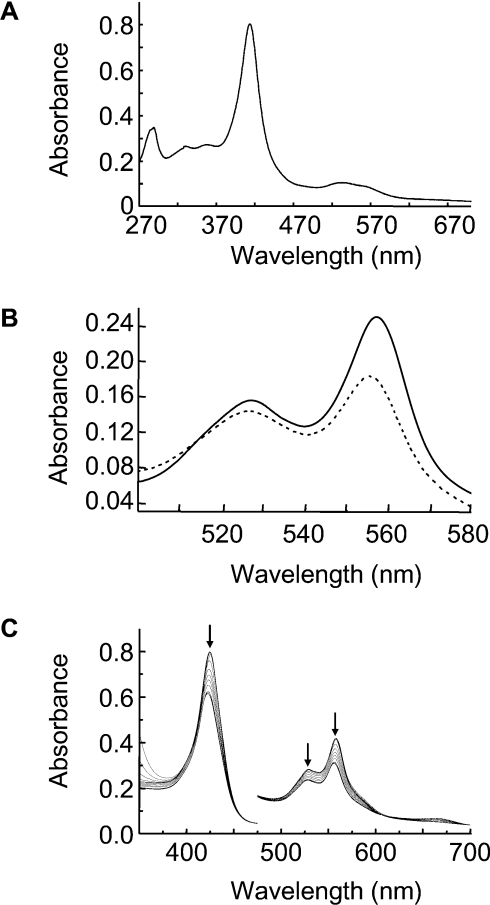
As explained in the Experimental section, S160C was obtained from E. coli cells in the apo form, but, after purification, was found to bind haem readily. The UV–visible spectrum (100 mM phosphate, pH 7.0, 25 °C) of ferric S160C (λmax=414, 535 and 559sh nm) (Figure 2A) showed increased formation of low-spin haem compared with rpAPX (λmax=406.5, 504 and 634.5 nm [10]).
Figure 2. UV–visible absorption spectra.
(A) UV–visible spectrum of ferric S160C (conditions=0.1 M potassium phosphate, pH 7.0). (B) Reduced pyridine haemochromagen spectrum of ferrous S160C immediately after (solid line) and 14 h after (broken line) reduction. The reduced haemochromagen spectrum shown in the solid line is identical with that obtained for haem extracted from ferric S160C (see the Results section). (C) UV–visible spectrum of ferrous S160C monitored every 1 h for 14 h (conditions=0.1 M potassium phosphate, pH 7.0).
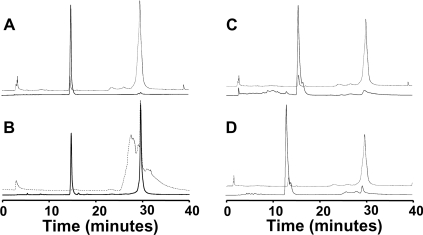
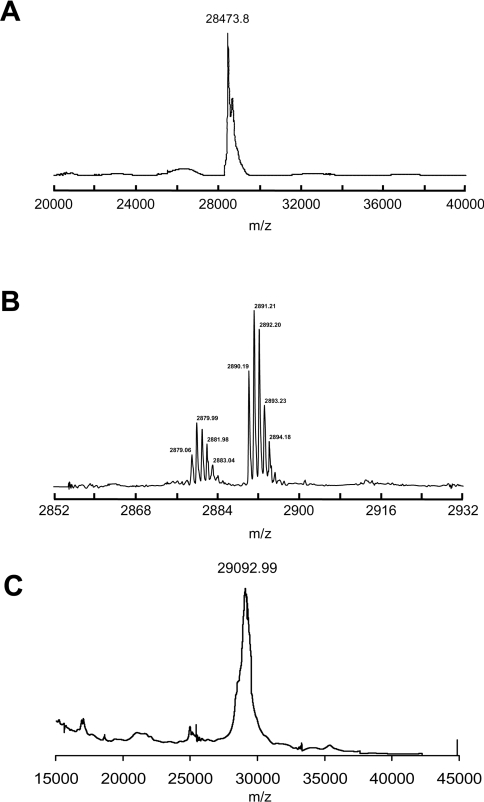
The haem in ferric S160C could be removed by acid-butanone extraction [17] (results not shown). This method is used as a diagnostic test for haem proteins and clearly indicates that haem is non-covalently attached to the protein. This was confirmed by HPLC (Figure 3A), which showed that the haem (retention time of 14.9 min, monitored at 398 nm) and the protein (retention time of 29.5 min, monitored at 280 nm) ran separately. MALDI–TOF MS (Figure 4A), of ferric S160C gave a peak with a molecular mass of 28473.8±8.7 Da, which is consistent with the calculated mass (28476.2 Da) for the apo-protein and which clearly indicates that the haem dissociates from the ferric protein under these conditions.
Figure 3. HPLC analyses.
HPLC chromatogram of S160C before (A) and after (B) reduction measured at 398 nm (solid line) and 280 nm (broken line). (C) HPLC chromatogram of S160C after oxidation with H2O2, measured at 398 nm (solid line) and 280 nm (broken line). (D) HPLC chromatogram of S160C reconstituted with mesoporphyrin after reduction, measured at 398 nm (solid line) and 280 nm (broken line).
Figure 4. Mass spectrometric analyses.
(A) MALDI–TOF mass spectrum of ferric S160C. (B) Expanded view (m/z=2850–2930 region) of the MALDI–TOF mass spectrum of S160C after reduction, showing the haem–peptide fragment at m/z=2880. (C) MALDI–TOF mass spectrum of S160C after reduction. Each spectrum was collected ten times to obtain the average mass and S.D.
The spectrum of the reduced pyridine haemochromagen derivative of ferric S160C (prepared following the method described in [14]), collected immediately after formation of the reduced haemochromagen derivative, showed complete extraction of the haem from the protein and had an α-band in the visible region at 556 nm (Figure 2B). This is consistent with a non-covalently bound haem structure, in which neither haem vinyl group is covalently modified [17].
Tryptic digestion of ferric S160C followed by MALDI–TOF MS was also carried out. In these experiments, good coverage of the protein sequence was observed, with most of the expected peptide fragments appearing in the mass spectrum (Table 1).
Table 1. MALDI–TOF MS data obtained after tryptic digestion of ferric and ferrous S160C.
Numbering is as used previously [10]. Fragments containing Cys160 are in bold. Fragments with a molecular mass <600 Da were not detected under the conditions used and are not indicated. nd, not detected.
| Residues | Peptide sequence | Calculated mass | Observed mass (ferric) | Observed mass (ferrous) |
|---|---|---|---|---|
| −9–3 (His6 tag) | GSHHHHHHGSGK | 1314.60 | 1314.86 | 1314.86 |
| 4–14 | SYPTVSPDYQK | 1284.61 | nd | 1284.86 |
| 32–38 | CAPLILR | 785.47 | 785.61 | 785.61 |
| 39–50 | LAWHSAGTFDSK | 1319.64 | 1319.90 | 1319.89 |
| 53–61 | TGGPFGTIK | 877.48 | nd | 877.64 |
| 62–79 | HQAELAHGANNGLDIAVR | 1885.96 | 1886.36 | 1886.35 |
| 80–85 | LLEPIK | 712.46 | nd | nd |
| 86–119 | EQFPIVSYADFYQLAGVVAVEITGGPEVPFHPGR | 3689.86 | 3690.59 | 3690.61 |
| 120–130 | EDKPEPPPEGR | 1250.60 | 1250.88 | 1250.85 |
| 137–142 | GSDHLR | 684.34 | 684.46 | 684.46 |
| 148–170 | AMGLSDQDIVALCGGHTIGAAHK | 2265.11 | 2265.56 | nd |
| 148–170+haem | AMGLSDQDIVALCGGHTIGAAHK | 2881.61 | nd | 2879.99 |
| 173–199 | SGFEGPWTSNPLIFDNSYFTELLTGEK | 3049.45 | 3050.17 | 3050.15 |
| 200–209 | DGLLQLPSDK | 1085.58 | 1085.88 | 1085.86 |
| 210–223 | ALLTDSVFRPLVEK | 1587.91 | 1588.25 | 1588.22 |
| 224–241 | YAADEDVFFADYAEAHLK | 2074.94 | 2075.35 | 2075.39 |
| 242–250 | LSELGFAEA | 936.47 | nd | nd |
Properties of ferrous S160C
Reduction of ferric S160C with sodium dithionite in the presence of dithiothreitol (5 mM) under strict anaerobic conditions gave a UV–visible spectrum (λmax=424.5, 527 and 557 nm) (Figure 2C) that was typical of a reduced b-type haem. Over time, however, subtle changes were observed in the spectrum of the ferrous S160C protein: the maxima shifted to 422, 526.5 and 555.5 nm. Similarly, the spectrum of the reduced pyridine haemochromagen derivative now had an α-band that had shifted from 556 to 553.5 nm (Figure 2B). These spectral changes are diagnostic of the covalent attachment to one of the two vinyl groups on the haem [8]. Reactions at higher pH (8.4), which would have allowed assessment of the importance or otherwise of the (deprotonated) thiolate form of the cysteine side chain, were not accessible as a result of protein precipitation over the timescale of the reaction.
HPLC and mass spectrometric experiments on the reduced protein provide further evidence in support of covalent haem attachment as a consequence of haem reduction. The HPLC chromatogram of S160C was altered upon reduction (Figure 3B), with most of the haem being eluted at 29.6 min instead of at 15 min as for ferric S160C. This haem peak now coincided with the protein peak (monitored at 280 nm) (Figure 3B). Co-elution of haem and protein is clear evidence for the presence of a covalent haem–protein interaction and has been used previously for characterization of other covalently linked haem proteins [3,10,18–21]. There was some protein degradation observed upon reduction of ferric S160C, resulting in the appearance of the broad peak eluted at 27 min in the 280 nm chromatogram (Figure 3B). This means that not all of the haem (≈31% in this case as assayed by HPLC) will form a covalent link, as the degraded protein is inactive towards the reaction (and hence the unbound haem peak is not expected to disappear totally). To remove this potential complication, samples derived from only the co-eluting 280/398 nm peak were used in subsequent mass spectrometry. The protein-bound haem peak was purified from free haem by HPLC, and MALDI–TOF analysis gave a molecular mass of 29092.99±6.76Da (Figure 4C), which is the mass expected for the covalently bound haem–S160C adduct [molecular mass (calculated) of 29092.63 Da].
Tryptic digestion/MS provided further evidence in support of covalent haem linkage formation. A sample of ferrous S160C after reduction with dithionite/dithiothreitol was subjected to the same trypsin digestion procedure as that described above for the ferric enzyme. In this case, good coverage of the overall sequence was observed again, with most of the expected peptide fragments detected (Table 1). However, in this case, the 148–170 fragment at molecular mass 2265 Da (which includes Cys160) was not seen (Table 1). Instead, there was an additional peak observed at molecular mass 2879.99 Da, which was assigned to the haem–peptide adduct AMGLSDQDIVALC160(haem)GGHTIGAAHK [molecular mass (calculated) of 2881.62 Da] (Table 1 and Figure 4B). In further experiments with the ferric S160C protein without reduction, it was found that this peak was not seen before reduction of ferric S160C and must therefore be a product of the reductive process (results not shown).
Reaction under oxidative conditions
In further experiments, it was shown that there was no covalent haem attachment when S160C was cycled with H2O2 (Figure 3C). This was in direct contrast with the S160M variant, which gave a covalent haem adduct under the same oxidative conditions and timescales [10].
Reactions with mesoporphyrin
Reactions with S160C reconstituted with iron(III) mesoporphyrin (in which alkyl groups replace the 2- and 4-vinyl groups) were also carried out. In this case, apo-S160C reconstituted with mesoporphyrin did not result in covalent attachment under reductive conditions (as is evident using HPLC; Figure 3D), with the haem being eluted at 15.5 min (this was verified by separate elution of free mesoporphyrin; results not shown). Oxidation of apo-S160C reconstituted with mesoporphyrin, by reaction with H2O2, also showed no evidence for covalent attachment (results not shown). In experiments with wild-type rpAPX, it was shown that no covalent haem modification occurred on reduction under conditions identical with those used for S160C (results not shown). Together, all these data indicate that a haem vinyl group, Cys160 and reductive conditions are all required for covalent attachment to occur.
DISCUSSION
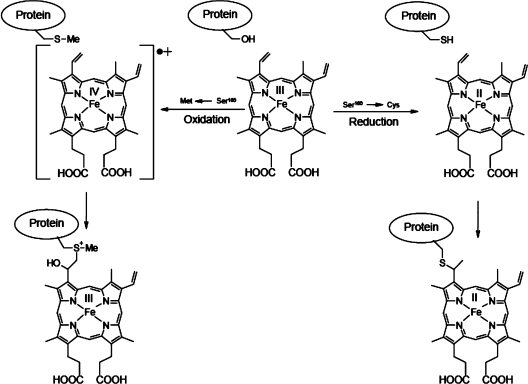
The present findings with the S160C variant of rpAPX, taken together with those made previously with the S160M variant [10], indicate that positioning of either a cysteine or a methionine side chain within potential bonding distance from a vinyl group of haem can lead to covalent bond formation. However, whereas oxidative conditions led to methionine–haem bond formation [10], only reducing conditions permitted thioether bond formation involving Cys160. The latter result is significant because thioether bond formation might, in principle, be achieved via a radical mechanism that depends upon the availability of an oxidant [22]. Although several lines of in vitro [8] and in vivo [23] evidence have suggested that reducing conditions promote thioether bond formation as seen in c-type cytochromes, these observations might relate to a requirement for ferrous haem and/or prevention of the two cysteine residues of the CXXCH motif becoming a disulfide, rather than a requirement for the bond formation itself. The fact that oxidative conditions (i.e. reaction of S160C with peroxide) failed to promote thioether bond formation can be taken as significant additional evidence that the thioether bond of haem proteins is not normally achieved through an oxidant-dependent radical reaction, either in vitro or in vivo where a post-translational modification apparatus is operative. Nevertheless, in some circumstances, covalent bonds between protein and haem (to give products other than the typical thioether bond of a c-type cytochrome) can be formed under oxidizing conditions. Thus Barker et al. [22] proposed a radical reaction involving consumption of molecular oxygen, to account for some of the products observed when a variant of cytochrome b5, bearing a cysteine residue in the haem-binding site, was expressed in the E. coli cytoplasm. Daltrop et al. [8] reported that reaction of apo-cytochrome c552 with ferric haem in vitro led to undefined mixed products. To what extent these non-physiological products depend upon complex factors such as the tendency of thiol groups (either of the protein or of dithiothreitol) to reduce ferric haem bound to different proteins, and thus with different reduction potentials, is not known. From a synthetic perspective, the generation of different covalent bonds within the same protein architecture under both oxidizing (using a ferryl intermediate in S160M) and reducing (using ferrous haem in S160C) conditions provides an interesting possibility of tuning the attachment mode of haem to protein with selective reactions. This idea is depicted in Scheme 1.
Scheme 1. How formation of a covalent haem–protein link may occur to different amino acid residues under both oxidizing and reducing conditions.
Although we did not make a detailed study of the kinetics of thioether bond formation in the S160C variant protein, we ascertained that the reaction required approx. 12 h to form significant amounts of protein with haem covalently attached. This is very similar to the time course observed previously for in vitro thioether bond formation in a bona fide c-type cytochrome [8]. Although this similarity might be coincidental, it could be that this represents the rate of reaction that is obtained as a consequence primarily of a proximity effect. It seems unlikely that a reaction on this timescale would be operative in vivo, and thus catalysed thioether bond formation, by any of the three characterized cytochrome c biogenesis systems [24,25], presumably must rely upon other factors so far unidentified to accelerate the reaction between thiol and vinyl. We have speculated previously that the histidine residue of the CXXCH unit might act as a proton donor in thioether bond formation, including during the in vitro formation of a thioether bond in a c-type cytochrome [26]. The equivalent of this histidine residue is not present in the S160C variant of APX, and inspection of its structure shows that the immediate environment of the introduced cysteine residue and the adjacent 2-vinyl group is hydrophobic. The nearest possible side chain capable of donating a proton is the side chain of His239, but this is 6 Å away and could only be involved in protonation via a bridging water. Thus proton donation from a nearby side chain may not be important for the reaction of the vinyl group with cysteine in this instance and therefore perhaps also not for thioether bond formation in cytochrome c552, where only a small dependence of rate of thioether bond formation on pH was detected [8].
An interesting question arises as to the generality with which covalent bonds between haem and polypeptide might form. In a separate work, Colas and Ortiz de Montellano [27] duplicated the ester link in horseradish peroxidase by incorporation of a glutamate residue close to the 3-methyl group (F41E variant of horseradish peroxidase) and also showed that the formation of the link was H2O2-dependent. A view has therefore emerged that any peroxidase, or perhaps any haem protein, that can react with H2O2 and that has a suitable amino acid side chain poised in the right place will, under the right conditions, form a covalent link to the haem. This view is reinforced by other protein engineering work on the CYP4 family of cytochrome P450s, which also contain a covalent link between a carboxy residue and a haem methyl group [2–4]. However, from the present study, we can conclude that cysteine can be inert to formation of a bond under oxidative conditions. Nevertheless, under reductive conditions, thioether bond formation might prove to be general. Thus the newly discovered single thioether bond between haem and polypeptide in the cytochrome b6f complex [28,29] could prove to be an additional in vivo example, while thioether bond formation has been seen recently following in vitro reaction of designer four-helix bundle proteins and haem [30]. Thus suitable mutagenesis, followed by either oxidizing or reducing reaction conditions as appropriate, may provide routes to a variety of engineered covalent links between polypeptide and haem. There is evidence that such links can enhance protein stability, which is a desirable feature for proteins that has possible applications in diagnostics or in facilitating enzyme-catalysed steps in chemical syntheses.
Acknowledgments
We thank Mr Kuldip Singh for technical assistance and Dr Martin Mewies for construction of the His6 tag vector. O. D. acknowledges a Junior Research Fellowship from Christ Church, Oxford. This work was supported by The Wellcome Trust (grants 062641/Z/00/Z and 063688/Z/01/Z to E. L. R.), BBSRC (Biotechnology and Biological Sciences Research Council) (grant C20071 to S.J.F.) and The Leverhulme Trust (grant RF/RFG/2005/0299 to E. L. R.).
References
- 1.Barker P. D., Ferguson S. J. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure. 1999;7:R281–R290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 2.Colas C., Ortiz de Montellano P. R. Autocatalytic radical reactions in physiological prosthetic heme modification. Chem. Rev. 2003;103:2305–2332. doi: 10.1021/cr0204303. [DOI] [PubMed] [Google Scholar]
- 3.LeBrun L. A., Xu F., Kroetz D. L., Ortiz de Montellano P. R. Covalent attachment of the heme prosthetic group in the CYP4 cytochrome P450 family. Biochemistry. 2002;41:5931–5937. doi: 10.1021/bi025527y. [DOI] [PubMed] [Google Scholar]
- 4.Henne K. R., Kunze K. L., Zheng Y.-M., Christmas P., Soberman R. J., Rettie A. E. Covalent linkage of prosthetic heme to CYP4 family of P450 enzymes. Biochemistry. 2001;40:12925–12931. doi: 10.1021/bi011171z. [DOI] [PubMed] [Google Scholar]
- 5.Hoy J. A., Kundu S., Trent J. T., III, Ramaswamy S., Hargrove M. S. The crystal structure of Synechocystis hemoglobin with a covalent heme linkage. J. Biol. Chem. 2004;279:16535–16542. doi: 10.1074/jbc.M313707200. [DOI] [PubMed] [Google Scholar]
- 6.Lee D., Pervushin K., Dischof D., Braun M., Thony-Meyer L. Unusual heme-histidine bond in the active site of a chaperone. J. Am. Chem. Soc. 2005;127:3716–3717. doi: 10.1021/ja044658e. [DOI] [PubMed] [Google Scholar]
- 7.Uchida T., Stevens J. M., Daltrop O., Harvat E. M., Hong L., Ferguson S. J., Kitagawa T. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J. Biol. Chem. 2004;279:51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 8.Daltrop O., Allen J. W. A., Willis A. C., Ferguson S. J. In vitro formation of a c-type cytochrome. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7872–7876. doi: 10.1073/pnas.132259099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojo S., Sano S. Mechanism of a novel synthesis of hemin-C from protohemin and L-cysteine: a Markownikoff-type radical-addition reaction. J. Chem. Soc. Perkin Trans. 1981;1:2864–2870. [Google Scholar]
- 10.Metcalfe C. L., Ott M., Patel N., Singh K., Mistry S. C., Goff H. M., Raven E. L. Autocatalytic formation of green heme: evidence for H2O2-dependent formation of a covalent methionine-heme linkage in ascorbate peroxidase. J. Am. Chem. Soc. 2004;126:16242–16248. doi: 10.1021/ja048242c. [DOI] [PubMed] [Google Scholar]
- 11.Tomlinson E. J., Ferguson S. J. Conversion of a c type cytochrome to a b type that spontaneously forms in vitro from apo protein and heme: implications for c type cytochrome biogenesis and folding. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5156–5160. doi: 10.1073/pnas.090089397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Woodyear T. D., Johnson C. M., Barker P. D. Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562. Biochemistry. 2000;39:1499–1514. doi: 10.1021/bi991831o. [DOI] [PubMed] [Google Scholar]
- 13.Dalton D. A., Diaz del Castillo L., Kahn M. L., Joyner S. L., Chatfield J. M. Heterologous expression and characterization of soybean cytosolic ascorbate peroxidase. Arch. Biochem. Biophys. 1996;328:1–8. doi: 10.1006/abbi.1996.0135. [DOI] [PubMed] [Google Scholar]
- 14.Antonini M., Brunori E. Amsterdam: North Holland Publishers; 1971. Hemoglobin and Myoglobin and their Reactions with Ligands. [Google Scholar]
- 15.Daltrop O., Ferguson S. J. Cytochrome c maturation: the in vitro reactions of horse heart apocytochrome c and Paracoccus denitrificans apocytochrome c550 with heme. J. Biol. Chem. 2003;278:4404–4409. doi: 10.1074/jbc.M211124200. [DOI] [PubMed] [Google Scholar]
- 16.Sharp K. H., Mewies M., Moody P. C. E., Raven E. L. The crystal structure of the ascorbate peroxidase/ascorbate complex. Nat. Struct. Biol. 2003;10:303–307. doi: 10.1038/nsb913. [DOI] [PubMed] [Google Scholar]
- 17.Teale F. W. J. Cleavage of the haem–protein link by acid methylethylketone. Biochim. Biophys. Acta. 1959;35:543. doi: 10.1016/0006-3002(59)90407-x. [DOI] [PubMed] [Google Scholar]
- 18.Daltrop O., Smith K. M., Ferguson S. J. Stereoselective in vitro formation of c-type cytochrome variants from Hydrogenobacter thermophilus containing only a single thioether bond. J. Biol. Chem. 2003;278:24308–24313. doi: 10.1074/jbc.M301967200. [DOI] [PubMed] [Google Scholar]
- 19.Rae T. D., Goff H. M. The heme prosthetic group of lactoperoxidase. J. Biol. Chem. 1998;273:27968–27977. doi: 10.1074/jbc.273.43.27968. [DOI] [PubMed] [Google Scholar]
- 20.Baer B. R., Schuman J. T., Campbell A. P., Cheesman M. J., Nakano M., Moguilevsky N., Kunze K. L., Rettie A. E. Sites of covalent attachment of CYP4 enzymes to heme: evidence for microheterogeneity of P450 orientation. Biochemistry. 2005;44:13914–13920. doi: 10.1021/bi051267j. [DOI] [PubMed] [Google Scholar]
- 21.Limburg J., LeBrun L. A., Ortiz de Montellano P. R. The P450cam G248E mutant covalently binds its prosthetic heme group. Biochemistry. 2005;44:4091–4099. doi: 10.1021/bi047686i. [DOI] [PubMed] [Google Scholar]
- 22.Barker P. D., Ferrer J. C., Mylrajan M., Loehr T. M., Feng R., Konishi Y., Funk W. D., Macgillivray R. T. A., Mauk A. G. Transmutation of a heme protein. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson D. W., Neupert W. Import of cytochrome-c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome-c. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4340–4344. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens J. M., Daltrop O., Allen J. W. A., Ferguson S. J. C-type cytochrome formation: chemical and biological enigmas. Acc. Chem. Res. 2004;37:999–1007. doi: 10.1021/ar030266l. [DOI] [PubMed] [Google Scholar]
- 25.Stevens J. M., Uchida T., Daltrop O., Ferguson S. J. Covalent cofactor attachment to proteins: cytochrome c biogenesis. Biochem. Soc. Trans. 2005;33:792–795. doi: 10.1042/BST0330792. [DOI] [PubMed] [Google Scholar]
- 26.Allen J. W. A., Leach N., Ferguson S. J. The histidine of the c-type cytochrome CXXCH haem-binding motif essential for haem attachment by the Escherichia coli cytochrome maturation (Ccm) apparatus. Biochem. J. 2005;389:587–592. doi: 10.1042/BJ20041894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colas C., Ortiz de Montellano P. R. Horseradish peroxidase mutants that autocatalytically modify their heme prosthetic group. J. Biol. Chem. 2004;279:24131–24140. doi: 10.1074/jbc.M401687200. [DOI] [PubMed] [Google Scholar]
- 28.Kurisu G., Zhang H. M., Smith J. L., Cramer W. A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 29.Stroebel D., Choquet Y., Popot J. L., Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 30.Ishida M., Dohmae N., Shiro Y., Oku T., Iizuka T., Isogai Y. Design and synthesis of de novo cytochromes c. Biochemistry. 2004;43:9823–9833. doi: 10.1021/bi049546e. [DOI] [PubMed] [Google Scholar]