Abstract
Mammalian IRPs (iron regulatory proteins), IRP1 and IRP2, are cytosolic RNA-binding proteins that post-transcriptionally control the mRNA of proteins involved in storage, transport, and utilization of iron. In iron-replete cells, IRP2 undergoes degradation by the ubiquitin/proteasome pathway. Binding of haem to a 73aa-Domain (73-amino-acid domain) that is unique in IRP2 has been previously proposed as the initial iron-sensing mechanism. It is shown here that recombinant IRP2 and the 73aa-Domain are sensitive to proteolysis at the same site. NMR results suggest that the isolated 73aa-Domain is not structured. Iron-independent cleavage of IRP2 within the 73aa-Domain also occurs in lung cancer (H1299) cells. Haem interacts with a cysteine residue only in truncated forms of the 73aa-Domain, as shown by a series of complementary physicochemical approaches, including NMR, EPR and UV–visible absorption spectroscopy. In contrast, the cofactor is not ligated by the same residue in the full-length peptide or intact IRP2, although non-specific interaction occurs between these molecular forms and haem. Therefore it is unlikely that the iron-dependent degradation of IRP2 is mediated by haem binding to the intact 73aa-Domain, since the sequence resembling an HRM (haem-regulatory motif) in the 73aa-Domain does not provide an axial ligand of the cofactor unless this domain is cleaved.
Keywords: EPR, haem-binding protein, haem-regulatory motif (HRM), iron regulatory protein 2 (IRP2), NMR spectroscopy, proteolysis
Abbreviations: 73aa-Domain, 73-amino-acid domain; DMEM, Dulbecco's modified Eagle's medium; DMF, dimethylformamide; DTT, dithiothreitol; EDAC, 1-ethyl-3-(3-dimethylaminopropyl)carbodi-imide; EMSA, electrophoretic mobility-shift assay; HA, haemagglutinin; HRM, haem-regulatory motif; IRE, iron-responsive element; IRP, iron regulatory protein; NOE, nuclear Overhauser effect
INTRODUCTION
IRPs (iron regulatory proteins) are ubiquitous regulators of cellular iron homoeostasis. They are cytosolic RNA-binding proteins that post-transcriptionally control translation of proteins involved in storage (ferritin), transport (e.g. transferrin receptor 1) and use (e.g. erythroid α-aminolaevulinic acid synthase) of iron. The two metazoan IRPs, IRP1 and IRP2, bind with high affinity to RNA stem–loop structures known as IREs (iron-responsive elements) located in the 3′- or in the 5′-untranslated regions of mRNA. Depending on the 3′ or 5′ position of IRE with respect to the coding sequence, the binding of IRP induces mRNA stabilization or translational inhibition respectively [1]. The relative importance of this regulatory system may change under different conditions and for different cell types [2]. For instance, IRPs play a prominent role in adjusting protoporphyrin synthesis to transferrin-bound iron availability in erythroblasts, as suggested by the zebrafish anaemia induced by sustained activation of IRP1 [3]. But synthesis of proteins participating in iron homoeostasis does not always fit with the IRE–IRP model of regulation along differentiation of the lineage [4,5]. Other regulatory events, including transcriptional ones, may combine with, or supersede, the translational control carried out by IRP.
Therefore the precise role of the IRP–IRE system in mammalian iron homoeostasis has received much attention over the last few years after knockout animals were generated. IRP2−/− mice develop microcytic anaemia [6–8], whereas IRP1−/− mice are asymptomatic. Inactivation of both IRP1 and IRP2 is lethal at an early stage during embryonic development [9]. The disruption of IRP2 has also been associated with brain iron overload and the development of a neurodegenerative movement disorder [6,9]. However, other IRP2−/− mice do not show obvious signs of neurodegeneration [10]. Thus, despite these significant progresses, the respective roles of IRP1 and IRP2 in cellular iron homoeostasis remain to be fully delineated. However, IRP2 is often proposed to be the main regulator under normal conditions, since IRP1 is mainly in a non-IRE-binding form in tissues and it does not always seem to be able to replace IRP2 in animals deficient in the gene of the latter protein [11,12].
The cellular concentration of IRP2 results from the equilibrium between de novo synthesis and degradation [13]. In iron-replete cells, IRP2 is rapidly degraded by the ubiquitin/proteasome pathway [14,15]. Early work led to the conclusion that a proline- and cysteine-rich stretch of 73 amino acids [73aa-Domain (73-amino-acid domain)] encoded by the unique IRP2-specific exon 5 is necessary and sufficient for IRP2 degradation by a mechanism involving oxidation of cysteine residues upon iron binding and polyubiquitination [14,16]. More recent results have proposed that the 73aa-Domain plays a role in IRP2 degradation in response to haem and that the cysteine residue of a Cys-Pro-Phe-His, haem regulatory-like, motif in the domain binds oxidized haem [17–19]. However, studies by independent groups showed that an IRP2 deletion mutant lacking the full-length domain remains sensitive to iron-dependent degradation, similar to wild-type IRP2 [20,21].
To get direct information about the role of the 73aa-Domain in IRP2 degradation, the recombinant protein and the 73aa-Domain were purified and their interaction with haem was studied with a series of complementary methods. We have discovered a highly susceptible proteolytic site in IRP2-specific 73aa-Domain. Such a cleavage site may have remained undetected in previous studies and it strongly influences the haem-binding properties of the protein. Its widespread occurrence, both in vitro and inside cells, may explain at least part of the previous conflicting results obtained with IRP2.
EXPERIMENTAL
Plasmids
IRP2 cDNA was extracted from the pET16b-IRP2 plasmid constructed at EMBL (European Molecular Biology Laboratory), Heidelberg, Germany. NdeI and ClaI sites were used to subclone the IRP2 cDNA into the pT7-7 bacterial expression plasmid [22] to give pTIRP2.
The DNA fragment encoding the 73aa-Domain, corresponding to exon 5 of the hIRP2 gene, was obtained by PCR amplification with the 5′-GACTTCAGCATATGGCAATACAGAATGCACC-3′ (forward) and 5′-CTTGATTTTACACGAATTCTCATTCAGGCACTGG-3′ (reverse) primers from the above pTIRP2 plasmid. The fragment was digested with NdeI and EcoRI, and cloned into the pTYB12 plasmid (IMPACT system; New England Biolabs, Beverly, MA, U.S.A.) to give pTYB12/73-D. This plasmid encodes the 73aa-Domain sequence N-terminally fused to the intein gene. The correct construction of pTYB12/73-D was checked by sequencing.
73aa-Domain production in Escherichia coli
Plasmid pTYB12/73-D was used to transform ER2566 competent cells (F− λ− fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10-TetS)2 R(zgb-210::Tn10)(TetS) endA1 [dcm] from New England Biolabs) and grown in LB (Luria–Bertani) medium (Gibco BRL, Paisley, Renfrewshire, Scotland, U.K.) supplemented with 100 μg/ml ampicillin at 37 °C until early exponential phase. Induction was carried out for 4 h at 22 °C after the addition of 0.5 mM IPTG (isopropyl β-D-thiogalactoside).
73aa-Domain purification
Cells were harvested at 5000 g for 30 min at 10 °C and the cell pellet was suspended in 50 mM potassium phosphate buffer (pH 8) with 250 mM NaCl (buffer A). Oxygen was removed from the gas phase by bubbling purified argon. Cell suspensions were broken by sonication and centrifuged at 20000 g for 30 min at 4 °C. The supernatant was loaded anaerobically on a chitin beads column (New England Biolabs) equilibrated with buffer A for affinity chromatography. After two washes with the same buffer, cleavage of the fused intein-73aa-Domain protein was carried out on the column with 3 vol. of the same buffer adjusted at pH 7.2 and containing 50 mM DTT (dithiothreitol). The resin was left either at room temperature (22 °C) for 40 h or at 4 °C in the presence of 5 μM PMSF, depending on the experiment (see the Results section). Excess buffer was removed to elute the 73aa-Domain fragment and the column was washed with 3 vol. of buffer A. Eluted fractions were analysed by SDS/PAGE on a 15% acrylamide gel. Fractions containing the expected peptide of approx. 8 kDa were pooled and concentrated on a YM3 membrane (3000 Da cut-off value; Amicon; Millipore, Danvers, MA, U.S.A.). The concentrated fraction was further purified by gel filtration through a BioGel P10 (Bio-Rad Laboratories, Hercules, CA, U.S.A.) column equilibrated with 50 mM potassium phosphate buffer at pH 6.2 containing 250 mM NaCl. A single protein fraction was eluted and the purification yield was 1.3 mg/litre of culture.
IRP2 production in E. coli
The pTIRP2 plasmid was used to transform K38 cells already containing the pGP1-2 plasmid [22]. The latter plasmid bears the T7 RNA polymerase gene under the control of the λpL promoter, and the cI-857 temperature-sensitive repressor. Cells were grown in Terrific broth (Gibco BRL) with 100 μg/ml ampicillin and 50 μg/ml kanamycin at 30 °C for 20 h. Induction was carried out by increasing the temperature to 42 °C for 1 h 30 min, prior to production at 30 °C for 6 h.
IRP2 purification
All purification steps were carried out under argon to avoid uncontrolled protein oxidation. IRP2 was monitored by EMSAs (electrophoretic mobility-shift assays) to detect IRP2 RNA-binding activity [23]. Cells were harvested at 4000 g for 30 min at 25 °C: the cell pellet was resuspended in 20 mM Tris/HCl and 5 μM PMSF (pH 7.4) and frozen. After thawing, lysates were prepared by sonication and centrifuged at 45000 g for 1 h at 20 °C. Ammonium sulfate (1.07 M, 30% saturation at 4 °C) was added to the supernatant prior to 30 min centrifugation at 12000 g and 4 °C. The resulting supernatant was brought to 3.2 M ammonium sulfate (60% saturation) and centrifuged at 12000 g for 90 min at 4 °C. The pellet was resuspended in 20 mM Tris/HCl, 5 μM PMSF and 1% (v/v) glycerol (pH 8) and dialysed for 2 h against the same buffer, followed by overnight dialysis against this buffer supplemented with 0.1 mM DTT. The dialysed supernatant was centrifuged at 45000 g for 30 min at 20 °C before being loaded on to a 50 ml DEAE-cellulose column (DE52; Whatman International, Maidstone, Kent, U.K.) equilibrated with dialysis buffer. Elution was carried out with a discontinuous gradient of NaCl in the same buffer: 2 column vol. of 0.15 M NaCl were followed by 3 column vol. of 0.3 M NaCl. Ammonium sulfate (0.4 M) was added to the eluted active fractions, followed by centrifugation at 18000 g for 20 min at 20 °C. The supernatant was loaded on to a 200 ml phenyl-Sepharose 6B column (GE Healthcare, Orsay, France) equilibrated with 20 mM Tris/HCl, 5 μM PMSF, 1% glycerol, 0.1 mM DTT and 0.4 M (NH4)2SO4 (pH 8). Elution was carried out by decreasing the ammonium sulfate concentration in the same buffer. The active fraction was desalted by overnight dialysis and loaded and filtered on to a 2L-Superdex 200 column (GE Healthcare) equilibrated with 50 mM Tris/HCl, 1% glycerol, 0.1 mM DTT and 50 mM NaCl (pH 8). The active fraction was loaded on to 50 ml of hydroxyapatite (Bio-Gel Hydroxyapatite HTP; Bio-Rad Laboratories) equilibrated with 20 mM Tris/HCl, 1% glycerol and 0.1 mM DTT (pH 7.4). Elution was carried out using increasing potassium phosphate concentrations between 5 and 200 mM with 1% glycerol and 0.1 mM DTT. The last step of IRP2 purification involved heparin-affinity HPLC (GE Healthcare) by applying a gradient of potassium chloride in 20 mM Hepes-OH and 0.1 mM DTT (pH 7.4).
Haem–agarose affinity gel resin synthesis
A volume of 2 ml of Affi-Gel 102 cross-linked agarose (Bio-Rad Laboratories) suspension was washed twice with water, twice with DMF (dimethylformamide) and resuspended in DMF. Then, 10 mg of haemin chloride (Sigma–Aldrich, Saint-Quentin Fallavier, France) was dissolved in 3 ml of DMF and added to the gel. EDAC [1-ethyl-3-(3-dimethylaminopropyl)carbodi-imide] was used as coupling reagent for carboxyl-containing ligands. EDAC (300 mg) was dissolved in 800 μl of 50% DMF in water. The EDAC solution (100 μl) was added to the haem–agarose mixture before acidification with 1 M HCl to pH 3. A further 400 μl of EDAC was added before re-acidification to pH 4. The mixture was left to react for 3 h at room temperature on a rotating rod and the pH was adjusted every 30 min. The remaining 700 μl of the EDAC solution was added to the reaction mixture and left rotating overnight. After 21 h, the reaction was stopped by adding 300 μl of 1 M NaOH to increase the pH to 7.5. The haem-linked resin was washed three times in DMF. The supernatant was colourless on the final rinse and the resin had a pale brown colour, indicating the presence of haem. The resin was resuspended in 1.5 ml of 0.1 M NaCl.
Coupling efficiency was calculated indirectly. Unchanged haem recovered in the resin washes was quantified using spectrophotometry and subtracted from haem added to the initial reaction mixture. A volume of 42 μl of recovered supernatant from resin washes was diluted 100-fold in water. Pyridine (1 ml) was added, followed by 0.5 ml of 1 M NaOH. Sodium dithionite (5 mg) was added and absorbance of this solution was measured at 419 nm. Coupling yield was 1.5 μmol of haem/ml of agarose resin.
Interaction between haem–agarose resin and purified proteins
The haem-bound resin was washed twice with water, twice with 8 M urea and twice with elution buffer (50 mM sodium phosphate and 0.5 M NaCl, pH 7.3) prior to incubation with purified proteins. To check for the trapping efficiency of the resin towards apo-haem proteins, 50 μl of resin, resuspended in elution buffer, was incubated at 22 °C for 2 h on a rotating rod with 20 μg of purified apomyoglobin. The same amount of apomyoglobin was also incubated with 50 μl of non-derivatized Affi-Gel as a control of binding specificity. The same procedure was applied to 5 μg of purified IRP2 with 30 μl of resin under identical conditions. Unbound proteins were first eliminated in the supernatant of a 2 min 10000 g centrifugation, then by washing the resin with 50 mM sodium phosphate and 0.5 M NaCl (pH 7.3). Bound proteins were eluted after thorough mixing with 8 M urea in the same buffer for 10 min. Protein fractions were precipitated using 10% (v/v) trichloroacetic acid and resuspended in sample buffer [10% (v/v) 2-mercaptoethanol, 6% (w/v) SDS, 20% glycerol and 0.2 mg/ml Bromophenol Blue) before being denatured and analysed by SDS/PAGE and Western blotting.
Culture of the H1299 lung cancer cell line
H1299 lung cancer cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine, 100 units of penicillin/ml and 0.1 mg of streptomycin/ml. The clones producing HA (haemagglutinin)-tagged wild-type human IRP2 or HA-tagged IRP2 with deletion of the specific 73aa-Domain, IRP2Δ73, under the control of a tetracycline-inducible promoter were maintained in DMEM containing 2 μg of tetracycline/ml, 2 μg of puromycin/ml and 250 μg of G418/ml. H1299 cells were plated for 48 h in tetracyclin-free media to express IRP2–HA or IRP2Δ73–HA cDNA. Tetracycline (2 μg/ml) was then added back to shut off transcription. No protease inhibitors were added to the culture medium.
Plasmids expressing the IRP2–HA and IRP2Δ73–HA cDNA were prepared and transfected in H1299 lung cancer cells as described elsewhere [21,24].
Proteins extraction from the H1299 lung cancer cell line
H1299 cells were harvested and washed twice with PBS. Cell pellets were resuspended in lysis buffer (1% Triton X-100, 25 mM Tris/HCl, pH 7.4, and 40 mM KCl) in the presence of protease inhibitors {a mixture of AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], pepstatin A, E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane], bestatin, leupeptin and aprotinin; Sigma} and incubated for 20 min in ice prior to 15 min centrifugation at 15000 g and 4 °C. Total protein concentrations were determined with the Bio-Rad Laboratories (Munich, Germany) protein assay.
Western blotting
Total proteins (50 μg) were resolved by SDS/PAGE on 8% gels, and proteins were transferred on to nitrocellulose membranes. The blots were saturated with 10% (w/v) non-fat milk in PBS/Tween 0.2% and probed overnight at 4 °C with antibodies against HA (Santa Cruz Biotechnologies, Santa Cruz, CA, U.S.A.) or β-actin (Sigma) at 1:1000 dilution in PBS-Tween 0.2% with 5% non-fat milk. To probe directly IRP2, antibodies against the QKAGKLSPLKVQPKKLP peptide (amino acids 151–167) of human IRP2 [a gift from Dr Cécile Bouton, ICSN (Institut de Chimie des Substances Naturelles), Gif-sur-Yvette, France] were used at 1:2000 dilution. Following three washes with PBS-Tween 0.2%, the blots were incubated with peroxidase-coupled goat anti-rabbit IgG at 1:5000 dilution for 2 h at room temperature, before detection by the enhanced chemiluminescence method (ECL®; GE Healthcare). Alternatively, the blots were incubated for 2 h with alkaline-phosphatase-coupled goat anti-rabbit IgG at 1:5000 dilution at room temperature, followed by colorimetric detection [Sigma Fast BCIP (5-bromo-4-chloroindol-3-yl phosphate)/NBT (Nitro Blue Tetrazolium)]. The results shown in the Figures are representative of several independent experiments.
NMR analysis
Peptide samples were prepared for NMR in 50 mM phosphate buffer and 250 mM NaCl (pH 6.2) in 1H2O/2H2O (9:1, v/v) under anaerobic conditions at concentrations ranging from 200 to 300 μM. NMR tubes were generally sealed with gas-tight caps, unless otherwise indicated.
1H NMR spectra were recorded at 25 °C on a Varian Inova 600 spectrometer equipped with a penta 1H/31P/13C/2H/15N resonance probe operating at 600 MHz on the proton channel. Water suppression on a 10 p.p.m. spectral width was achieved using a classical watergate sequence [25] after optimization of water-selective Gaussian pulses. A 2.5 s recycle delay was used to collect these data. During processing, residual water was further suppressed using a frequency filter, and truncation artefacts were removed using a standard Gaussian window function over the 6913 Hz spectral width. Chemical shifts were externally referenced to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt at 25 °C. To improve sensitivity, two-dimensional 1H,1H-spectra were collected at 25 °C on a Varian Inova 800 spectrometer equipped with a triple 1H/13C/15N resonance unlabelled probe operating at 800 MHz on the proton channel. NOESY and TOCSY spectra were recorded with 150 and 70 ms mixing times respectively using excitation sculpting for water suppression [26] and a 1.1 s relaxation delay. Efficient water suppression was detrimental to the detection of backbone amide protons exchanging with water, but beneficial for the detection of aromatic residues such as phenylalanine residues. A HET-SOFAST 1H-spectrum was also collected with acquisition parameters suitable to assess peptide compactness [27].
Other methods
Absorption spectra, EPR spectra [28] and N-terminal sequences [29] were obtained as previously described.
RESULTS
The purified 73aa-Domain of IRP2 is susceptible to proteolysis
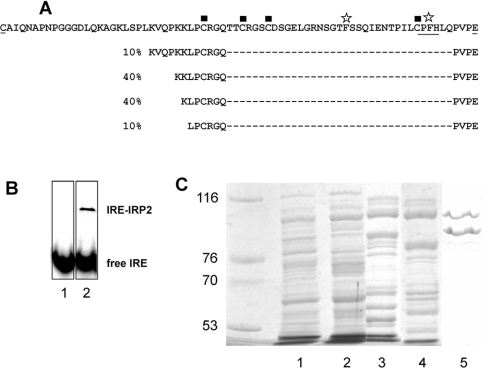
The 73aa-Domain of IRP2 was produced in E. coli using the intein-tag system and purified after on-column cleavage under reducing conditions (see the Experimental section). The final fraction was loaded on to a 15% acrylamide gel and analysed by Western blotting using antibodies raised against IRP2. Fast-migrating immunoreactive material was detected, as expected for the 73aa-Domain of IRP2 (results not shown). However, analysis by N-terminal Edman sequencing gave several sequences, corresponding to four truncated forms of the 73aa-Domain (Figure 1A). Proteolysis occurred near closely spaced lysine residues, resulting in truncated peptides of approx. 45 amino acids (and a minor one of 51), containing the C-terminus of the 73aa-Domain and encompassing all cysteine residues encoded by IRP2 exon 5. In the following, this C-terminal part of the peptide with heterogeneous N-termini will be called the ‘truncated peptide’.
Figure 1. Cleavage of purified 73aa-Domain and IRP2.
(A) N-terminal sequencing of the purified 73aa-Domain of IRP2 reveals the presence of truncated products. The 73aa-Domain sequence is indicated on the first line. The underlined amino acids at the N- and C-termini are those encoded across the splice junctions and the first cysteine does not belong to the construction used. The phenylalanine residues discussed in the text are indicated with star symbols, the cysteine residues are indicated with squares, and the HRM-like sequence is underlined. The N-termini of other detected products are shown on the other lines, with their respective proportions. (B) EMSA of purified recombinant IRP2. Lane 1, labelled IRE probe alone; lane 2, recombinant IRP2 incubated with an excess of IRE probe. Two wells from the same gel were put side by side for the Figure. (C) Two major bands were obtained after the last step of recombinant IRP2 purification. Active fractions from each purification step (1, crude extract; 2, DE52; 3, phenyl-Sepharose; 4, Superdex; 5, heparin-affinity) were separated by SDS/8%-PAGE and stained with Coomassie Blue. The left lane displays molecular-mass markers.
In order to reduce proteolysis, a similar purification procedure was carried out at low temperature by minimizing proteolytic activities. N-terminal sequencing revealed the presence of one major sequence starting at the expected intein cleavage site, indicating purification of full-length IRP2 73aa-Domain.
These results indicate that IRP2 73aa-Domain is highly sensitive to proteolytic cleavage, but that proteolysis in bacterial lysates can be largely avoided by adding the serine protease inhibitor PMSF during the purification steps. Therefore PMSF was added in all samples obtained by breaking cells, except in the experiments aiming at isolating cleaved proteins or peptides.
Recombinant IRP2 is also efficiently cleaved at the same site
It was of interest to know whether the 73aa-Domain cleavage was limited to the isolated peptide, or if the same site in the complete IRP2 protein was also prone to proteolysis. Following expression in E. coli and purification, the final fraction of human IRP2 was active (Figure 1B) and it was analysed by electrophoresis (Figure 1C). Two major bands of ∼106 and ∼80 kDa were detected and subjected to N-terminal Edman sequencing. The 106 kDa band was found to be the full-length purified IRP2, whereas the N-terminus of the 80 kDa band was a mixture of two sequences beginning with Lys-Val-Gln-Pro-Lys-Lys-Leu-Pro- and Lys-Leu-Pro-Cys-Arg-Gly-Gln-Thr. In agreement with the results of Figure 1(A), these fragments correspond to cleavage sites located in the 73aa-Domain. They indicate that two truncated forms are significantly produced from recombinant IRP2, despite the addition of PMSF during the purification steps. These results show that the 73aa-Domain is particularly susceptible to proteolysis in the whole molecule and indicate that this region is probably highly accessible, as predicted from the three-dimensional structure of the homologous IRP1 protein [30].
Thus analysis of the degradation products of both purified 73aa-Domain and IRP2 points to the specific susceptibility of this domain to proteolytic cleavage.
Proteolytic cleavage of IRP2 occurs in H1299 cells producing IRP2–HA
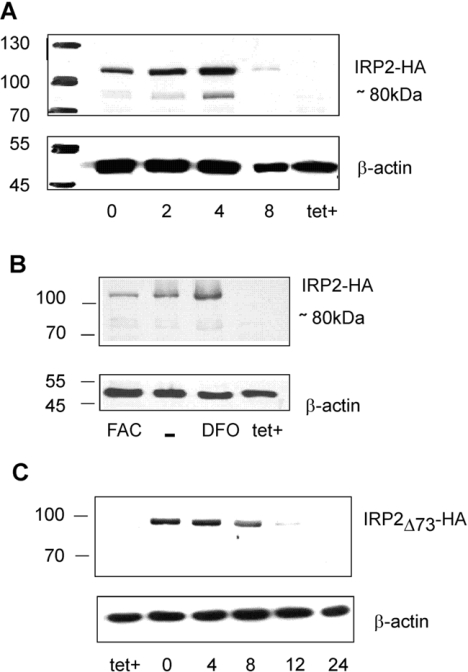
The cleavage sites in the 73aa-Domain of IRP2 were evidenced above with purified proteins. To address whether cellular IRP2 is also sensitive to cleavage at the same sites, H1299 lung cancer cells producing HA-tagged IRP2 (IRP2–HA) or a version devoid of the 73aa-Domain (IRP2Δ73–HA) were studied.
The cells were plated for 48 h in tetracycline-free medium to accumulate IRP2–HA. Tetracycline was then added to shut off IRP2–HA synthesis, and IRP2–HA levels were monitored at different time intervals by Western blotting with HA antibodies. A second band of approx. 80 kDa was clearly detected in addition to the 106 kDa band corresponding to full-length IRP2 (Figure 2A). The approx. 80 kDa size matches that of the truncated recombinant IRP2 determined in vitro. The intensity of the 80 kDa band changed in parallel with the major 106 kDa band in all experiments, indicating that the amount of the 80 kDa band was related to that of IRP2. Beyond 8 h after stopping IRP2 production, the protein concentration was largely decreased and the 80 kDa band became undetectable, indicating further degradation.
Figure 2. Cleavage of IRP2 in H1299 cells.
(A) IRP2–HA turnover in H1299 cells. After induction for 48 h, tetracycline was added back to cells for different times, indicated in hours below each lane. The tet+ label corresponds to extracts of control cells kept in the presence of tetracycline, i.e. without induction of IRP2–HA cDNA expression. Total cell lysates (50 μg) were analysed by SDS/PAGE on 8% gels. Western blots were probed using anti-HA (upper part) or anti-β-actin (lower part) antibodies on two pieces of the membrane. In some cases, only a few wells of the same gel are presented, as these gels contained more samples than needed for the considered Figure. (B) IRP2–HA-specific cleavage is iron-independent. After induction for 48 h, tetracycline was added back to cells for 4 h (lane –), with 50 μg/ml of iron ammonium citrate (FAC) or with 100 μM desferrioxamine (DFO). Total cell lysates (50 μg) were analysed as in (A). The second faint band above the approx. 80 kDa product is non-specific, as it can be seen in the tet+ (uninduced) well in the absence of IRP2–HA. (C) IRP2Δ73–HA is not specifically cleaved in H1299 cells. After 48 h induction of IRP2Δ73–HA by removal of tetracycline, cells were put back in the tetracycline-containing medium for 0, 2, 4, 8, 12 or 24 h and analysed as above. Molecular masses in kDa are shown on the left.
In order to establish whether proteolysis occurred before or after cell lysis, the experiments were carried out either in the presence or in the absence of protease inhibitors during the preparation of extracts (see the Experimental section). Similar results to those shown in Figure 2(A) were obtained by Western-blot analysis under both sets of experimental conditions, indicating that IRP2–HA proteolysis was probably not occurring upon sample processing after cell lysis.
We next examined whether iron had some effect on the 80 kDa cleavage product. After induction of IRP2 for 48 h, tetracycline was added back and 50 μg/ml ferric ammonium citrate or 100 μM of the iron chelator desferrioxamine was added after 1 h. The cells were lysed 7 h later, and the band at 80 kDa was detectable in Western blots under all conditions in proportion to the IRP2 amount (Figure 2B). Therefore the specific cleavage of IRP2 was not sensitive to these treatments.
To assess further whether the cleavage site evidenced in vitro (Figure 1) is responsible for the appearance of the truncated form of IRP2 in cells, the same experiments were carried out with H1299 cells producing HA-tagged IRP2Δ73 in which the 73aa-Domain is absent (Figure 2C). Up to 8 h after the end of the induction period, a band of 98 kDa corresponding to intact IRP2Δ73 was observed. No smaller bands were detected, suggesting that degradation of IRP2Δ73 does not generate sufficiently stable intermediates that can be detected on these gels. Comparison of Figures 2(A) and 2(C) indicates that the IRP2-specific cleavage occurs in the 73aa-Domain.
It has been established that iron accelerates IRP2 degradation [14,31] and it has been proposed that IRP2 detects intracellular iron levels by binding haem via the 73aa-Domain [17–19] in a process triggering its degradation [19,32]. Since the cleavage sites shown in Figure 1(A) leave truncated fragments containing all four cysteine residues and the histidine of the 73aa-Domain, but none of the lysine residues, it was decided to investigate haem and metal binding to these peptides.
1H-NMR does not evidence folding of either truncated or full-length 73aa-Domain, even in the presence of iron or haem
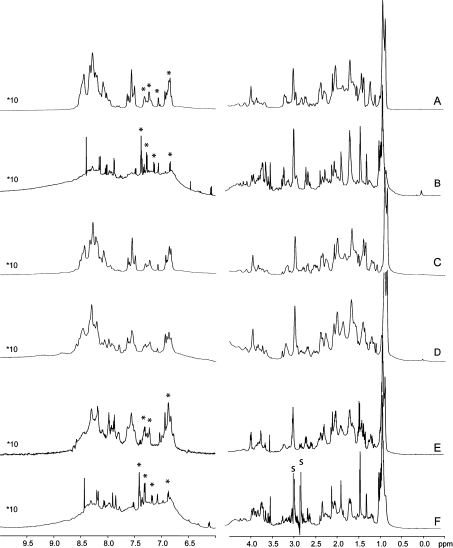
Purification of the 73aa-Domain was carried out under argon and in the presence of DTT to avoid oxidative damage to the peptide. The starting material of the following experiments is thus the reduced peptide in which cysteine residues occur as free thiol groups. These experiments were carried out in relatively dilute solutions, 300 μM at most, since purified 73aa-Domain is precipitated at higher concentrations. The 1H-watergate spectrum of the truncated peptide is reported in Figure 3(A). In what follows, ‘truncated peptide’ refers to the mixture of cleaved peptides identified in Figure 1(A). All of the detected signals fall into the 0.8–8.6 p.p.m. range. A narrow and intense signal centred around 1 p.p.m. accounts for the methyl groups of the molecule, and the spectra display a particularly low ratio of protons in the amide/aromatic region between 6 and 9 p.p.m. as compared with the aliphatic region. These features are indicative of the absence of secondary- and tertiary-structure elements in the peptide. This is further confirmed by the absence of any inter-residue correlation in an extended series of NOESY [and ROESY (rotating-frame Overhauser enhancement spectroscopy)] experiments, as well as by a λnoe=0.80 value in the 1D-HETnoe-SOFAST NMR spectrum [27] collected at 800 MHz (results not shown). These results strongly suggest that the truncated peptide of the 73aa-Domain does not fold under these experimental conditions.
Figure 3. 1H-watergate spectra of the 73aa-Domain.
Spectra were recorded for the truncated peptide (A), for full-length 73aa-Domain (B), for the truncated peptide in the presence of ZnCl2 (C), FeSO4 (D) or haemin (E), and for the full-length peptide in the presence of haemin (F). Peaks labelled with an asterisk have been identified as non-exchangeable protons on spectra recorded in 2H2O and are considered in the TOCSY spectra discussed in the text. In (F), ‘s’ stands for peaks of residual DMF used to dissolve the added haemin. The intensity of the signals in the amide region on the left is multiplied by ten as compared with that in the aliphatic region on the right.
Qualitatively similar results were obtained with full-length 73aa-Domain (Figure 3B): the region spanned by the 1H-NMR signals is as narrow as in the case of the truncated peptide, and no inter-residue NOEs (nuclear Overhauser effects) were detected. Therefore the full-length peptide does not display secondary-structure elements, and the additional presence of the lysine- and proline-rich N-terminal sequence of the 73aa-Domain (Figure 1A) does not help folding of this domain. Furthermore, the 1H-watergate spectrum in Figure 3(B) reveals a hump below sharp resonances in the aromatic/amide region, suggesting that the full-length peptide has a tendency to aggregate or to sample a series of different conformations in solution at the concentrations used for the NMR study.
The truncated peptide was then oxidized either by bubbling oxygen into the sample or by addition of an excess of ferricyanide. Neither of these treatments significantly changed the initially recorded 1H-NMR spectrum of the truncated peptide (results not shown). Therefore oxidation of the truncated domain did not favour peptide folding, or the 1H-NMR spectra of the folded fraction, if any, escaped detection under these experimental conditions.
If the 73aa-Domain acts as a sensor, addition of metals may contribute to some conformational changes and folding. Addition of zinc chloride did not induce any major changes in the 1H-watergate spectrum of the full-length (results not shown) or the truncated peptide (Figure 3C). The 1H-spectra of the truncated peptide (Figure 3A) show displacement of amide proton resonances from the 8–8.5 p.p.m. range to the 7.5–8 p.p.m. region, as well as broadening of some resonances, in the presence of ferrous iron (Figure 3D). These small effects are assigned to the paramagnetism of Fe(II) ions at room temperature and local perturbations arising from slight pH changes or non-specific iron binding. Despite these minor changes, bivalent cations, including Fe(II), do not induce folding of the two studied forms of the 73aa-Domain.
Next, reduced truncated 73aa-Domain was incubated with excess of haemin and gel-filtered to remove unbound cofactor. The filtered protein fraction was coloured, indicating that some haem-containing compound co-eluted. However, no major changes, such as extensive signal spreading or overall significant signal intensity alteration, were observed in the 1H-watergate spectra (compare Figures 3A and 3E). The same experiment carried out with full-length 73aa-Domain gave qualitatively similar results (see Figures 3B and 3F).
Conformations of truncated or full-length 73aa-Domain studied by TOCSY
The information content of one-dimensional NMR is nevertheless scanty, and the 73aa-Domain samples at their low solubility limits are not likely to display strong NOE. Therefore these samples were compared by a NMR method circumventing part of these problems, namely by recording TOCSY spectra. Since haemin-reacted and gel-filtered full-length 73aa-Domain samples retained the colour of the cofactor, some interaction must occur. Indeed, TOCSY spectra of such samples displayed slightly less off-diagonal signals in the amide and aliphatic regions than spectra of the same peptide without haemin, although most of the spin systems were clearly conserved between the two samples (see Supplementary Table 2 at http://www.BiochemJ.org/bj/408/bj408ppppadd.htm. In contrast, only signals of two spin systems, that were not present in spectra of the 73aa-Domain without haem, were observed in the spectra of the haemin-reacted material (see Supplementary Table 3 at http://www.BiochemJ.org/bj/408/bj408429add.htm). These results are not in favour of a clear, strong and specific binding of haemin to the full-length 73aa-Domain peptide, an event that should change a subset of spin systems corresponding to residues in the vicinity of the binding site.
Comparison was then made between truncated and full-length peptides. The number of off-diagonal signals over the full-range TOCSY spectra of haemin-reacted truncated 73aa-Domain was not significantly smaller than that detected with the corresponding full-length sample (results shown for only part of the aromatic region in Figure 4). Only a few correlations in the full-length peptide, such as those annotated b4–b6 in Figure 4(B), were absent from truncated 73aa-Domain spectra (Figure 4A). However, many spin systems were clearly shifted between these two samples (see Supplementary Tables 2 and 4 at http://www.BiochemJ.org/bj/408/bj4080429add.htm). In most cases, the signals could not be sequence-specifically assigned, because the 73aa-Domain contains many occurrences of the same amino acids. One important exception is for phenylalanine residues: the 73aa-Domain sequence has two phenylalanine residues in its C-terminal part, so that both are present in truncated 73aa-Domain. The two phenylalanine side-chain proton signals can be identified in the TOCSY spectra of the truncated (Figure 4A, insets A1 and A2) and full-length (Figure 4B, insets B1 and B2) peptides. Both phenylalanine rings appear as intense and narrow signals, in the truncated peptide with spectroscopic signatures a1i and a2i (Figure 4A, insets A1 and A2, and Figure 4C) and in the full-length peptide with spectroscopic signatures b1i and b2i (Figure 4B, insets B1 and B2, and Figure 4D). Detailed analysis of these phenylalanine signals can be found in the Supplementary material (http://www.BiochemJ.org/bj/408/bj4080429add.htm). It indicates that haem interaction perturbs the NMR signatures of these residues in different ways for the two forms of the 73aa-Domain.
Figure 4. Analysis of haem binding on full-length and truncated 73aa-Domain peptides through two-dimensional TOCSY experiments.
Aromatic region of the TOCSY spectra collected with extensive water suppression and with 70 ms mixing time on truncated (A) and full-length (B) 73aa-Domain peptide. Samples in 50 mM phosphate buffer in 1H2O/2H2O (90:10, v/v) at pH 7 were made to react with haemin as described in the Experimental section. Corresponding one-dimensional spectra collected with the same water suppression scheme on the truncated and full-length peptide samples are reported in (C) and (D) respectively. Circles emphasize differences between the two two-dimensional spectra. The two phenylalanine residues found in both truncated (a) and full-length (b) 73aa-Domain are arbitrarily labelled with subscripts 1 and 2. As they occur in different local conformations, the corresponding spin systems are labelled i, ii and iii (see Supplementary material at http://www.BiochemJ.org/bj/408/bj4080429add.htm). The correlations are drawn with dashed lines. Additional unassigned spin systems are labelled with subscripts 4, 5 and 6.
Furthermore, some of the TOCSY signals that were found to shift or disappear upon association of haemin with full-length 73aa-Domain were recovered for truncated 73aa-Domain in the presence of haemin (Supplementary Tables 1 and 2 at http://www.BiochemJ.org/bj/408/bj4080429add.htm). One likely explanation for these results is that haemin interaction with full-length 73aa-Domain occurs through non-specific association of the high-spin ferric cofactor with the peptide, in agreement with other spectroscopic data (see below). As a result, the relaxation properties of the signals for protons close enough to haem change, and the corresponding signature becomes invisible in conventional TOCSY experiments. In the case of truncated 73aa-Domain, haemin mainly binds to a specific site and paramagnetic relaxation does not perturb the same signals. Therefore the signals lost in haemin-reacted full-length 73aa-Domain are recovered in haemin-reacted truncated 73aa-Domain as they occurred for the peptide in the absence of haem. Support for this interpretation is provided in the following, by implementation of complementary methods.
Additional spectroscopic evidence for interaction between haem and the truncated peptide
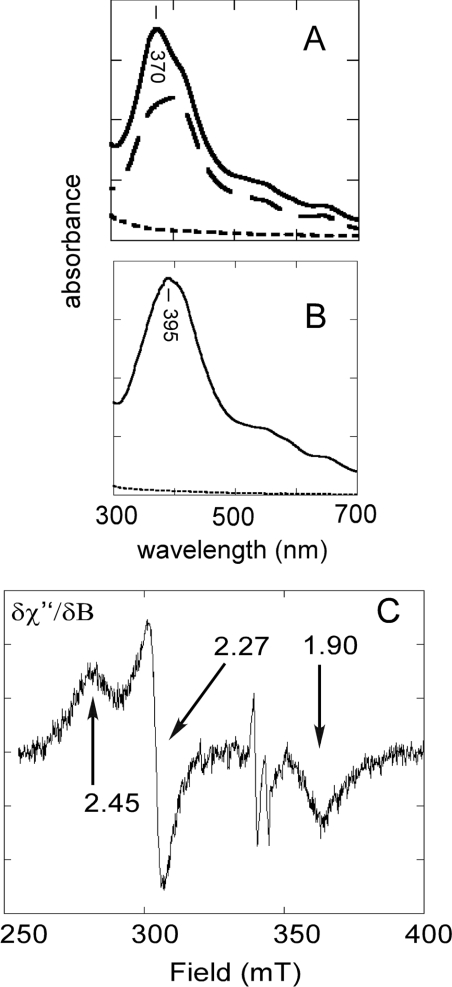
In order to characterize further the interaction between haem and the 73aa-Domain, absorption spectra were recorded. The absorption spectra of haemin solutions displayed overlapping intense bands between 350 and 450 nm, with a maximum at 398 nm (Figure 5A, dashed lines) as already observed in [33]. The truncated 73aa-Domain was added and, after reaction for 10 min in the absence of oxygen, the sample was gel-filtered to remove unbound haem. The visible absorption band of haemin sharpened and its maximum shifted to 370 nm (Figure 5A, full line). No increases of the baseline towards shorter wavelengths were apparent, excluding any aggregation problem detected at higher concentrations (see above). The hypsochromic shift of the Soret band has already been observed by interaction between haem and a thiolate axial ligand [34,35].
Figure 5. Interaction of haem with the IRP2 73aa-Domain studied by absorption spectroscopy and EPR.
(A) Absorption spectra of the truncated peptide in 50 mM phosphate buffer and 0.25 M NaCl (pH 8) in the absence (dotted line) and in the presence of one equivalent of haem (continuous line). The haemin absorption spectrum is shown for reference (discontinuous line). (B) Absorption spectra of the intact peptide with symbols as in (A). Maxima for the haem–peptide complexes are indicated. (C) EPR spectrum in the g≈2 region of the complex between haem and the truncated form of IRP2 73aa-Domain. Microwave frequency 9.655 GHz; microwave power 4 mW; receiver gain 4×105; modulation amplitude 1 mT, temperature 13 K.
Similar experiments were carried out with full-length 73aa-Domain. The maximum of the haemin absorption spectrum in the presence of the full-length domain did not shift significantly as compared with free haemin (Figure 5B). This observation is a strong indication that no residues of the 73aa-Domain contribute an axial ligand to haem that would be detected by a change in the electronic properties of the cofactor.
To evaluate further the interaction between haem and the truncated form of the 73aa-Domain, EPR measurements were carried out. The truncated 73aa-Domain was made to react with a 5-fold molar excess of haem, followed by gel filtration (Figure 5C). A low-spin ferric iron signal in the g≈2 region was detected, with g values at 2.45, 2.27 and 1.90. This pattern has already been observed for ferric haem with a thiolate axial ligand [36,37]. Integration of the signal indicated that only part of the iron present in the sample contributed to the spectrum, suggesting that binding of haem to truncated 73aa-Domain was not quantitative. Alternatively, haem may occur in a mixture of spin systems contributed by ligation in slightly different modes, as borne out by the observation that, in addition to the low-spin signal at g≈2, a high-spin signal was recorded at g≈6. Since the sample had been filtrated prior to EPR measurements, it is unlikely that the g≈6 signal corresponds to unbound haem, and a proportion of haem is probably bound to the sample as a penta-co-ordinated species contributing a high-spin signal.
The g values of the low-spin signal indicate the presence of a weak sixth ligand. These results are similar to those of hexa-co-ordinated haem iron of cytochrome P450 binding to the cysteine residue of the enzyme active site and to the oxygen atom of a water molecule. These are different from cytochrome P450 experiencing bisthiolate axial ligation [38]. Our EPR results thus exclude that two truncated domains bind to the haem iron in each axial position. The sixth co-ordination position of haem bound to the truncated peptide is most probably H2O or OH−, since the low-spin ferric iron g values shifted from 2.45, 2.27 and 1.90 to 2.36, 2.27 and 1.92 when the pH of the sample was increased from 6.8 to 8.
The EPR spectrum of full-length 73aa-Domain in the presence of haemin was strikingly different from that of the truncated peptide. A high-spin signal at g≈6 dominated the spectra (results not shown). Therefore haem does associate with the full-length, peptide, since it is not separated by gel filtration, but this interaction does not display signs of strong axial ligation with specific residues of the peptide. This conclusion agrees with those obtained from the NMR and UV–visible spectra.
Purified IRP2 does not bind to the synthesized haem–agarose resin
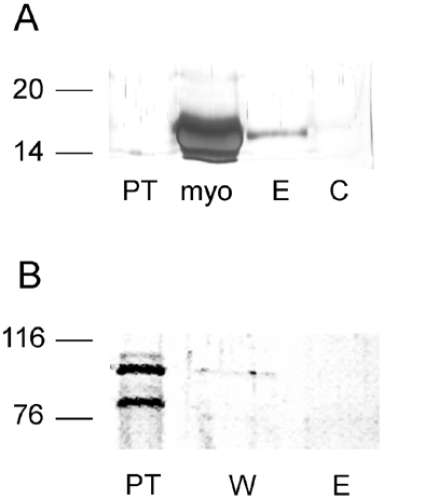
In order to discriminate between the full-length 106 kDa and the truncated 80 kDa purified IRP2 (Figure 1C) with respect to haem binding, a haem–agarose resin was synthesized. The efficiency of the haem–agarose resin to bind haemoproteins was probed with purified apomyoglobin. A band of 17 kDa corresponding to apomyoglobin interacted with the haem-containing resin, but not with non-derivatized agarose (Figure 6A).
Figure 6. Apomyoglobin binds to haem–agarose resin, whereas purified IRP2 does not.
(A) Interaction between purified apomyoglobin and the haem–agarose resin or non-derivatized agarose resin. Fractions were analysed by SDS/PAGE on a 12% gel, followed by silver nitrate staining. The loaded fractions were: haem–agarose resin pass-through after loading (PT), purified apomyoglobin as a control (myo), apomyoglobin eluted from the haem–agarose resin with 8 M urea (E), and control sample eluted after washing from the interaction between the non-derivatized agarose resin and apomyoglobin (C). (B) Interaction between purified IRP2 and the haem–agarose affinity resin: pass-through fraction (PT), first rinse with washing buffer (W) and the material eluted using 8 M urea (E). Molecular masses in kDa are shown on the left.
The purified mixture of truncated and full-length IRP2 (Figure 1C) was made to react with the haem–agarose resin to determine whether any of them bound haem. Neither the 106 kDa band, corresponding to full-length IRP2, nor the 80 kDa band, corresponding to the truncated forms of the protein (see above), interacted with the haem–agarose resin (Figure 6B). The same experiment was not carried out with the 73aa-Domain, since all studied molecular forms of this peptide were found above in NMR and EPR experiments to interact with haem in different ways, and no useful information was expected from such studies.
EPR analyses reveal that purified IRP2 does not provide an axial ligand to haem
The EPR spectra of purified IRP2 and haemin showed a high-spin signal at g≈6 that was quite similar to the one observed with full-length 73aa-Domain. No low-spin ferric signal was detected. Therefore EPR measurements of purified IRP2, containing the cleaved products of approx. 80 kDa (Figure 1C), did not show any interaction with haem similar to that characterized with truncated 73aa-Domain (Figure 5C). The haem–agarose resin experiment (Figure 6), together with EPR results, showed no specific interactions between haem and purified IRP2, as it was concluded for full-length 73aa-Domain. From these analyses, it appears that purified IRP2 does not provide a specific axial ligand to the cofactor, although weak non-specific binding cannot be excluded by our experiments.
DISCUSSION
Production in E. coli and purification of IRP2 73aa-Domain afforded several versions of the peptide: the expected full-length domain and truncated forms lacking N-terminal amino acids including most of the lysine residues, but retaining all cysteine residues (Figure 1A). The potential interactions of these peptides with haem were studied with a series of complementary methods.
1H-NMR spectra did not provide any evidence that full-length 73aa-Domain or its truncated ∼45–50-amino-acid forms are folded. The presence of the N-terminal amino acids of the 73aa-Domain in the full-length form did not trigger the building of secondary-structure elements or the occurrence of long-range interactions between parts of the domain. Moreover, the addition of iron and zinc salts or haem did not promote folding of the peptide, whether intact or truncated. However, haem addition to the 73aa-Domain shifted some proton signals. Evidence for interaction between haem and the truncated peptide was provided by the approx. 25 nm hypsochromic shift of the haem Soret band and by the appearance and characteristics of a low-spin ferric EPR haem signal. In contrast with globins or cytochromes that provide non-cysteine axial ligands and induce a bathochromic shift of the Soret band of free haemin, the shift observed in our experiments with the truncated peptide, but not with the full-length domain, agrees with cysteine axial ligation to ferric haem. Such a cysteine may be contributed by the HRM (haem-regulatory motif)-like sequence (-CPFH-) of the peptide [34,35,39]. These results are in agreement with results of TOCSY experiments. Whereas haem interaction with full-length 73aa-Domain obscured some spin systems, probably through relaxation effects, ligation of haem by truncated 73aa-Domain selectively shifted a subset of signals, including a set assigned to one of the two phenylalanine residues. The latter may belong to the HRM-like sequence (Cys-Pro-Phe-His) close to the C-terminus of the 73aa-Domain. It is also important to point out that haem binding to this specific site of truncated 73aa-Domain has never been quantitative, i.e. a fraction of our peptide preparations never specifically interacted with haem as deduced from EPR and 1H-NMR results.
The different haem-binding properties of truncated and full-length 73aa-Domain may be related to different accessibilities of the potential haem-binding site. 1H-NMR results showed that none of the peptide forms fold under any of the implemented conditions. Therefore both of them are most probably subject to extensive conformational dynamics with a few favoured conformations, as deduced from the several 1H-NMR signatures assigned to the same amino acid (Figure 4). The relatively large size of the full-length peptide may not then stabilize axial co-ordination to haem. This interpretation is not limited to the situation with isolated peptides, since recombinant IRP2 also failed to bind haem immobilized on cross-linked agarose (Figure 6) and to produce a low-spin EPR signal with the cofactor. It is noteworthy that not all sequences containing a potential HRM have the ability to bind haem, as shown for the C-terminal portion of the haem-regulated α-subunit of eukaryotic initiation factor-2 kinase [40].
Collectively, the 1H-NMR, EPR and UV–visible absorption results demonstrate a different behaviour of truncated and full-length 73aa-Domain towards haem. Clear evidence was obtained indicating a specific, but probably not quantitative, interaction between haem and a cysteine residue of the truncated peptides, possibly Cys201 of the Cys-Pro-Phe-His motif. In contrast, full-length 73aa-Domain does not provide specific ligands to haem, in apparent contradiction to previous conclusions [16–19]. Yamanaka et al. [17] showed that the absorption spectrum of haem incubated with purified His-tagged IRP2 had a maximum at 420 nm; this result was used as evidence for haem binding to IRP2, but was later dismissed as being contributed by the His tag [19]. Jeong et al. [18] reported that the absorption spectrum of a peptide from the specific IRP2 domain showed two maxima upon incubation with haemin, at 370 and 420 nm. The material used in the present study was a ∼63-amino-acids peptide spanning the first residue encoded by exon 5 of the human IRP2 gene up to Leu200. Thus this peptide did not contain the Cys-Pro-Phe-His fragment of the HRM-like sequence from Cys201 to His204, but it had the other three cysteine residues (Figure 1A). The modified form in which all these cysteine residues were replaced by alanine residues still showed two major peaks at 370 and 420 nm with haemin, but of lower intensity than for the wild-type peptide. This would indicate that the peptide still binds haem in the absence of all cysteine residues; otherwise the spectrum should be blank in the visible region (Figure 5, dotted spectra). Instead of showing that one of the three cysteine residues of the peptide is a haem-binding site [18], these results are rather indicative of weak interactions between haem and the 63-amino-acid peptide, the specificity of which remains to be established.
The interaction between untagged purified IRP2 and haem has been previously addressed [19]. The maximum of the absorption spectrum was found at 370 nm, as determined here with truncated 73aa-Domain. This preparation was devoid of His tag, a potential interaction site with haem. The same spectrum also showed a shoulder at 420 nm, as for the bacterial protein Irr (iron response regulator) [34]. This shoulder is said to indicate a second haem-binding site [34], without any further details. It was concluded that IRP2 degradation is triggered by haem binding [19], but the material used in these experiments was not thoroughly characterized for possible proteolysis. It is thus difficult to reconcile these findings with our results. Yet it is now clear that IRP2 proteolysis close to the lysine residues of the 73aa-Domain is a very efficient process that must be taken into account in any studies addressing the turnover mechanism of this protein.
In the present study, both full-length and truncated 73aa-Domain peptides contain the exact number of cysteine residues encoded by exon 5 of the IRP2 gene, and the sequences were confirmed prior to analysis. Our results indicate that intact 73aa-Domain of IRP2 does not display signs of site-specific axial binding to haem. Importantly, even though the isolated truncated form of IRP2 73aa-Domain can bind haem in vitro, we showed that purified untagged IRP2 does not bind haem immobilized on cross-linked agarose. It may thus be suspected that haem binding to truncated 73aa-Domain has little biological meaning and that haem interaction with IRP2 is an experimental artefact.
The data in Figure 2(A) show that, as concluded from in vitro experiments, IRP2 is readily cleaved in mammalian cells, presumably near the lysine residues of the 73aa-Domain, to yield a relatively stable intermediate. A cleavage product originating from truncation in the 73aa-Domain has already been reported in rat liver extracts, with the IENTPVL… N-terminal sequence [31]. This cleavage occurs very close to the N-side of the HRM-like sequence. It is different from the fragment characterized herein, but (i) different biological materials were used in both studies, (ii) the Ser-Ser-Gln↓Ile-Glu-Asn cleavage site is not one of a conventional endoprotease, and (iii) the Ile-Glu-Asn… starting sequence may result from exoproteolytic processing of a longer product. In both the previous [31] and the present studies (Figure 2), cleavage occurs in the 73aa-Domain and it is iron-independent. The approx. 80 kDa C-terminal end of IRP2 does not bind to haem–agarose (Figure 6). Altogether, these results do not support a function for this degradation intermediate as an efficient haem sensor in vivo. Nevertheless, the demonstration of the occurrence of this intermediate reveals an as yet unsuspected mechanistic hint of IRP2 degradation, in which endoproteases cleave the protein at closely spaced specific sites located within the 73aa-Domain.
The results presented herein indicate that IRP2 may be degraded in cells by additional pathways to the one leading to the proteasome [14,41–43]. The mechanism by which this protein senses intracellular iron awaits further investigation. It is conceivable that intracellular iron or haem levels may control IRP2 abundance in an indirect manner, possibly involving iron- or haem-dependent properties of accessory partners.
Online data
Acknowledgments
We acknowledge the contribution of Laura Udakis to some of the reported experiments. We thank Dr Cécile Bouton for the gift of the anti-IRP2 antibody. This work was supported by a travel grant for C. D. from the Centre Jacques Cartier (Lyon, France).
References
- 1.Hentze M. W., Muckenthaler M. U., Andrews N. C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 2.Wallander M. L., Leibold E. A., Eisenstein R. S. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., Weber G. J., Dooley K., Davidson A. J., Schmid B., et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 4.Sposi N. M., Cianetti L., Tritarelli E., Pelosi E., Militi S., Barberi T., Gabbianelli M., Saulle E., Kühn L., Peschle C., Testa U. Mechanisms of differential transferrin receptor expression in normal hematopoiesis. Eur. J. Biochem. 2000;267:6762–6774. doi: 10.1046/j.1432-1033.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 5.Schranzhofer M., Schifrer M., Cabrera J. A., Kopp S., Chiba P., Beug H., Müllner E. W. Remodeling the regulation of iron metabolism during erythroid differentiation to ensure efficient heme biosynthesis. Blood. 2006;107:4159–4167. doi: 10.1182/blood-2005-05-1809. [DOI] [PubMed] [Google Scholar]
- 6.LaVaute T., Smith S., Cooperman S., Iwai K., Land W., Meyron-Holtz E., Drake S. K., Miller G., Abu-Asab M., Tsokos M., et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 7.Cooperman S. S., Meyron-Holtz E. G., Olivierre-Wilson H., Ghosh M. C., McConnell J. P., Rouault T. A. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schumann K., Hentze M. W. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 9.Smith S. R., Ghosh M. C., Ollivierre-Wilson H., Hang Tong W., Rouault T. A. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol. Dis. 2006;36:283–287. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Galy B., Holter S. M., Klopstock T., Ferring D., Becker L., Kaden S., Wurst W., Grone H. J., Hentze M. W. Iron homeostasis in the brain: complete iron regulatory protein 2 deficiency without symptomatic neurodegeneration in the mouse. Nat. Genet. 2006;38:967–970. doi: 10.1038/ng0906-967. [DOI] [PubMed] [Google Scholar]
- 11.Meyron-Holtz E. G., Ghosh M. C., Rouault T. A. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 12.Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T. A. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samaniego F., Chin J., Iwai K., Rouault T. A., Klausner R. D. Molecular characterization of a second-responsive element binding protein, iron regulatory protein 2. Structure, function and post-transcriptional regulation. J. Biol. Chem. 1994;269:30904–30910. [PubMed] [Google Scholar]
- 14.Iwai K., Klausner R. D., Rouault T. A. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 1995;14:5350–5357. doi: 10.1002/j.1460-2075.1995.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwai K., Drake S. K., Wehr N. B., Weissman A. M., LaVaute T., Minato N., Klausner R. D., Levine R. L., Rouault T. A. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang D. K., Jeong J., Drake S. K., Wehr N. B., Rouault T. A., Levine R. L. Iron regulatory protein 2 as iron sensor. Iron-dependent oxidative modification of cysteine. J. Biol. Chem. 2003;278:14857–14864. doi: 10.1074/jbc.M300616200. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka K., Ishikawa H., Megumi Y., Tokunaga F., Kanie M., Rouault T. A., Morishima I., Minato N., Ishimori K., Iwai K. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 2003;5:336–340. doi: 10.1038/ncb952. [DOI] [PubMed] [Google Scholar]
- 18.Jeong J., Rouault T. A., Levine R. L. Identification of a heme-sensing domain in iron regulatory protein 2. J. Biol. Chem. 2004;279:45450–45454. doi: 10.1074/jbc.M407562200. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa H., Kato M., Hori H., Ishimori K., Kirisako T., Tokunaga F., Iwai K. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell. 2005;19:171–181. doi: 10.1016/j.molcel.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Hanson E. S., Rawlins M. L., Leibold E. A. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 2003;278:40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Chen G., Muckenthaler M., Galy B., Hentze M. W., Pantopoulos K. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol. Cell. Biol. 2004;24:954–965. doi: 10.1128/MCB.24.3.954-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabor S. Current Protocols in Molecular Biology. In: Ausubel F. A., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: Greene Publishing and Wiley-Interscience; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 23.Martelli A., Moulis J.-M. Zinc and cadmium specifically interfere with RNA-binding activity of human iron regulatory protein 1. J. Inorg. Biochem. 2004;98:1413–1420. doi: 10.1016/j.jinorgbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Pantopoulos K. Conditional derepression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol. Cell. Biol. 2002;22:4638–4651. doi: 10.1128/MCB.22.13.4638-4651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 26.Hwang T. L., Shaka A. J. Water suppression that works – excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A. 1995;112:275–279. [Google Scholar]
- 27.Schanda P., Forge V., Brutscher B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 2006;44:S177–S184. doi: 10.1002/mrc.1825. [DOI] [PubMed] [Google Scholar]
- 28.Brazzolotto X., Gaillard J., Pantopoulos K., Hentze M. W., Moulis J.-M. Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J. Biol. Chem. 1999;274:21625–21630. doi: 10.1074/jbc.274.31.21625. [DOI] [PubMed] [Google Scholar]
- 29.Martelli A., Salin B., Dycke C., Louwagie M., Andrieu J.-P., Richaud P., Moulis J.-M. Folding and turnover of human iron regulatory protein 1 depend on its subcellular localization. FEBS J. 2007;274:1083–1092. doi: 10.1111/j.1742-4658.2007.05657.x. [DOI] [PubMed] [Google Scholar]
- 30.Dupuy J., Volbeda A., Carpentier P., Darnault C., Moulis J.-M., Fontecilla-Camps J. C. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Guo B., Yu Y., Leibold E. A. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J. Biol. Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]
- 32.Goessling L. S., Mascotti D. P., Thach R. E. Involvement of heme in the degradation of iron-regulatory protein 2. J. Biol. Chem. 1998;273:12555–12557. doi: 10.1074/jbc.273.20.12555. [DOI] [PubMed] [Google Scholar]
- 33.de Villiers K. A., Kaschula C. H., Egan T. J., Marques H. M. Speciation and structure of ferriprotoporphyrin IX in aqueous solution: spectroscopic and diffusion measurements demonstrate dimerization, but not mu-oxo dimer formation. J. Biol. Inorg. Chem. 2007;12:101–117. doi: 10.1007/s00775-006-0170-1. [DOI] [PubMed] [Google Scholar]
- 34.Qi Z., Hamza I., O'Brian M. R. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13056–13061. doi: 10.1073/pnas.96.23.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki H., Tashiro S., Hira S., Sun J., Yamazaki C., Zenke Y., Ikeda-Saito M., Yoshida M., Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23:2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson J. H., Andersson L. A., Sono M. The diverse spectroscopic properties of ferrous cytochrome P-450–CAM ligand complexes. J. Biol. Chem. 1983;258:13637–13645. [PubMed] [Google Scholar]
- 37.Stuehr D. J., Ikeda-Saito M. Spectral characterization of brain and macrophage nitric oxide synthases. Cytochrome P-450-like hemeproteins that contain a flavin semiquinone radical. J. Biol. Chem. 1992;267:20547–20550. [PubMed] [Google Scholar]
- 38.Sono M., Dawson J. H. Formation of low spin complexes of ferric cytochrome P-450–CAM with anionic ligands. Spin state and ligand affinity comparison to myoglobin. J. Biol. Chem. 1982;257:5496–5502. [PubMed] [Google Scholar]
- 39.Zhang L., Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafie-Kolpin M., Chefalo P. J., Hussain Z., Hahn J., Uma S., Matts R. L., Chen J. J. Two heme-binding domains of heme-regulated eukaryotic initiation factor-2α kinase. N terminus and kinase insertion. J. Biol. Chem. 2000;275:5171–5178. doi: 10.1074/jbc.275.7.5171. [DOI] [PubMed] [Google Scholar]
- 41.Guo B., Phillips J. D., Yu Y., Leibold E. A. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 42.Kim S., Wing S. S., Ponka P. S-nitrosylation of IRP2 regulates its stability via the ubiquitin–proteasome pathway. Mol. Cell. Biol. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Pantopoulos K. The pathway for IRP2 degradation involving 2-oxoglutarate-dependent oxygenase(s) does not require the E3 ubiquitin ligase activity of pVHL. Biochim. Biophys. Acta. 2005;1743:79–85. doi: 10.1016/j.bbamcr.2004.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.