Abstract
The CFTR (cystic fibrosis transmembrane conductance regulator) gene is a tightly regulated and differentially expressed transcript in many mucosal epithelial cell types. It appears that DNA sequence variations alone do not explain CFTR-related gastrointestinal disease patterns and that epigenetic modifiers influence CFTR expression. Our aim was to characterize the native chromatin environment in cultured cells for intestinal CFTR expression by determining the relationship between histone acetylation and occupation of CFTR by multiple transcription factors, through a common regulatory element. We used HDAC (histone deacetylase) inhibition and ChIP (chromatin immunoprecipitation) analyses to define regions associated with acute acetylation of histone at the CFTR locus. We identified a region within the first intron associated with acute acetylation of histone H4 as an epigenetic signature corresponding to an intestine-specific enhancer element for CFTR. DHS (DNase I-hypersensitivity) assays and ChIP were used to specify control elements and occupation by regulatory factors. Quantitative ChIP procedures indicate that HNF1α (hepatic nuclear factor 1α) and Cdx2 (caudal homeobox protein 2) occupy and regulate through a novel intronic enhancer element of CFTR and that Tcf4 (T-cell factor 4) overlaps the same DNA element. RNAi (RNA interference) of Tcf4 and HNF1α decreased intestinal cell CFTR expression, identifying these as positive regulatory factors and CFTR as a target for Wnt signalling. We have linked the acetylation signature of nucleosomal histones to active intestinal CFTR expression and occupation by transcription factors HNF1α, Cdx2 and Tcf4 which converge to modify chromatin architecture. These studies suggest the therapeutic potential of histone modification strategies, such as inhibition of HDAC activity, to treat CFTR-associated disease by selectively enhancing CFTR expression.
Keywords: acetylation, caudal homeobox protein 2 (Cdx2), cystic fibrosis transmembrane conductance regulator (CFTR), hepatocyte nuclear factor 1α (HNF1α), histone, T-cell factor 4 (Tcf4)
Abbreviations: AP, activator protein; Cdx2, caudal homeobox protein 2; CFTR, cystic fibrosis transmembrane conductance regulator; ChIP, chromatin immunoprecipitation; DHS, DNase I-hypersensitivity; HAT, histone acetyltransferase; HDAC, histone deacetylase; HEK-293T, human embryonic kidney; HNF1α, hepatic nuclear factor 1α; HPRT, hypoxanthine phosphoribosyltransferase; LEF, lymphoid enhancer factor; PCK1, phosphoenolpyruvate carboxykinase 1; qChIP, quantitative ChIP; RNAi, RNA interference; siRNA, small interfering RNA; Tcf, T-cell factor; TSA, trichostatin A
INTRODUCTION
Cystic fibrosis is a common human genetic disease trait associated directly with mutations in the CFTR (cystic fibrosis transmembrane conductance regulator) gene. Despite the characterization of virtually all common human sequence variations within the CFTR locus, the link between mutation and disease outcome is not clear and rather appears to be related to the expression of CFTR. It is suspected that epigenetic modifiers of CFTR transcription may profoundly influence disease expression beyond that associated with CFTR protein mutations. CFTR is a tightly regulated and differentially expressed transcript in many mucosal epithelial cell types. Although there have been some important advances in identifying the DNA elements controlling CFTR transcription [1–3], the link between expression of CFTR and the native chromatin environment of the CFTR locus remains poorly understood. CFTR expression appears, paradoxically, to be both a marker of gastrointestinal cellular differentiation and proliferation. Despite the presumption that expression of CFTR is associated with terminal mucosal cell fate, the transcription of CFTR is retained in many aggressive gastrointestinal tumour cell lines [4]. CFTR expression varies throughout fetal gut development [5], and during epithelial regeneration. CFTR itself also appears to be a regulator of fetal intestinal development. Interestingly, the underlying basis for dysregulation of CFTR transcription in disease settings, such as pancreatitis and cholestatic disorders, suggests that CFTR expression maybe an important indicator for predicting the cellular fate of many gastrointestinal cell types. Alterations from the normal epigenetic signature within the CFTR locus may carry the potential for gastrointestinal disease expression in patients without the typical CFTR mutations [6].
The characterization of chromatin surrounding the CFTR locus may therefore serve as an important predictor of cellular fate for specific intestinal cell types and provide a model to determine whether the epigenetic basis of gene dysfunction correlates with human disease. Using histone acetylation as a measure of the dynamic alterations within chromatin that influence gene expression, we identified an element in the first intron that gives rise, in part, to cell-type-specific organization of chromatin structure. We used ChIP (chromatin immunoprecipitation) assays to identify regions that were associated with the acute acetylation of histone upon treatment with the HDAC (histone deacetylase) inhibitor, TSA (trichostatin A). The aim of the present study was to characterize the native chromatin environment for CFTR expression by determining the correlation between histone acetylation and occupation of CFTR by multiple transcription factors.
We have shown previously that histone acetylation, acting via the inverted CCAAT signal (Y box) within the proximal CFTR promoter [3], contributes to an accessible state of chromatin structure and promotes the initiation of CFTR transcription. A stoichiometric relationship between co-regulatory factors which acetylate nucleosomal histones, such as the SAGA [Spt/Ada/GCN5 (general control non-derepressible 5)]-associated human HAT (histone acetyltransferase) GCN5, and those which regulate histone deacetylation, such as HDAC1, located in the vicinity of the CFTR promoter, appear to reflect the transcriptional activity of CFTR [7]. Other studies also support a genetic model whereby cell-type-specific regulation of CFTR is influenced by distal DNA regulatory elements, separated by several kilobases from the proximal promoter region [8]. These distal elements are also likely to modulate the chromatin environment surrounding the transcriptional initiation region within the CFTR promoter. Therefore the organization of sites that are responsive to changes in nucleosomal histone acetylation may underlie the control of CFTR expression in specific cell types. Previous studies with transgenic mice implicate a segment of the promoter and first intron in intestine-specific regulation of CFTR ([9], and F. Wang and M.J. Walsh, unpublished work). These regions correspond to the hypersensitivity to nuclease in native chromatin. In accordance with numerous models that show a relationship between histone acetylation and gene activity [10], we demonstrate that the nuclease hypersensitivity of a specific region in the first intron selectively corresponds to the hyperacetylated state within the native chromatin context. We also show that this DHS (DNase I-hypersensitivity) region corresponds to the acute deposition of histone H3 and H4 acetylation upon inhibition of histone deacetylase activity, whereas other regions do not. These finding show that the restrictive boundaries of histone acetylation may define specific regions of acute hyperacetylation, poised for the recruitment of transcription factors. Furthermore, we have characterized this minimal region of the first intron as a regulatory site for the possible combinatorial actions of HNF1α (hepatocyte nuclear factor 1α), Cdx2 (caudal homeobox protein 2) and Tcf (T-cell factor) 4. These studies demonstrate that boundaries of histone acetylation in the CFTR first intron correlate with transcription factor occupation in vivo in a native chromatin context. Overall, these studies provide a molecular basis for possible therapeutic approaches such as HDAC inhibition that utilize histone modification to treat CFTR-associated disease, by enhancing selective expression of CFTR.
MATERIALS AND METHODS
Cell culture
Colon carcinoma cell lines Caco-2 and T84 and pancreatic adenocarcinoma cell line CF-PAC1 were obtained from the A.T.C.C. (Manassas, VA, U.S.A.). HEK-293T (human embryonic kidney) cells were a gift from Dr Edward Johnson (Mount Sinai School of Medicine). Cell lines were maintained in DMEM (Dulbecco's modified Eagle's medium) with high glucose (4.5 g/litre) and supplemented with 10% newborn calf serum (Hyclone) and 1× gentamycin (Invitrogen). Cell cultures were maintained in an incubator at 5% CO2 and 37°C.
DHS analysis of genomic DNA
Assays were essentially performed as described in [7], with the minor modifications described here. Nuclei from TSA-treated and untreated cell lines from HEK-293T, Caco-2 and CF-PAC1 were isolated following treatment with TSA (0.1 μM) for 6 h. Nuclei were isolated and treated with DNase I. DNase I-treated DNA was extracted and then digested with EcoRI at 37°C for 8 h. DNA fragments were fractionated by agarose gel electrophoresis, blotted on to membranes and hybridized to a 32P-labelled DNA fragment as described previously [7], corresponding to the human CFTR locus 3.33 kb EcoRI fragment of the first intron.
DNase I protection analysis
The DNase I protection experiment was carried out as described previously by us [3] with modifications. Essentially, a double-stranded DNA fragment was synthesized using PCR with the forward primer (5′-TTATATGCAACCTTCTCC-3′) end-labelled by the addition of [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). Addition of the 32P-labelled primer to the PCR product was used to generate the 32P-labelled fragment with the unlabelled reverse primer (5′-GTGATATGATACAGGAACAATC-3′). To confirm the location of protection from DNase I hydrolysis corresponding chemical degradation by Maxam–Gilbert sequencing procedure was conducted on the 32P-labelled fragment (results not shown).
ChIP analysis
ChIP assays were performed as described previously [11], with the exception that specific antibodies against human antigens of acetylated histone H4 (AbCam), acetylated histone H3 (Upstate Biotechnology), total histone H4 (Upstate Biotechnology), total histone H3 (Upstate Biotechnology), Cdx2 (BL-3195, Bethyl Laboratories), HNF1α (Novus Biologicals), Tcf4 (clone 6H5-3, AbCam), β-catenin (clone 10H8, Calbiochem) and SP1 (Upstate Biotechnology) were used. Primers that were used to generate the PCR fragments shown in Figure 1 are designated as follows: cftr1-forward, 5′-GGATGATTTGATTAGAAGCAG-3′; cftr1-reverse, 5′-CTCCAAAGAGGATCATAGAA-3′; cftr2-forward, 5′-GCCTGGTGCTGGGCGGTAAG-3′; cftr2-reverse, 5′-TGCTCTTTCCCCCGCCTTCACTG-3′; cftr3-forward, 5′-GTATAGCCTACTACACACCTAGGCAAT-3′; cftr3-reverse, 5′-CCTAATCAACAGAGAAATTCCACG-3′; cftr4-forward, 5′-CTCTTTCAGTACGTGTCCTAAG-3′; cftr4-reverse, 5′-CCAGGACCGGCTGCCACCAGCT-3′; cftr5-forward, 5′-AGCTTTCACATCTCTCTTATGTT-3′; and cftr5-reverse, 5′-ACTGGTTGGCATTTAGGTTCCTC-3′.
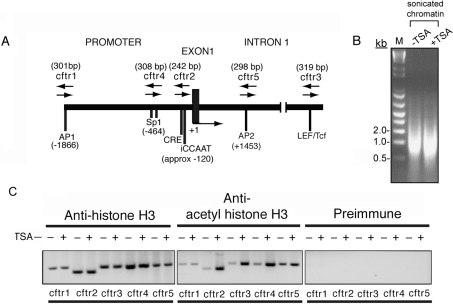
Figure 1. Mapping of histone acetylation through the 5′ region of the CFTR locus.
(A) Schematic representation of the human CFTR within the 5′-upstream region. PCR primers corresponding to sequences of the CFTR 5′-region (recognized as putative transcription factor-binding sequences positioned relative to the major transcriptional start site) were used to amplify specific fragments of CFTR following the ChIP procedure, and are indicated as cftr1, cftr2, cftr3, cftr4 and cftr5. PCR fragments corresponding to putative transcription factor-binding sites for iCCAAT [inverted CCAAT (Y box) element], CRE (cAMP-regulatory element), Sp1 [Sp1 (specificity protein 1)-binding elements], AP1 [AP (activator protein) 1-binding elements], AP2 (AP2-binding elements) and LEF/Tcf are shown relative to the position of transcript start sites. Relative sizes of the predicted PCR products are indicated above the PCR products. Input DNA represents 1% of total DNA from chromatin of immunoprecipitated samples. (B) Analysis of sonicated chromatin using 1% agarose gel electrophoresis. M, markers (sizes indicated in kb). (C) Soluble chromatin was immunoprecipitated with ChIP-grade anti-(histone H3) and anti-(acetylated histone H3) antibodies from T84 colonic carcinoma cells cultured in the absence (−) or presence (+) of 0.1 μM TSA for 6 h. The PCR products, corresponding to the DNA fragments shown, were amplified from the immunoprecipitated chromatin material. The PCR fragments represented in (A) are indicated below the gels in (C).
ChIP studies shown in Figure 3 were conducted with separate sets of primers spanning the first intron of CFTR as described in Supplementary Table 1 (http://www.BiochemJ.org/bj/408/bj4080317add.htm).
Figure 3. ChIP of acetylated histone H4 and analysis of the human CFTR first intron.
Caco-2 cells were mapped for acetylation of histone H4 following treatment with TSA. (A) Agarose (1%) gel electrophoresis of sheared chromatin. Sizes are indicated in kb. (B and C) Chromatin was immunoprecipitated with antibodies against acetylated histone H4, and DNA extracted from immunoprecipitated chromatin was amplified by PCR using specific primer pair sets shown relative to the human CFTR first intron. Primer pairs were designed to span the entire DHS site and neighbouring region within the first intron of human CFTR as described in Supplementary Table 1.
Sequence analysis
Examination of genomic sequences for putative cognate elements within the CFTR locus was performed with TRANSFAC, MacVector (Accelyrs) and BLAST (NCBI) sequence databases.
RNAi (RNA interference)
Synthetic siRNA (small interfering RNA) directed against Tcf4 (Ambion, Silencer™ siRNA sequences, id 107026) and HNF1α (Dharmacon, Ontarget plus® siRNA) were purchased and prepared for cell culture transfection according to the respective manufacturer's recommendations. Transfection of approx. 2×107 Caco-2 cells using Oligofectamine® reagent (Invitrogen) or Oligofect (Qiagen) was performed using the respective manufacturer's instructions. At 36 h post-transfection, total RNA was recovered from transfected Caco-2 cells following TSA treatment with RNAeasy (Qiagen) and 5 μg of RNA was used to reverse-transcribe CFTR, PCK1 (phosphoenolpyruvate carboxykinase 1) and HPRT (hypoxanthine phosphoribosyltransferase) mRNA. PCR was carried out in an Applied Biosystems 7900HT sequence detection systems analyser with 1, 2 or 3 μl of the first-strand cDNA mixture with the Quantitect SYBR Green QPCR kit (Qiagen). For CFTR, PCK1 and HPRT mRNA, the cycling parameters were as follows: (i) 95°C for 15 min; and (ii) 36 cycles consisting of 95°C for 15 s, 55°C for 30 s and 72°C for 30 s. Normalization was performed with 18S rRNA levels using the Applied Systems riboprobe quantification kit according to the manufacturer's instructions. All results were analysed with Applied Biosystems SDS software version 2.2.
qChIP (quantitative ChIP)
qChIP was performed as described previously [11], with the following modifications. Essentially, 2×107 cells were transfected. ChIP was conducted using the procedure described above; however, amplification of specific regions of the human CFTR locus was performed by PCR with DNA recovered from immunoprecipitation and performed using three dilutions of DNA containing 2, 1 or 0.5 μl with 28 cycles of amplification. The primers selected for PCR are described in Supplementary Table 1 for the PCR sites pcr1 and pcr7. PCR products were analysed by on 1.5% agarose gels and were visualized by ethidium bromide staining and UV transillumination. For quantitative real-time PCR, DNA was dissolved in 50 μl of water and 2 μl was used from each sample with Brilliant SYBR Green QPCR kit (Applied Biosystems). All chromatin immunoprecipitates were analysed by real-time PCR at least three times, and the difference for the mean was less than 15%. Each PCR product (3 μl) was extracted at 22, 28 and 34 complete cycles for visualization on agarose gels and stained with ethidium bromide. To account for both nucleosome density surrounding individual genomic locations and specific acetylation of histone H4, comparison was made of the signal generated with antisera against acetylated histone H4 and acetylated histone H3 compared with that generated with antisera for total histone H4 and total histone H3 respectively. Quantitative PCR signal generated with antibodies against HNF1α and Cdx2 post-TSA treatment relative to input control was calculated. All values were corrected for non-specific signal using a rabbit pre-immune serum control for each input value. Results are means±S.D.
Immunoblotting/Western blotting
Caco-2 cells transfected with siRNAs against HNF1α and Tcf4 were monitored for the presence of HNF1α and Tcf4, compared with controls. Approx. 10 μl (20%) from the chromatin fraction before immunoprecipitation (ChIP) was taken to evaluate the presence of HNF1α and Cdx2. Immunoblots of Caco-2 cell lysates were analysed on SDS/12.5% polyacrylamide gels. Immunoblotting was carried out with antisera against HNF1α and Cdx2. Antigens were detected by horseradish-peroxidase-conjugated secondary antibody with a commercial chemical luminescent kit (Pierce).
RESULTS
Hyperacetylation of human CFTR in the colonic epithelial cell line T84 corresponds to intronic control elements
To determine whether the human CFTR locus is organized into chromatin domains enriched with the deposition of acetylated histone species, we characterized different upstream regions of the CFTR locus according to putative transcription factor recognition sites and analysis of histone acetylation. This was performed in a cell line (T84) known to carry moderate levels of CFTR transcription. We examined regions associated with histone hyperacetylation upon inhibition of histone deacetylase activity within a 5′-upstream segment containing the promoter (−2.2 kb upstream and 3.6 kb downstream) within the first intron of CFTR (Figure 1). Using ChIP analysis, we tested whether induction of histone acetylation by TSA, an HDAC inhibitor [12,13], corresponds to specific regions within the CFTR locus. To do this, we generated several pairs of PCR primers corresponding to sites overlapping putative recognition sequences for known transcription factors (as shown in Figure 1A). These elements included: cftr1=AP1, AP (activator protein) 1-binding elements; cftr2=iCCAAT, inverted CCAAT (Y box) element, and CRE, cAMP-regulatory element, cftr3=LEF (lymphoid enhancer factor)/Tcf-binding elements; cftr4=Sp1, Sp1 (specificity protein 1)-binding elements; cftr5=AP2, AP2-regulatory element (Figure 1A). The results of the ChIP procedure showed three of these regions: cftr2, cftr3 and cftr4 to be clearly more susceptible to changes in histone acetylation compared with regions cftr1 and cftr5. Results presented in Figure 1(C) indicate that chromatin overlapping the CFTR locus is differentially organized within domains that are preferentially susceptible to changes in histone acetylation induced by TSA and which overlap putative targets for transcription factor binding.
Cell-type-specific DHS regions overlap putative HNF1α-, Cdx2- and Tcf4-binding elements within the first intron
Previously, we have shown that the accessibility of chromatin to nucleases corresponds to active regions for transcription factor binding within the proximal CFTR promoter for basal promoter activity [3]. However, these studies did not take in to account distal elements of the CFTR locus that may co-operate with the CFTR promoter in the cell-type control of transcription. In the present study, DHS mapping revealed a cell-type-specific DHS (DHS1) at approx. 2.1 kb into the EcoRI fragment of the first intron segment (Figure 2A). The hypersensitive site was detected in the gastrointestinal cell lines CF-PAC1 and Caco-2, but not in the HEK-293T cells. These results are consistent with previous analysis [9], but which had failed to map nuclear protein binding.
Figure 2. DHS sites within the first intron of human CFTR overlaps putative HNF1α, Cdx2 and LEF/Tcf4 sites.
(A) DHS analysis was performed on the EcoRI fragment within the human CFTR first intron in different cell lines: HEK-293T (293T), Caco-2 and CFPAC. Sizes are indicated in kb. (B) A representative map corresponding to genomic DNA flanking exon 1 of human CFTR between the EcoRI fragment. DNA sequence shown indicate the sequence of CFTR from a PCR used in the DNase I footprint experiment (C) in the presence of nuclear extract from CFPAC, Caco-2 and control HEK-293T cells (293T), with the amount of nuclear extract used as shown. The outlined (boxed and greyed) sequence (B) indicates the putative protected footprint from the nuclear extracts shown in (C). The locations of the putative HNF1α, Cdx2 and LEF/Tcf4 sites within the DNA fragment are as indicated. Arrows depict the primer locations used for subsequent footprinting experiments. (C) DNase I footprint analysis shows the region of protection from DNase I digestion. The PCR fragment shown in (B) was 5′-end-labelled with [γ-32P]ATP and used in all subsequent DNase I footprint reactions. Increasing amounts of nuclear extract was used (5, 10 and 50 μg) from cell lines CF-PAC-1 and Caco-2, and 50 μg from control HEK-293T cell lines.
In order to confirm whether the DHS site in the first CFTR intron may be associated with transcription factor occupation, we performed DNA footprinting analysis using Caco-2, HEK-293T and CF-PAC1 nuclear extracts with a DNA fragment corresponding to the putative binding sites for Cdx2 and HNF1α in close proximity to the DHS site. As shown in Figure 2(C), DNase I footprint experiments reveal a cell-type-specific region of protection from DNase I digestion. Sequence analysis of this region carried out in parallel revealed that protection from DNase I degradation contains overlapping consensus sites for Cdx2 and HNF1α (Figure 2B). Furthermore, embedded within the same element was a weak consensus for LEF/Tcf protein binding that corresponds with the same protection from DNase I. Therefore data shown here indicate that the DHS site investigated by in vivo methods to detect nuclease-hypersensitive sites in Figure 2(A) correspond to a more sensitive in vitro assay that demonstrates protection from DNase by nuclear protein binding.
Histone acetylation of the CFTR first intron sequence corresponds to specific occupation by Cdx2 and HNF1α in colonic cells
As a measure of response to the inhibition of HDAC activity with TSA in native chromatin, the EcoRI fragment encompassing a 3.3 kb fragment within the first intron of the human CFTR locus was characterized further by mapping of histone H4 acetylation (Figure 3) in the DHS1 region found in Caco-2 and CF-PAC1 cells (Figure 2). ChIP analysis to determine the boundaries of histone acetylation demonstrated an increase in acetylated histone H4 approaching DHS1, in Caco-2 cells. As shown in Figure 3, ChIP studies corresponding to sites of pcr5, pcr6 and pcr7reflect the greatest change in histone H4 acetylation compared with sites pcr1, pcr3, pcr4 and pcr2, with pcr7 overlapping the DHS1 site. The results of the DHS assays corroborate previous reports [9] identifying that the first intron corresponds to cell-type-specific expression. However, data from these ChIP assays advance further the paradigm that the molecular basis for the DHS at this site involves histone acetylation. Consistent with the notion that alteration of chromatin structure through histone acetylation corresponds to the occupation of transcription factors HNF1α and Cdx2, we directed ChIP analysis with antisera directed against both Cdx2 and HNF1α. As shown in Figures 4(B) and 4(C), qChIP studies indicate that the induction of histone H4 and histone H3 acetylation directly overlaps the occupation of pcr7 site by Cdx2 and HNF1α. Quantitative real-time PCR indicates more prominent induction of H4 acetylation than H3 acetylation, relative to total H4 and H3 respectively at this site. There is also increased association of both HNF1α and Cdx2 on induction of histone H4 and H3 acetylation. The weaker binding signal of acetylated histone H3 in Figure 4(A) may not indicate differences in relative acetylation, but rather reflect differences in affinity of antibodies against acetylated histone H3 and acetylated histone H4 used in this assay.
Figure 4. Occupation of the human CFTR first intron by HNF1α, Cdx2, acetylated H4 and acetylated H3 following TSA induction in native chromatin.
(A) ChIP analysis performed on Caco-2 cells that express endogenous CFTR was used to evaluate the occupation of HNF1α, Cdx2 and acetylated H3 and H4 within the first intron of the CFTR locus. An internal control for the immunoprecipitation of chromatin was performed with anti-(histone H3) antibodies. Antibody against Sp1 (specificity protein 1) (Upstate Biotechnology) was used as a negative control. Immunoprecipitation of chromatin with anti-HNF1α (α HNF1α), Cdx2 (α cdx2), acetylated histone H3 (α Ac H3) and acetylated histone H4 (α Ac H4) total histone H4 (α histone H4) and total histone H3 (α histone H3) are shown. (B) Quantitative PCR signal generated with antibody against HNF1α and Cdx2 post-TSA treatment, relative to input control. (C) The values shown represent the signal generated with the antisera against acetylated histone H4 and acetylated histone H3 compared with that generated with antisera for total histone H4 and total histone H3 respectively.
Reduction of HNF1α correlates with the loss of H4 acetylation in the CFTR DHS1 regulatory site in TSA-treated cells
To determine further the role of HNF1α to regulate the acetylation of histone H4 in vivo, we used RNAi of human HNF1α in the Caco-2 cell line and monitored the extent of histone H4 acetylation induced by TSA. One consequence could have been that loss of HNF1α occupation may not have altered the deposition of acetylated histone H4, as seen in some cases for HNF1α. Conversely, acetylation of histone H4 may in fact be dependent on the local presence of HNF1α, as shown previously [14]. In order to address both of these possibilities we used siRNA strategies to reduce HNF1α expression in the Caco-2 cell line and then used qChIP [11] to measure acetylated histone H4 in vivo. Analysis was by qChIP of Caco-2 cells treated with siRNA and scrambled RNA, and untreated cells were monitored. The results of our study indicate that siRNA of HNF1α achieved an approximate reduction of >70% when measured by Western blot analysis and densitometry (Figure 5). RNAi of HNF1α did not affect the viability of the Caco-2 cells or proliferation rates when compared with the scrambled control (results not shown). Representative agarose gel images of amplified DNA fragment pcr7 indicate that a decrease in HNF1α by RNAi (Figure 3B) resulted in a significant loss in histone H4 acetylation when compared with the pcr1 product, where no difference from baseline H4 acetylation was seen. As a positive control, ChIP with the human albumin gene, a known target for occupation by HNF1α, in the Caco-2 cell line was performed (results not shown). To determine the selective nature of histone acetylation throughout the intronic segment, each amplified fragment (pcr1–pcr7) was interrogated by qChIP and demonstrates that HNF1α occupation influences the fate of histone acetylation encompassing the region occupied by HNF1α. The absence of HNF1α by RNAi diminishes the acetylation of histone H4 upon TSA treatment only in the amplified fragments represented by pcr6 and pcr7. Therefore we confirm that the ‘restrictive’ acetylation of the pcr7 PCR product in the colonic Caco-2 cells was likely to be due to the presence of HNF1α in that native chromatin context.
Figure 5. Loss of HNF1α in the Caco-2 cell line by RNAi results in depletion of acetylated H4 deposition.
(A) Western blot of endogenous levels of HNF1α and Tcf4 48 h after transfection with siRNAs, denoted as si HNF1α or si Tcf4, against endogenous HNF1α and Tcf4 nuclear protein, and control mock scrambled siRNAs is shown. Endogenous levels of HNF1α and Tcf4 are shown before transfection with siRNA and indicated from the Caco-2 cells. Input amount of total cellular protein was compared with the lamin B2 protein as shown in the lower panels. (B) Examination by ChIP of Caco-2 cells treated with TSA, following transfection with siRNAs directed against HNF1α mRNA, with rabbit antisera against acetylated histone H4. Representative ChIP was performed and illustrated from an agarose gel comparing the signals generated for pcr7 and pcr1 products (see also Supplementary Table 1 at http://www.BiochemJ.org/bj/408/bj4080317add.htm and Figure 3). (C) qChIP was performed on each of the fragments illustrated in Figure 3(B). Values of acetylated histone H4 were plotted relative to the antisera against the total signal generated for all species of histone H4. Results are means±S.E.M. (n>3).
RNAi of HNF1α and Tcf4 transcription results in loss of CFTR expression, and indicates that HNF1α and Tcf4 act as positive regulatory factors of CFTR
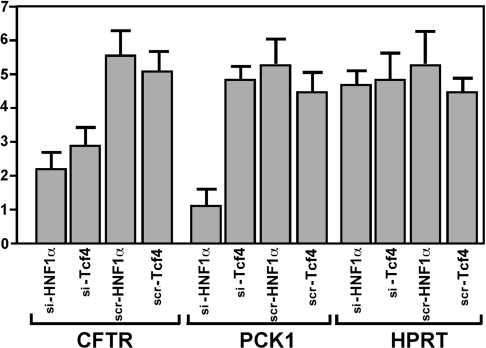
In an effort to confirm a genetic role for the loss of histone H4 acetylation associated with suppression of HNF1α in the Caco-2 cell line, we evaluated the presence of transcripts of putative targets for HNF1α: CFTR, PCK1 and HPRT. Suppression of HNF1α by siRNA-directed transfection against HNF1α resulted in decreased CFTR and PCK1 transcript levels (Figure 6), consistent with a previous report [15]. When we compared the expression of CFTR, PCK1 and HPRT upon the suppression of Tcf4, we found that CFTR expression alone was diminished (Figure 6). Our results therefore support the role of HNF1α and Tcf4 to act as positive regulatory factors of CFTR.
Figure 6. Depletion of HNF1α and Tcf4 by transfection with siRNAs corresponds with loss of endogenous CFTR expression in Caco-2 cells.
Quantitative PCR studies of CFTR, PCK1 and HPRT were conducted with cells using synthetic siRNAs directed against target sequences corresponding to the antisense strand of human HNF1α (si-HNF1α) and Tcf4 (si-Tcf4) mRNAs. Controls were used for each of the targeted messages for human HNF1α and Tcf4 as scrambled (scr) siRNAs. siRNAs were transfected using Oligofect reagent. At 36 h post-transfection, total RNA was recovered from transfected Caco-2 cells with RNAeasy and 5 μg of RNA was used to reverse-transcribe CFTR, PCK1 and HPRT mRNA. PCR was carried out with 1, 2 or 3 μl of the first-strand cDNA mixture with the Brilliant SYBR Green QPCR kit, as detailed in the text. Normalization was performed with 18S ribosomal RNA levels using the riboprobe quantification kit. All results were analysed using Applied Biosystems SDS software version 2.2.
Tcf4 is a novel transcription factor for CFTR
An unanticipated finding of the present study was the potential of the related LEF/Tcf transcription factor Tcf4 to associate with CFTR, based on identifying a weak consensus within the DHS site and protected footprint region (Figure 2). Previous studies have indicated that ablation of the Tcf4 gene corresponds with the proliferation of crypt cells within the small bowel epithelium in mice [16–18]. Interestingly, homologous deletion of Tcf4 corresponds to the lack of CFTR expression from samples taken from the intestinal epithelium from tcf4−/− mice. However, it remains paradoxical that levels of CFTR transcripts are often present in many tumour cells derived from pancreatic and colonic adenocarcinoma cell lines, with some retaining some of the protein function of CFTR. The underlying molecular genetic basis for this, however, remains unknown. Furthermore, Tcf4 also appears to also impose expression on other genes that, like CFTR, are also associated with terminal and differential function of specialized colonic enterocytes [18,19]. To address the potential of Tcf4 to act on CFTR expression, we examined the occupation of Tcf4 on the CFTR first intron first by electrophoretic mobility-shift analysis from nuclear extracts from gastrointestinal cell lines T84, Caco-2 and CF-PAC1. Then ChIP analysis was used to determine the occupation of CFTR by Tcf4 in vivo as shown in Figure 7. Supershift assays with monoclonal antibodies directed at mouse and human Tcf4 and β-catenin confirmed specific LEF/Tcf4 and β-catenin association with the DNA-binding complex to the sequence (Figure 7A). Negative controls with nuclear extract and non-specific IgG confirmed specificity of Tcf4 and β-catenin association for the cognate DNA sequence. Immunoprecipitation of chromatin with monoclonal antibodies against human Tcf4 demonstrated in vivo occupation of Tcf4 at the CFTR intronic segment (pcr7 product in Figure 3) in Caco-2, T84 and CF-PAC1 cells (Figure 7B).
Figure 7. LEF/Tcf-related Tcf4 transcription factor binds to the CFTR first intron sequence in vivo and is cell-type-specific.
(A) Nuclear extracts were prepared from colon adenocarcinoma cell line Caco-2. Using the radiolabelled fragment of the first intron of human CFTR (5′-TATTTAGTGGACATGTACCAA-3′), nuclear extracts were tested for binding to the labelled oligonucleotide. Monoclonal antibodies directed against human Tcf4 and β-catenin were used in an electrophoretic supershift assay. Unlabelled competing oligonucleotides from the human CFTR intron fragment and the consensus target for Tcf4 were used to determine the specificity of binding to the first intron of human CFTR. Immunoprecipitation of chromatin with anti-Tcf4 (αTcf4) and anti-β-catenin antibodies are shown. (B) ChIP analysis, performed on cell lines that express endogenous CFTR, were used to evaluate the occupation of Tcf4 within the first intron of the CFTR locus. Chromatin from gastrointestinal cell lines CF-PAC-1, T84 and Caco-2 were used for comparison with that from HEK-293T cells (293T). An internal control for the immunoprecipitation of chromatin was performed with anti-(histone H3) antibodies and anti-HNF1α as a positive control.
DISCUSSION
Chromatin is thought to be the rate-limiting barrier for access to the underlying genome, influencing all aspects of cellular control that utilize DNA as a template, such as transcription, replication and DNA repair. The current belief is that the covalent modification of histones and the chromatin environment can convey cellular ‘memory’, which instructs transcriptional mechanisms to recognize imprinted epigenetic marks for executing gene expression programmes, as they relate to developmental processes involving cellular differentiation and proliferation. Among the well-studied covalent modifications detected within nucleosomal histones is acetylation [20], with this covalent mark being most closely associated with gene activity. However, histone acetylation and active transcription through RNA polymerase II are unlikely to overlap completely. Instead of the entire locus becoming completely acetylated, discrete regions of acetylation are sufficient to confer access to genomic DNA for transcriptional activation [21,22]. Directed acetylation is also known to be associated with cell-type-specific regulation of target genes. The expression of CFTR clearly requires the acetylation of nucleosomal histones in chromatin proximal to the site of transcription initiation [7]. Many activating transcription factors have the capacity to recruit HATs as co-activators that target specific DNA elements for acetylation [22]. Yet, studies evaluating CFTR transcription fail to account for the role that transcription factors have in the recruitment of HATs that acetylate histones surrounding the immediate area of protein occupation in CFTR. It is unclear, however, whether access to genomic DNA is granted through the action of transcription factors directly, or whether regions of the genome are made susceptible to transcription factor access by this so-called ‘memory’ through histone acetylation. Additionally, it is also possible that acetylation may become cumulative and correlate with the level of CFTR expression in a cell-type-dependent manner.
We aimed to determine the boundaries of histone acetylation within a 5′-upstream segment of the CFTR locus that is susceptible to HDAC inhibition in intestinal cells. We found nucleosomal acetylation to be relatively restricted and discrete within this segment of the CFTR locus likely to create a post-translational signature in chromatin necessary to convey cell-type-specific signals for gene activation. Furthermore, these ‘hyperacetylation’ sites correspond in large part to overlapping binding sites for transcription factors. It is therefore possible that certain regions of chromatin that are more susceptible to HDAC inhibition may be poised for hyperacetylation and transcription factor recruitment and activation over others. The complexity of covalent histone modification through several post-translational modifications make it plausible that lysine acetylation may be competing with other histone marks, possibly on the same lysine residue of the histone species [23], therefore limiting the number of lysine residues available for acetylation by HATs.
We conclude that covalent modification may be a consequence of the local concentration of histone acetyltransferases present compared with the actions of repressive co-regulatory activities and the availability of cognate DNA regulatory elements. It is interesting to consider that requirements for nucleosomal histone acetylation in vivo maybe rate-limiting and unnecessary in areas not governing transcription at the expense of conserving biochemical resources in the nucleus. Therefore we may account for our results (Figures 1, 2 and 3) that acetylation along the genomic axis of an active CFTR locus may carry the mosaic organization for the acetylation of nucleosomal histones concentrated on areas of transcription factor targeting. In support of this notion is the fact that a relatively small number of genes (<3%) were affected by the treatment with the HDAC inhibitor TSA [24], suggesting that histone acetylation is only one part of a complex signature in chromatin that ultimately regulates specific gene expression patterns.
Our results identify three specific transcription factors, HNF1α, Cdx2 and Tcf4, which appear to converge on a regulatory element overlapping the DHS1 site of CFTR (Figures 2, 3, 4 and 6) that carries a significant degree of sequence conservation. The occupation of multiple transcription factors at a single regulatory DNA motif is consistent with the notion of conservation of transcriptional remodelling mechanisms in chromatin [25]. A question raised by the present study was whether HNF1α and Cdx2 co-operate to enhance CFTR transcription, as described previously with Cdx2 and HNF1α in other target genes [26]. A consistent theme emerges with HNF1α and Cdx2 occupation of many of the same genetic targets to orchestrate intestinal cell differentiation [27]. HNF1α appears to be active at multiple sites within CFTR [15]. However, the present study demonstrates the occupation by and the potential coordinating functions of Cdx2, HNF1α and Tcf4 acting via a common regulatory region identified through mapping of HDAC inhibition induced histone acetylation.
The existence of a weak consensus for LEF/Tcf binding within the DNA footprint region of CFTR was an unexpected finding (Figures 2B and 2C). However, upon further examination of the DNA element within the footprint region, Tcf4 and β-catenin binding was identified with antisera from Caco-2 cell nuclear extract (Figure 7A). Our results confirm that Tcf4 occupies the CFTR region overlapping the DHS site and the footprint region (Figure 7B). Our studies also indicate that suppression of endogenous Tcf4 in Caco-2 cells correlates with the loss of the CFTR transcript, with the control PCK1 transcripts remaining unaffected (Figure 6). These results suggest that Tcf4 is restricted to CFTR regulation. Tcf4 is a terminal effector of the Wnt pathway. These data therefore identify CFTR to be a downstream target gene for Wnt signalling through Tcf4. Additionally, Tcf4 and β-catenin probably modulate transcription of additional potential transcriptional regulators for CFTR expression [28,29]. It appears that CFTR may be regulated by both proliferation and differentiation signalling. Several studies indicate a role for Wnt via Tcf4 and β-catenin to co-ordinate an important balance between cell proliferation and differentiation in the gastrointestinal tract. Wnt signalling appears to be critical for maintaining intestinal crypt cell proliferation [16,17,30,31] and for terminal cell differentiation and organization in non-neoplastic intestinal epithelial cells [16,32]. It is possible that our results with Tcf4 may be specific to the adenocarcinoma cell lines tested. However, the fact that Wnt appears to play dual physiological roles in regulating gene expression in non-neoplastic intestinal epithelium suggests that Tcf4 can be a bona fide physiological effector for CFTR expression [17,31]. Recent evidence also identifies antagonists of Wnt signalling that may link membrane ion fluxes to the regulation of the potency, duration or distribution of Wnt signals [33]. Interestingly, an earlier report in murine colonocytes suggests enhancement of chloride transport during pre-neoplastic transformation by chemical carcinogenesis [34]. Whether this is specifically linked with Wnt signalling was not addressed and remains unclear.
Gene occupation by HNF1α and Cdx2 was also determined by ChIP studies through PCR amplification of the same region occupied by Tcf4 (Figure 4) The murine caudal homeobox genes Cdx1 and Cdx2, members of the caudal-related homeobox family of transcription factors, are known target genes of Wnt signalling in the control of intestinal cell proliferation and differentiation [28,35–39]. Like CFTR, they are also primarily expressed both in the mature intestinal epithelium and at specific stages during embryonic development [38,40]. Interestingly, Cdx2 has also been shown to play dual roles in the intestine, stimulating crypt cell proliferation [38], but also associated with inducing terminal cell differentiation [41,42]. The Cdx1 promoter possesses LEF/Tcf-binding motifs [43], and it appears that Cdx1 and Cdx2 can differentially target the outcome of Tcf4/β-catenin transcriptional activity [28,29]. However, the precise mechanism of this interaction remains unclear.
Combined activity at the intronic site of HNF1α, Cdx2 and Tcf4, factors that are known to participate in both cell differentiation and proliferation signalling, indicate that the fate of cell-specific CFTR expression may be determined by the balanced interaction of these factors, organized around a specific chromatin environment. Previous studies that have shown distinct and disparate sequences to be involved in the long-range control of CFTR [1] also suggest that multiple regulatory factors are likely to be involved in the transcriptional regulation of CFTR.
Our findings illustrate the complex relationship between factors that regulate CFTR, in this case via histone modification, and their native chromatin environment. We provide a novel perspective on therapeutic approaches such as HDAC inhibition that may be utilized to correct diseases associated with CFTR dysfunction by selectively enhancing CFTR expression.
Online data
Acknowledgments
We gratefully acknowledge the advice and support from Dr Hitomi Nishio for help conducting ChIP experiments. This work was supported by an individual investigator award to M. J. W. from the Cystic Fibrosis Foundation and by Public Health Service award HL067099 to M. J. W. Support for T. P. was through The Department of Pediatrics Chief's Fund, The Division of Pediatric Gastroenterology, Hasbro Children's Hospital/Rhode Island Hospital for completing the experiments.
References
- 1.Phylactides M., Rowntree R., Nuthall H., Ussery D., Wheeler A., Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur. J. Biochem. 2002;269:553–559. doi: 10.1046/j.0014-2956.2001.02679.x. [DOI] [PubMed] [Google Scholar]
- 2.Vuillaumier S., Dixmeras I., Messai H., Lapoumeroulie C., Lallemand D., Gekas J., Chehab F. F., Perret C., Elion J., Denamur E. Cross-species characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene reveals multiple levels of regulation. Biochem. J. 1997;327:651–662. doi: 10.1042/bj3270651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pittman N., Shue G., LeLeiko N. S., Walsh M. J. Transcription of cystic fibrosis transmembrane conductance regulator requires a CCAAT-like element for both basal and cAMP-mediated regulation. J. Biol. Chem. 1995;270:28848–28857. doi: 10.1074/jbc.270.48.28848. [DOI] [PubMed] [Google Scholar]
- 4.Neugut A. I., Jacobson J. S., Suh S., Mukherjee R., Arber N. The epidemiology of cancer of the small bowel. Cancer Epidemiol. Biomarkers Prev. 1998;7:243–251. [PubMed] [Google Scholar]
- 5.Mouchel N., Broackes-Carter F., Harris A. Alternative 5′ exons of the CFTR gene show developmental regulation. Hum. Mol. Genet. 2003;12:759–769. doi: 10.1093/hmg/ddg079. [DOI] [PubMed] [Google Scholar]
- 6.Groman J. D., Meyer M. E., Wilmott R. W., Zeitlin P. L., Cutting G. R. Variant cystic fibrosis phenotypes in the absence of CFTR mutations. N. Engl. J. Med. 2002;347:401–407. doi: 10.1056/NEJMoa011899. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Moy L., Pittman N., Shue G., Aufiero B., Neufeld E. J., LeLeiko N. S., Walsh M. J. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J. Biol. Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 8.Nuthall H. N., Vassaux G., Huxley C., Harris A. Analysis of a DNase I hypersensitive site located −20.9 kb upstream of the CFTR gene. Eur. J. Biochem. 1999;266:431–443. doi: 10.1046/j.1432-1327.1999.00872.x. [DOI] [PubMed] [Google Scholar]
- 9.Rowntree R. K., Vassaux G., McDowell T. L., Howe S., McGuigan A., Phylactides M., Huxley C., Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001;10:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Ananthanarayanan M., Li S., Balasubramaniyan N., Suchy F. J., Walsh M. J. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J. Biol. Chem. 2004;279:54348–54357. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y. B., Yoshida M., Horinouchi S. Selective induction of cyclin-dependent kinase inhibitors and their roles in cell cycle arrest caused by trichostatin A, an inhibitor of histone deacetylase. Ann. N.Y. Acad. Sci. 1999;886:200–203. doi: 10.1111/j.1749-6632.1999.tb09416.x. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida M., Horinouchi S. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N.Y. Acad. Sci. 1999;886:23–36. doi: 10.1111/j.1749-6632.1999.tb09397.x. [DOI] [PubMed] [Google Scholar]
- 14.Parrizas M., Maestro M. A., Boj S. F., Paniagua A., Casamitjana R., Gomis R., Rivera F., Ferrer J. Hepatic nuclear factor 1-α directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol. Cell. Biol. 2001;21:3234–3243. doi: 10.1128/MCB.21.9.3234-3243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouchel N., Henstra S. A., McCarthy V. A., Williams S. H., Phylactides M., Harris A. HNF1α is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P. J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 17.van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 18.Barker N., Huls G., Korinek V., Clevers H. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am. J. Pathol. 1999;154:29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Idogawa M., Yamada T., Honda K., Sato S., Imai K., Hirohashi S. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/β-catenin complex. Gastroenterology. 2005;128:1919–1936. doi: 10.1053/j.gastro.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Gray S. G., Teh B. T. Histone acetylation/deacetylation and cancer: an “open” and “shut” case? Curr. Mol. Med. 2001;1:401–429. doi: 10.2174/1566524013363537. [DOI] [PubMed] [Google Scholar]
- 21.Brownell J. E., Allis C. D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 22.Wolffe A. P., Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 23.Berger S. L. Molecular biology: the histone modification circus. Science. 2001;292:64–65. [PubMed] [Google Scholar]
- 24.Van Lint C., Emiliani S., Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 25.Wiren M., Silverstein R. A., Sinha I., Walfridsson J., Lee H. M., Laurenson P., Pillus L., Robyr D., Grunstein M., Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory P. A., Lewinsky R. H., Gardner-Stephen D. A., Mackenzie P. I. Coordinate regulation of the human UDP-glucuronosyltransferase 1A8, 1A9, and 1A10 genes by hepatocyte nuclear factor 1α and the caudal-related homeodomain protein 2. Mol. Pharmacol. 2004;65:953–963. doi: 10.1124/mol.65.4.953. [DOI] [PubMed] [Google Scholar]
- 27.Walters J. R. Cell and molecular biology of the small intestine: new insights into differentiation, growth and repair. Curr. Opin. Gastroenterol. 2004;20:70–76. doi: 10.1097/00001574-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Domon-Dell C., Freund J. N. Stimulation of Cdx1 by oncogenic β-catenin/Tcf4 in colon cancer cells: opposite effect of the CDX2 homeoprotein. FEBS Lett. 2002;518:83–87. doi: 10.1016/s0014-5793(02)02650-9. [DOI] [PubMed] [Google Scholar]
- 29.Domon-Dell C., Wang Q., Kim S., Kedinger M., Evers B. M., Freund J. N. Stimulation of the intestinal Cdx2 homeobox gene by butyrate in colon cancer cells. Gut. 2002;50:525–529. doi: 10.1136/gut.50.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batlle E., Henderson J. T., Beghtel H., van den Born M. M., Sancho E., Huls G., Meeldijk J., Robertson J., van de Wetering M., Pawson T., Clevers H. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 31.Roose J., Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta. 1999;1424:M23–M37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman J., Kuhnert F., Davis C. R., Kuo C. J. Wnts as essential growth factors for the adult small intestine and colon. Cell Cycle. 2004;3:554–557. [PubMed] [Google Scholar]
- 33.Zeng W., Wharton K. A., Jr, Mack J. A., Wang K., Gadbaw M., Suyama K., Klein P. S., Scott M. P. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 34.Fraser G. M., Portnoy M., Bleich M., Ecke D., Niv Y., Greger R., Schwartz B. Characterization of sodium and chloride conductances in preneoplastic and neoplastic murine colonocytes. Pflügers Arch. 1997;434:801–808. doi: 10.1007/s004240050468. [DOI] [PubMed] [Google Scholar]
- 35.Lickert H., Domon C., Huls G., Wehrle C., Duluc I., Clevers H., Meyer B. I., Freund J. N., Kemler R. Wnt/β-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 36.Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J. N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallo G. V., Soubeyran P., Lissitzky J. C., Andre F., Farnarier C., Marvaldi J., Dagorn J. C., Iovanna J. L. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J. Biol. Chem. 1998;273:14030–14036. doi: 10.1074/jbc.273.22.14030. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian V., Meyer B., Evans G. S. The murine Cdx1 gene product localises to the proliferative compartment in the developing and regenerating intestinal epithelium. Differentiation. 1998;64:11–18. doi: 10.1046/j.1432-0436.1998.6410011.x. [DOI] [PubMed] [Google Scholar]
- 39.Silberg D. G., Furth E. E., Taylor J. K., Schuck T., Chiou T., Traber P. G. CDX1 protein expression in normal, metaplastic, and neoplastic human alimentary tract epithelium. Gastroenterology. 1997;113:478–486. doi: 10.1053/gast.1997.v113.pm9247467. [DOI] [PubMed] [Google Scholar]
- 40.Duprey P., Chowdhury K., Dressler G. R., Balling R., Simon D., Guenet J. L., Gruss P. A mouse gene homologous to the Drosophila gene caudal is expressed in epithelial cells from the embryonic intestine. Genes Dev. 1988;2:1647–1654. doi: 10.1101/gad.2.12a.1647. [DOI] [PubMed] [Google Scholar]
- 41.Soubeyran P., Andre F., Lissitzky J. C., Mallo G. V., Moucadel V., Roccabianca M., Rechreche H., Marvaldi J., Dikic I., Dagorn J. C., Iovanna J. L. Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology. 1999;117:1326–1338. doi: 10.1016/s0016-5085(99)70283-0. [DOI] [PubMed] [Google Scholar]
- 42.Ren P., Silberg D. G., Sirica A. E. Expression of an intestine-specific transcription factor (CDX1) in intestinal metaplasia and in subsequently developed intestinal type of cholangiocarcinoma in rat liver. Am. J. Pathol. 2000;156:621–627. doi: 10.1016/S0002-9440(10)64766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beland M., Pilon N., Houle M., Oh K., Sylvestre J. R., Prinos P., Lohnes D. Cdx1 autoregulation is governed by a novel Cdx1–LEF1 transcription complex. Mol. Cell. Biol. 2004;24:5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.