Abstract
X-ray crystallography studies, as well as live-cell fluorescent imaging, have recently challenged the traditional view of protein kinase CK2. Unbalanced expression of catalytic and regulatory CK2 subunits has been observed in a variety of tissues and tumours. Thus the potential intersubunit flexibility suggested by these studies raises the likely prospect that the CK2 holoenzyme complex is subject to disassembly and reassembly. In the present paper, we show evidence for the reversible multimeric organization of the CK2 holoenzyme complex in vitro. We used a combination of site-directed mutagenesis, binding experiments and functional assays to show that, both in vitro and in vivo, only a small set of primary hydrophobic residues of CK2β which contacts at the centre of the CK2α/CK2β interface dominates affinity. The results indicate that a double mutation in CK2β of amino acids Tyr188 and Phe190, which are complementary and fill up a hydrophobic pocket of CK2α, is the most disruptive to CK2α binding both in vitro and in living cells. Further characterization of hotspots in a cluster of hydrophobic amino acids centred around Tyr188–Phe190 led us to the structure-based design of small-peptide inhibitors. One conformationally constrained 11-mer peptide (Pc) represents a unique CK2β-based small molecule that was particularly efficient (i) to antagonize the interaction between the CK2 subunits, (ii) to inhibit the assembly of the CK2 holoenzyme complex, and (iii) to strongly affect its substrate preference.
Keywords: CK2 protein kinase, cyclic peptide, hotspot, peptide disruptor, protein–protein interaction, substrate specificity
Abbreviations: BiFC, bimolecular fluorescence complementation; CK2β-F, F190A single mutant of CK2β; CK2β-M, M166A single mutant of CK2β; CK2β-Y, Y188A single mutant of CK2β; CK2β-YF, Y188A/F190A double mutant of CK2β; CK2β-MYF, M166A/Y188A/F190A triple mutant of CK2β; EYFP, enhanced yellow fluorescent protein; GFP, green fluorescent protein; GST, glutathione transferase; GST-β C-ter, GST–CK2β-(182–215); HBS, Hepes-buffered saline; MBP, maltose-binding protein; RP, reverse phase; TFA, trifluoroacetic acid; YC-CK2, CK2 with C-terminal EGYP tag; YN-CK2, CK2 with N-terminal EGYP tag
INTRODUCTION
All but a handful of the protein kinase family members are monomeric enzymes which are often regulated by reversible phosphorylation. In contrast, the regulation and the molecular architecture of multisubunit serine/threonine protein kinases such as phosphorylase kinase [1], cAMP-dependent protein kinase [2], cyclin-dependent protein kinases [3], DNA-dependent protein kinase [4], 5′-AMP-activated protein kinase [5] and IκB (inhibitory κB) kinase [6] rely on the reversible binding of regulatory subunits to the catalytic subunits of these enzymes. The presence of the regulatory subunit either can play a negative role on the catalytic subunit (e.g. cAMP-dependent protein kinase [7]) or is a prerequisite for activation (e.g. cyclin-dependent protein kinases [3]).
Protein kinase CK2 shares with few other protein kinases a quaternary structure consisting of two catalytic subunits (CK2α and CK2α') and two regulatory subunits (CK2β) [8]. However, as compared with other multisubunit protein kinases, CK2 exhibits notable distinguishing structural and functional features [9,10]. CK2 catalytic subunits possess constitutive activity [11,12], but CK2β is a central component of the tetrameric CK2 complex [13], controlling CK2 substrate specificity, cellular localization and enzymatic activity. In this respect, CK2β operates as a targeting subunit and/or a docking platform affecting the accessibility to the catalytic site of binding substrates whose phosphorylation is either stimulated or prevented by the CK2β subunit [14,15]. In this context, the spontaneous high-affinity association of recombinant CK2 subunits in a stable heterotetrameric complex (dissociation constant, Kd, of 5.4 nM), as well as numerous complementary biochemical observations led to the long-held tenet that CK2 is a strong obligate complex. However, this traditional view was challenged when X-ray crystallography studies revealed that the CK2α/CK2β interface is smaller than the surface contacts that are usually observed in stable protein complexes [13]. The possible intersubunit flexibility suggested by these studies was reinforced further by the observation of independent movement of CK2α and CK2β in living cells [16]. In addition, the presence in mouse brain and testes of a CK2β fraction devoid of CK2α [17] and a pool of free CK2α' in prostate tumour cells [18] have been reported. Collectively, these observations raise the likely prospect that the CK2 holoenzyme complex is subject to disassembly and reassembly [19]. As CK2 substrates localize to many different subcellular compartments, a dynamic interaction of CK2 subunits should increase the kinase specificity, and the assembly of the complex is a likely point of regulation [20]. The ability to interfere with specific protein–protein interactions has already provided powerful means of influencing the functions of selected proteins within the cell [21,22]. Thus the potential intersubunit flexibility of the CK2 holoenzyme complex makes it amenable to drug-discovery efforts aimed at inhibiting this protein–protein interaction.
In the present paper, we show evidence for the reversible multimeric organization of the CK2 holoenzyme complex in vitro. Site-directed mutagenesis, binding experiments and functional assays provided further insights into the molecular interaction between the CK2 subunits, demonstrating that, both in vitro and in vivo, only a small set of primary hydrophobic residues of CK2β which make contacts at the centre of the interface dominate affinity. Characterization of hotspots led us to a structure-based design of small peptide inhibitors derived from the CK2β C-terminal domain. One conformationally constrained 11-mer peptide can efficiently antagonize the interaction between the CK2 subunits, representing the first small molecule that binds to this hydrophobic interface. The present study represents the first step of a systematic approach to set the framework for the discovery and development of chemical inhibitors of this interaction.
EXPERIMENTAL
Materials
[γ-32P]ATP (3000 Ci/mmol) was purchased from MP Biosciences. The peptide substrate (RRREDEESDDEE) for CK2 kinase assay was obtained from NeoMPS. The purity of this peptide (89%) was determined by HPLC on a Nucleosil C18 column using a linear triethanolamine phosphate/acetonitrile gradient. Solvents were of analytical grade. Cyclic peptides, biotinylated cyclic peptides and alanine-mutated cyclic peptides were synthesized by Eurogentec. The pEYFPc1 vector was purchased from Clontech Laboratories. EcoRI and BamHI restriction enzymes were from Invitrogen, and SacI and AgeI were from New England Biolabs.
Chemicals, peptide synthesis and purification
All chemical reagents and solvents were purchased from Sigma–Aldrich, Acros or Carlo-Erba and were used without further purification. All protected amino acids were obtained from Advanced ChemTech Europe, Bachem Biochimie SARL and France Biochem S.A. RP (reverse-phase)-HPLC analyses were performed using Waters equipment. The analytical [Nucleosil 120 Å (1 Å=0.1 nm) pore size, 3 μm diameter C18 particles, 30 mm long×4.6 mm2 cross-sectional area] column was operated at 1.3 ml/min, and the preparative (Delta-Pak 300 Å pore size, 15 μm diameter C18 particles, 200 mm long×25 mm2 cross-sectional area) column at 22 ml/min with UV monitoring at 214 and 250 nm using a linear buffer A–B gradient [buffer A: 0.09% TFA (trifluoroacetic acid) in water; buffer B: 0.09% TFA in 90% acetonitrile]. Mass spectra were obtained by ESI (electrospray ionization) MS on a VG Platform II in the positive mode.
The protected linear peptides were assembled manually on solid-phase using the standard Fmoc (fluoren-9-ylmethoxycarbonyl)/tBu (t-butyl) strategy with PyBOP® (benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate) as a coupling reagent on the acid-labile SASRIN® resin. They were cyclized directly without further purification, as reported previously [23]. For all peptides, glycine was chosen as the C-terminal end to secure the subsequent cyclization step from epimerization. The protected cyclic peptides were finally treated with a solution of TFA/TIS (tri-isopropylsilane)/water (95:2.5:2.5, by vol.) for 2 h. After evaporation, the cyclic peptides were purified by RP-HPLC. The purified cyclic peptides were homogeneous and showed expected primary ion molecular masses by MS.
Proteins
Human recombinant His6-tagged CK2α, GST (glutathione transferase)–CK2α and GFP (green fluorescent protein)–CK2α were obtained as described previously [16,24]. Expression and purification of chicken recombinant MBP (maltose-binding protein)–CK2β were performed as described previously [25]. GST–CK2β-(182–215), referred to as GST-β C-ter, was obtained by cloning by PCR the 33 C-terminal amino acids of CK2β in pGEX4T1. Two recombinant proteins, MBP–CDC25B (kindly provided by Bernard Ducommun, Université Paul Sabatier, Toulouse, France) and GST–Olig2-(1–177) (kindly provided by Thierry Buchou, iRTSV/LTS), were used as CK2β-dependent CK2 substrates.
In vitro kinase assay
CK2 kinase assays were performed in a final assay volume of 18 μl containing 3 μl of CK2α (36 ng) and a mixture containing 1 mM peptide substrate, 10 mM MgCl2 and 1 μCi of [γ-32P]ATP. The final concentration of ATP was 100 μM. Assays were performed under linear kinetic conditions for 5 min at room temperature (22 °C) before termination by the addition of 60 μl of 4% trichloroacetic acid [26].
In vitro CK2α–CK2β interaction assay
The CK2α–CK2β interaction assay involved competition between plate-bound MBP–CK2β and various soluble peptides for binding to soluble [35S]methionine-labelled CK2α. The assay was performed in Reacti-Bind streptavidin-coated high-binding-capacity 96-well plates (Pierce) in which each well was coated with 250 ng of biotinylated MBP–CK2β {using sulfo-NHS-LC-LC-biotin [sulfosuccinimidyl-6′-(biotinamido)-6-hexanamido hexanoate] with a spacer arm of 30.5 Å length (Pierce)} in 50 mM Tris/HCl, pH 7.2, and 0.4 M NaCl buffer for 1 h at room temperature. After three washes with 50 mM Tris/HCl, pH 7.2, 0.15 M NaCl and 0.05% Tween 20, the wells were blocked with 50 mM Tris/HCl, pH 7.2, 0.15M NaCl and 3% BSA for 1 h at room temperature. After three washes, competing peptides were added to each well in 50 mM Tris/HCl, pH 7.2, and 0.4 M NaCl, along with [35S]methionine-labelled CK2α (105 c.p.m.) synthesized in vitro by using the TNT® Quick coupled transcription/translation system (Promega). The plates were incubated for 1 h at room temperature, and, after three washes, the radioactivity or the fluorescence of each well in the plate was determined using a scintillation counter. Positive control (100% competition) was determined with a 10-fold molar excess of untagged CK2α, and negative control (0% competition) was performed in the absence of competitor. The IC50 is defined as the concentration of peptide necessary to inhibit 50% of the CK2α–CK2β complex formation.
Pull-down assays
GST-tagged proteins were immobilized on glutathione–Sepharose 4 Fast Flow beads (Amersham Biosciences), for 1 h at 4 °C, in 10 mM Tris/HCl, pH 7.5. Beads were then incubated with CK2α for 1 h at 4 °C. After four washes, CK2α activity was measured as described above.
MBP–CK2β (5 μg) was immobilized on amylose beads (New England Biolabs) for 1 h at 4 °C in 10 mM sodium phosphate buffer, pH 7.2, 0.5 M NaCl, 10 mM 2-mercaptoethanol, 1 mM EGTA and 0.05% Tween 20. Buffer was replaced by 10 mM Tris/HCl, pH 7.5, 0.15 M NaCl and 0.05% Tween 20. Then, increasing amounts of GST-β C-ter were added along with 5 μg of GST–CK2α for 20 min at 4 °C. After four washes, one-tenth of the beads was used for CK2 activity assay, and the remaining beads were used for CK2α detection by Western blotting.
Size-exclusion chromatography
An Ultrogel ACA34 gel-filtration column (0.5 cm×30 cm) was equilibrated in 50 mM Tris/HCl, pH 7.5, and 0.4 M NaCl and was calibrated using aldolase (molecular mass of 158 kDa), BSA (molecular mass of 68 kDa) and carbonic anhydrase (molecular mass of 29 kDa) as standards. CK2α (50 μg) alone or CK2α (50 μg) incubated with GST-β C-ter (30 μg) for 30 min at 4 °C was loaded on to the column. Eluted fractions (0.25 ml) were collected and assayed for the presence of CK2α and GST-β C-ter by Western blotting.
Site-directed mutagenesis
Site-directed mutagenesis of CK2β was performed with the pMALc2-CK2β vector using the QuikChange® site-directed mutagenesis kit (Stratagene) and specific primers from Eurogentec to generate different mutant MBP-CK2β proteins. Primers were M166A sense, 5′-GTTCCCCCATGCGCTCTTCATGGTG-3′, Y188A sense, 5′-GTGCCCAGGCTGGCTGGGTTCAAGATCCACCCTATGG-3′, and F190A sense, 5′-GTGCCCAGGCTGTATGGGGCCAAGATCCACCCTATGG-3′. MBP–CK2β mutant proteins were expressed in BL21 Escherichia coli cells and were purified as described previously [25] and stored at −80 °C in 10 mM sodium phosphate buffer, pH 7.0, 0.5 M NaCl, 10 mM 2-mercaptoethanol, 1 mM EGTA, and protease inhibitors (Sigma).
Surface plasmon resonance spectroscopy
Surface plasmon resonance measurements were performed using a BIAcore 3000 instrument. The running buffer was HBS (Hepes-buffered saline: 10 mM Hepes, pH 7.3, 0.15 M NaCl, 3 mM EDTA and 0.005% polysorbate 20). The carboxymethylated dextran surface of a CM5 sensor chip (BIAcore AB) was activated by injecting a coupling solution of N-hydroxysuccinimide and N-ethyl-N-(dimethylaminopropyl)carbodi-imide hydrochloride (BIAcore AB). Anti-GST antibody was diluted to 30 μg/ml in 10 mM acetate buffer, pH 5.0, and injected over the surface of the sensor chip for 7 min at a flow rate of 5 μl/min. The unreacted sites of the sensor chip surface were then quenched by injection of 1 M ethanolamine, pH 8.5. GST–CK2α (50 μg/ml) was immobilized on the surface at a flow rate of 5 μl/min in HBS. Different MBP–CK2β mutants were diluted to 50 μg/ml in HBS and injected over the surface at a flow rate of 5 μl/min. Regeneration of the surfaces was achieved by injection of 10 mM glycine/HCl, pH 2.2.
Plasmid constructs, cell transfections and BiFC (bimolecular fluorescence complementation) analysis
Full-length chicken CK2α sequence was cloned into the pEYFPc1 vector using EcoRI restriction sites, then oriented with BamHI digestion to generate the pEYFPc1-CK2α construct. Full-length mouse CK2β sequence was cloned into the same pEYFPc1 vector using BamHI restriction sites, then oriented with EcoRI digestion to generate the pEYFPc1-CK2β construct. Subsequently, the EYFP (enhanced yellow fluorescent protein) sequence was replaced with the 1–154 or the 155–238 fragment of EYFP. These fragments were PCR-amplified and inserted using SacI/AgeI to generate BiFC plasmid constructs. The resulting N- and C-terminal EYFP–CK2 fusions are referred to as YN-CK2α, YN-CK2β, YC-CK2α and YC-CK2β respectively. Primers (Eurogentec) for YN-CK2α and YN-CK2β were 5′-GCTACCGGTCGCCACCATGGTGAGCAAG-3′ and 5′-CACAACGTCTATATCATGCGAGCTCAGGCTTCGAATTCTGC-3′ respectively. Primers for YC-CK2α and YC-CK2β were 5′-CCGTCAGATCCGCTCGCGCTACCGGTCATGGCCGACAAGCAGAAGAACGGC-3′ and 5′-CGAAGCTTGAGCTCGAGATCTGAGTCCGG-3′ respectively. All of the constructs were verified by sequencing.
HeLa cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% fetal calf serum. At 1 day before transfection, cells were seeded in Lab-Tek® chambers (Nunc) and transfected with the different BiFC plasmid constructs using Lipofectamine™ 2000 reagent (Invitrogen) according to the manufacturer's instructions. At 4 h after transfection, cells were washed with PBS and incubated with fresh medium for 24 h at 37 °C and then switched to 30 °C for 2 h to promote fluorophore maturation [27]. Preliminary experiments have shown that a fraction of YN-CK2α or YC-CK2α and YN-CK2β or YC-CK2β could interact with endogenous CK2β and CK2α, trapping a part of EYFP devoted to fluorescence complementation [28]. Therefore, to enhance the signal, immunostaining of EYFP was performed by indirect immunofluorescence using the mouse anti-GFP antibody (Abcam) which recognizes only full-length EYFP, but neither N-terminal (1–154) nor C-terminal (155–238) EYFP (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/408/bj4080363add.htm). Under similar transfection conditions, polyclonal anti-GFP antibody (Abcam) did not discriminate between full-length and EYFP half-molecules (results not shown). Thus cells were fixed with 4% (w/v) paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100 for 10 min, pre-incubated with 5% goat serum for 30 min at room temperature, and incubated with the mouse anti-GFP as primary antibody for 1 h at room temperature, washed with PBS and incubated with goat anti-mouse IgG-conjugated Cy3 (indocarbocyanine) (Molecular Probes) for 45 min at room temperature in the dark. Nuclei were stained with 2 μg/ml Hoechst 33342 (Sigma) and coverslips were mounted with Dako (Dakocytomation), and examined using a Zeiss Axiovert 200 microscope and a 40× 1.3 Plan-Neofluar® objective. The results were expressed as the percentage of cells that were above the fluorescence threshold observed in cells expressing only YN-CK2α or YN-CK2β. Because the amount of expressed proteins is critical to the interpretation of results, the expression levels of the different chimaeras were also analysed in transfected cells by Western blotting using anti-CK2α or -CK2β antibodies as described previously [16,29].
RESULTS
Reversibility of the CK2 subunit multimeric organization in vitro
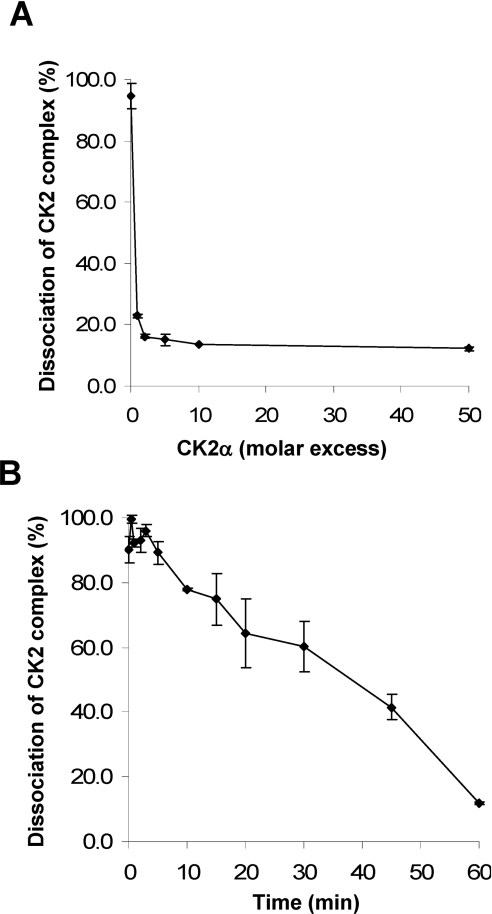
Surface plasmon resonance measurements with purified recombinant CK2α and CK2β subunits showed that they assemble rapidly (t½=60 s) into a stable heterotetrameric complex with nanomolar affinity (Kd=5.4 nM) with a ka (association rate constant) and kd (dissociation rate constant) of 6.6×104 M−1·s−1 and 3.6×10−4 s−1 respectively [30]. However, X-ray crystallography has revealed that the CK2α/CK2β interface was relatively small (832 Å2) and flexible, raising the possibility that CK2 tetramers may be subjected to disassembly and reassembly [13]. To test this hypothesis, we performed a competition binding assay in which complexes between MBP–CK2β and [35S]methionine-labelled CK2α were incubated with an increasing molar excess of CK2α. As shown in Figure 1(A), [35S]methionine-labelled CK2α was efficiently displaced from the pre-formed complex by the CK2α subunit. The residual radioactivity (15%) may represent [35S]methionine-labelled CK2α present in misfolded CK2 complexes that are resistant to dissociation. In the presence of a 10-fold molar excess of CK2α, the t½ of CK2 complex dissociation was evaluated to be ≈38 min, showing that, unlike the fast association of CK2 subunits (t½=1 min), the kinetics of disassembly were rather slow (Figure 1B). These data are consistent with the association/dissociation rate constants determined previously [30].
Figure 1. Reversibility of the CK2 subunit assembly in vitro.
(A) Exchange of CK2α in the subunit complex. Pre-formed complexes between immobilized MBP–CK2β and [35S]methionine-labelled CK2α were incubated for 1 h with increasing concentrations of unlabelled CK2α. After washing, the amount of [35S]methionine-labelled CK2α present in the complex was determined by radioactivity counting. (B) Kinetics of dissociation of the CK2 subunit complex. Pre-formed complexes between [35S]methionine-labelled CK2α and MBP–CK2β were incubated for different times with a 10-fold molar excess of unlabelled CK2α. After washing, the amount of [35S]methionine-labelled CK2α remaining in the complex was determined by radioactivity counting. Results are the means±S.D. (n=3).
Molecular and functional analysis of the CK2β C-terminus
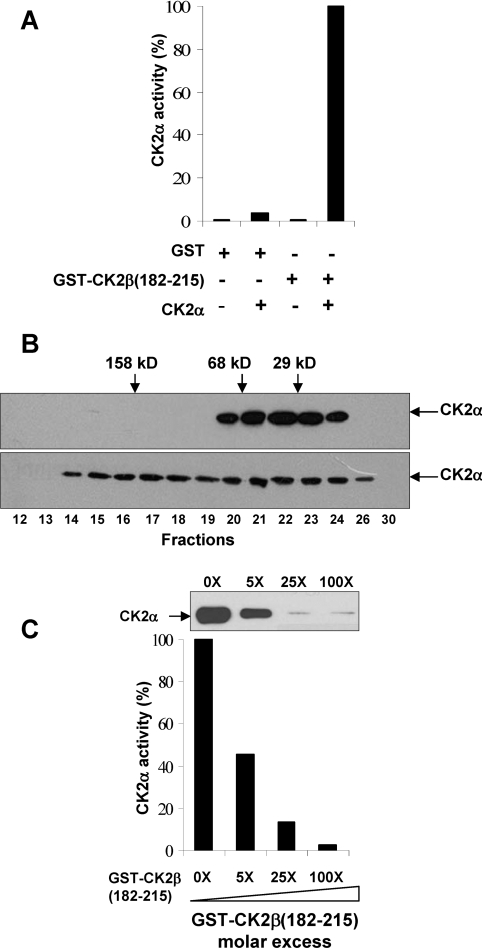
Analysis of the crystal data has revealed the major contribution of the α/β tail contact for the stability of the CK2 holoenzyme [13]. In addition, CK2β mutants in which the zinc-finger motif has been disrupted failed to dimerize and to recruit CK2α, showing that this dimerization is essential for the proper docking of CK2α [31,32]. A CK2α-interacting domain was delineated by residues 181–203 in the CK2β C-terminus [33]. Thus we have expressed and purified from bacteria a CK2β fragment encompassing residues 182–215 fused to GST (GST-β C-ter). Size-exclusion chromatography analysis indicated that this fusion protein behaves as a dimeric protein through GST–GST interaction (results not shown). Reconstitution experiments showed that this mini-protein was sufficient for a productive high-affinity interaction with CK2α in a pull-down assay (Figure 2A). Size-exclusion chromatography analysis showed that the free CK2α was eluted as a monomer (42 kDa). In contrast, CK2α incubated with GST-β C-ter was eluted as free CK2α as well as a CK2α–GST-β C-ter complex with an apparent molecular mass of 150 kDa (Figure 2B). These data indicate that this interaction generates a stable heterocomplex bridging two catalytic subunits. Thus the region delineated by residues 182–215 in GST-β C-ter contains an autonomous CK2α-interacting domain. We then examined the effect of this CK2β domain on the formation of the CK2 holoenzyme complex in a solid-phase competition assay. As illustrated in Figure 2(C), the interaction between CK2α and CK2β was strongly impaired by the presence of increasing concentrations of the GST-β C-ter protein. These results show that the 33-amino-acid C-terminal region of CK2β binds tightly to CK2α and behaves as an antagonist of the CK2 subunit interaction.
Figure 2. Interaction of a CK2β C-terminal fragment with CK2α.
(A) Stable association of CK2α with a GST-β C-ter fragment. Agarose beads containing different GST-tagged proteins were incubated with or without CK2α as indicated. After washing, the amount of associated CK2α was evaluated by a CK2 kinase assay as described in the Experimental section. (B) Size-exclusion chromatography of either free CK2α (upper panel) or CK2α–GST-β C-ter complex (lower panel). Collected fractions were assayed for the presence of CK2α by Western blotting. Elution of molecular mass standards is indicated (sizes are in kDa). (C) Antagonist effect of the GST-β C-ter fragment. Immobilized MBP–CK2β was incubated with the indicated fold excess of GST-β C-ter and a fixed amount of CK2α. After washing, the amount of associated CK2α was assessed by a CK2 kinase assay and Western blotting.
Identification and validation of CK2β hotspots for high-affinity CK2α binding
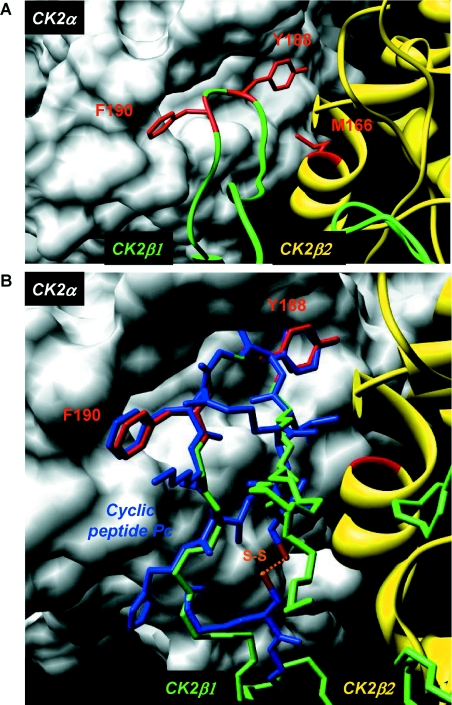
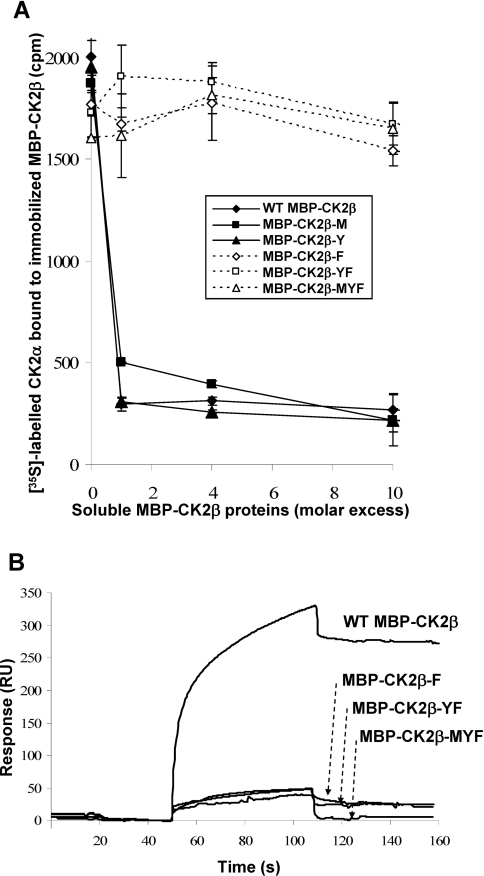
The crystal structure of the CK2 holoenzyme has suggested the predominant contribution of specific residues for the interfacial contacts of the CK2β tail with CK2α, which were localized in a cluster of hydrophobic amino acids centred around Tyr188. As already pointed out by Niefind et al. [13], this highly conserved segment of CK2β forms a structural element containing a β-hairpin loop with Tyr188 at its top (Figure 3A). This analysis provided a general outline of the CK2α-binding site, but did not identify experimentally the specific residues involved in the binding. Guided by the high-resolution structure of the CK2 holoenzyme, we used alanine mutagenesis to target CK2β residues (Met166, Tyr188 and Phe190) in this CK2α-docking site that make the most significant contacts with CK2α. Residues within CK2α that are involved in interactions with these CK2β residues are mainly hydrophobic residues such as Ile69, Pro104, Tyr39, Leu41, Val42, Ile57, Ile59 and Val105. In addition, carbonyl oxygen of Tyr188 and Lys191 CK2β backbone are making hydrogen bonds with Nϵ of Gln36 and Leu41 of CK2α respectively. However, it is known that the holoenzyme structure is stable under high ionic strength, suggesting that these hydrogen bonds may be barely involved in the subunit interaction [10]. Therefore we tested M166A, Y188A and F190A mutants by quantitative binding analysis. Figure 4(A) shows that the CK2α-binding activity of the single mutant F190A (CK2β-F) was significantly reduced, and this activity was completely abrogated for the Y188A/F190A double mutant (CK2β-YF) and for the M166A/Y188A/F190A triple mutant (CK2β-MYF). To validate these data, the binding properties of the CK2β mutants were also analysed by surface plasmon resonance spectroscopy. As shown in Figure 4(B), the binding activity of the CK2β-F single mutant was markedly decreased, and the CK2β-YF double or the CK2β-MYF triple mutations virtually abolished interaction.
Figure 3. Structure and mode of binding of Pc peptide on CK2α.
(A) Surface representation of the binding pocket of CK2α interacting with a short C-terminal loop of CK2β. Crystal structure of the CK2 holoenzyme shows that a C-terminal fragment of the CK2β1 chain encompassing residues 186–193 (green) forms a loop inserting into a deep hydrophobic pocket of CK2α [14]. Surface representation of the CK2α cleft in grey highlights its pocket-like characteristics. The phenyl and phenol rings of the two non-polar and aromatic CK2β residues Tyr188 and Phe190 respectively (red) are in quasi-planar opposite orientation and fit tightly into it. Met166 (red) located on the second CK2β2 chain (yellow) is labelled. (B) Molecular model of Pc peptide in CK2α pocket. A rough molecular model of Pc peptide (blue) was obtained from the CK2β1 structure determined previously (PDB code 1JWH [13]). In detail, the CK2β1 structure segment (R186LYGFKIH193 in green) was taken as template for the corresponding central peptide segment, and the terminal amino acids were added manually using the program Coot [39]. A basic geometric idealization of peptide bonds was then performed using the program Refmac 5 [40] from CCP4 [41]. For clarity, only key interacting residues of CK2β1 are shown (in red). The cysteine disulfide bridge is coloured orange. The superimposition of Pc peptide with the CK2β C-terminal loop demonstrates that Pc emulates the interaction of the natural ligand CK2β with CK2α. Figures were prepared with the Chimera software [42] and the 1JWH PDB structure [14].
Figure 4. CK2α-binding activity of CK2β mutant proteins.
(A) Solid-phase CK2α-binding assay of different CK2β mutants. Immobilized wild-type (WT) CK2β was incubated with [35S]methionine-labelled CK2α in the presence of increasing concentrations of soluble wild-type MBP–CK2β or MBP–CK2β mutants as indicated. After washing, the amount of associated CK2α was evaluated by radioactivity counting. Results are the means±S.D. (n=3). (B) Real-time binding kinetics between immobilized GST–CK2α and different MBP–CK2β mutant proteins measured by surface plasmon resonance. The interaction between immobilized GST–CK2α and MBP–CK2β mutants was recorded as described in the Experimental section. RU, response units.
Functional properties of CK2β mutants
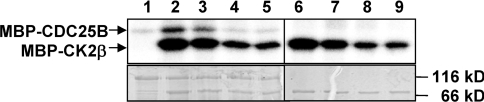
Since it is clear that, in many cases, CK2β mediates the interaction between the catalytic subunits and its cellular substrates, thereby modulating their phosphorylation, we assumed that any mutation perturbing the CK2 subunit interaction would affect the phosphorylation of CK2β-dependent substrates. A functional analysis of wild-type CK2β and CK2β-M, CK2β-YF and CK2β-MYF mutants was performed, testing their effect on the CK2-mediated phosphorylation of CDC25B [34]. The data show that, although the phosphorylation of CDC25B was weakly affected in the presence of the single CK2β-M mutant, the double CK2β-YF and triple CK2β-MYF mutants were inefficient for CK2-mediated phosphorylation of CDC25B (Figure 5, lanes 1–5). Similarly, autophosphorylation of the CK2β-YF and CK2β-MYF mutants was strongly affected (Figure 5, lanes 6–9), indicating that these mutants are also defective for the supramolecular organization of the CK2 holoenzyme [35]. Thus Tyr188 and Phe190 represent key CK2β hotspot residues for a functional interaction with CK2α in vitro.
Figure 5. Phosphorylation of CDC25B by CK2α in presence of CK2β or CK2β mutants.
MBP–CDC25B (1.5 μM) was pre-incubated for 15 min with CK2α (0.6 μM) in the presence of 0.35 μM of wild-type MBP–CK2β or the indicated MBP–CK2β mutants. Phosphorylation reaction was carried out for 5 min at room temperature in the presence of 25 μM [γ-32P]ATP/MgCl2. Phosphorylated proteins were separated by SDS/PAGE and analysed by autoradiography (upper panel) or Coomassie Blue staining (lower panel). Lane 1, CDC25B; lane 2, wild-type MBP–CK2β+CDC25B; lane 3, MBP–CK2β-M+CDC25B; lane 4, MBP–CK2β-YF+CDC25B; lane 5, MBP–CK2β-MYF+CDC25B; lane 6, wild-type MBP–CK2β; lane 7, MBP–CK2β-M; lane 8, MBP–CK2β-YF; lane 9, MBP–CK2β-MYF. Molecular masses are indicated in kDa.
In vivo validation of CK2β hotspots
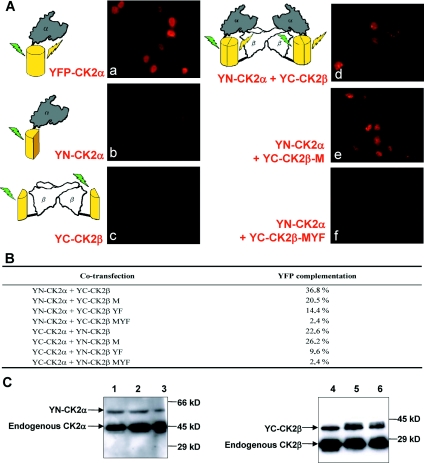
To visualize the CK2 subunit interaction in living cells, we applied a BiFC assay, which allows the investigation of interacting molecules in vivo [27]. The BiFC assay is based on the formation of a fluorescent complex by fragments of the EYFP brought together by the association of two interaction partners fused to non-fluorescent EYFP half-molecules. This approach enables visualization of the proteins interaction under conditions that closely reflect the normal physiological environment. Previous work from our laboratory has shown that visualization of CK2 subunit interaction by FRET (fluorescence resonance energy transfer) analysis required that the fluorochromes should be positioned on the N-terminal region of CK2α and CK2β [36]. Thus the (1–154; YN) and (155–238; YC) fragments of EYFP were fused to the N-terminus of CK2α, CK2β or different CK2β mutants. The corresponding constructs were co-transfected in HeLa cells, and the BiFC assay was performed using immunostaining of EYFP to enhance the signal as described in the Experimental section (see also Supplementary Figure 1). Cells transfected with full-length EYFP fused to CK2α (Figure 6A, panel a) or to CK2β (not shown) exhibit a strong fluorescence after EYFP immunostaining. As expected, cells separately transfected with YN-CK2α and YC-CK2β expression vectors (Figure 6A, panels b and c) or YN-CK2β and YC-CK2α (not shown) did not exhibit any detectable fluorescence even after immunostaining. Co-transfection of YN-CK2α and YC-CK2β yielded reconstitution of EYFP upon specific binding in 36.8% of the cells (Figures 6A, panel d, and 6B). When the fusion partners YN-CK2α and YC-CK2β were cross-exchanged, reconstitution was also observed (results not shown). Similarly, co-expression of YN-CK2α and YC fused to CK2β-M, CK2β-Y or CK2β-F single mutants allowed the observation of a characteristic fluorescent signal (Figures 6A, panel e, and 6B). In contrast, a weak complementation (14.4% of the cells above the fluorescence threshold) was observed with CK2β-YF double mutant, and EYFP reconstitution was barely detected (2.4%) for YN fused to CK2β-MYF triple mutant (Figure 6A, panel f), despite strong protein expression as determined on immunoblots (Figure 6C, lanes 3 and 6). These results indicate that mutations of CK2β residues Tyr188 and Phe190 strongly influence the efficiency of bimolecular complex formation, supporting the hypothesis that their mutation inhibits interaction with CK2α in the cellular context.
Figure 6. Visualization of interactions between CK2α and CK2β or CK2β mutants in living cells by BiFC.
(A) Immunofluorescence images of HeLa cells transfected with plasmids expressing the EYFP fragments fused to CK2α, CK2β or CK2β mutants as indicated in each panel. At 24 h after transfection, immunostaining of EYFP was performed to enhance the signal as described in the Experimental section. The schematic diagrams on the left of the images represent the experimental strategies used. (B) Quantification of the BiFC signal in cells co-transfected with different plasmid constructs. Results are expressed as the percentage of cells that were above the fluorescence threshold observed in cells expressing only YN-CK2α or YN-CK2β. (C) Western blot analysis of the levels of protein expression. Cells corresponding to panels d, e and f in (A) that expressed the indicated proteins were harvested, and the cell extracts were analysed by Western blotting using anti-CK2α (lanes 1–3) and CK2β (lanes 4–6) antibodies. Lanes 1 and 4, YN-CK2α+wild-type YC-CK2β; lanes 2 and 5, YN-CK2α+YC-CK2β-M; lanes 3 and 6, YN-CK2α+YC-CK2β-MYF. Molecular masses are indicated in kDa.
Constrained cyclic peptides as antagonists of CK2 subunit interaction
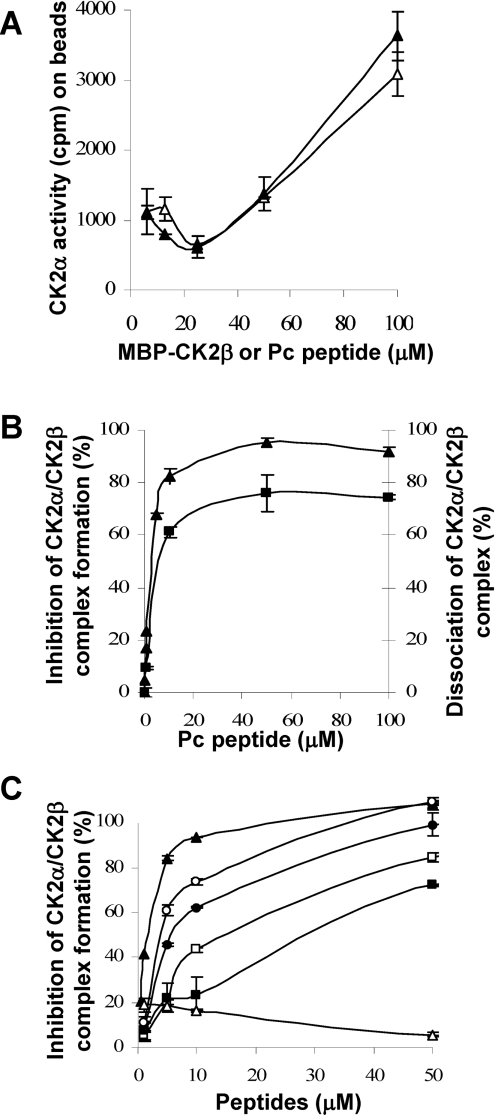
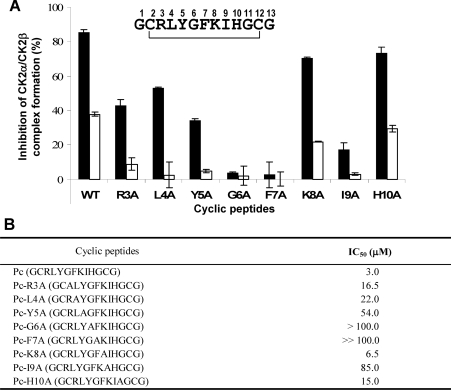
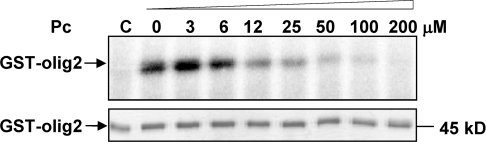
The dramatic effects of the two hotspot mutants on CK2β binding observed both in vitro and in the cellular context suggest that the interaction domain could be narrowed down to a short contiguous sequence. The high-resolution structure of the CK2 holoenzyme has revealed that a cluster of well-defined hydrophobic residues present on the CK2β chain face hydrophobic residues located N-terminally at the outer surface formed by the juxtaposition of the antiparallel β4/β5 sheets of CK2α [13,37]. This cluster, which contains the identified hotspots Tyr188 and Phe190, represents a segment of highly conserved residues (R186LYGFKIH193), exhibiting a specific structural feature: it points away from the protein core, and forms a 90° β-hairpin loop with Tyr188 at its top which binds into a shallow hydrophobic groove present in the β4/β5 sheets of CK2α [13]. The interface relies on the steric complementarity between this CK2α groove and the hydrophobic face of the CK2β hairpin loop and, in particular, on a triad of CK2β amino acids, Tyr188, Gly189 and Phe190, which inserts deep into the CK2α groove (Figure 3A). To assess whether peptides derived from this CK2β region could antagonize the CK2 subunit interaction, we developed a binding assay in which MBP–CK2β was bound on streptavidin-coated microtitre plates and incubated with [35S]methionine-labelled CK2α. Compounds that disrupt the CK2α–CK2β complex thus show reduced radioactivity relative to the background. We then used the X-ray structure of CK2β in the holoenzyme complex as a template for the design of conformationally constrained peptides derived from the CK2β C-terminal domain and centred around the Tyr188 and Phe190 hotspots. An eight-residue peptide (Arg186–His193) which contained the cluster of hydrophobic residues (Leu187, Tyr188 and Ile192) and three additional glycine residues was cyclized via two cysteine residues to ‘staple’ its conformation and to mimic the binding face of CK2β with CK2α (Figure 3B). The biological activity of this peptide, referred to as Pc peptide, was first examined in a CK2α pull-down assay. Figure 7(A) shows that the same amount of CK2α activity could be found associated with biotinylated CK2β or biotinylated Pc, indicating that this cyclic peptide stably interacts with CK2α. This interaction could be also visualized by surface plasmon resonance spectroscopy (results not shown). To assess whether the Pc peptide could antagonize the CK2 subunit interaction, a CK2 subunit binding assay was performed in the presence of increasing concentrations of Pc peptide. Representative results are shown in Figure 7(B). The presence of the Pc peptide strongly antagonized the formation of the CK2 complex (IC50=3 μM). Importantly, an almost complete disassembly of the pre-formed complex was observed in the presence of the Pc peptide (IC50=6.5 μM). The effect of varying the length from eight to 14 amino acids in a series of head-to-tail cyclized peptide analogues was examined. Although all length variants significantly blocked the interaction, the 11-residue Pc peptide was the most effective (Figure 7C). Noteworthily, a linear form of Pc was much less active in this binding assay (IC50=30 μM), and a linear form of Pc with an inverted sequence was without effect, showing that both the sequence and the constrained conformation of the peptide are essential for its antagonist activity. To explore the functional impact of each amino acid side chain on the Pc peptide antagonist activity, we synthesized a series of cyclic peptides in which each amino acid in the sequence RLYGFKIH was subjected to a systematic single-site replacement strategy. These derivatives were first tested at two fixed peptide concentrations (1.5 and 15 μM) in the CK2 subunit interaction assay. As shown in Figure 8(A), the largest effects of single alanine mutants were observed for residues Phe7 or Gly6 and to a lesser extent for residues Tyr5 and Ile9. Alanine substitutions of neighbouring positions (Arg3, Leu4, Lys8 and His10) caused marginal reductions in binding, indicating that these residues serve a more passive role. Quantitative determination of the IC50 values shows further that the most disruptive alanine substitutions are confined in a small patch that extends from Tyr5 to Phe7, and the largest reduction in binding was for Phe7 (Figure 8B). In accord with the definition of the corresponding CK2β hotspots, these data highlight the importance of these residues for the biological activity of peptide Pc. A functional analysis of this peptide was performed testing its effect on the phosphorylation of the Olig-2 transcription factor, which is catalysed exclusively by the tetrameric form of CK2 (T. Buchou, personal communication). Figure 9 shows that the presence of increasing concentrations of Pc led to a strong decrease of the original Olig-2 phosphorylation, reflecting a Pc-induced progressive dissociation of the catalytically active CK2 holoenzyme complex.
Figure 7. Disruption of CK2 subunit interaction by CK2β-derived cyclic peptides.
(A) Binding of Pc peptide or CK2β to CK2α. Increasing concentrations of biotinylated Pc peptide (▲) or biotinylated MBP–CK2β (Δ) were immobilized on streptavidin beads and incubated with CK2α (0.3 μM) for 15 min at 4 °C. After washing, the amount of associated CK2α was evaluated by a CK2 kinase assay. (B) Inhibition of CK2α–CK2β complex formation by Pc peptide (▲). Immobilized MBP–CK2β was incubated with increasing concentrations of Pc peptide, followed by the addition of [35S]methionine-labelled CK2α (105 c.p.m.). After washing, bound CK2α was determined by radioactivity counting. Dissociation of pre-formed CK2α–CK2β complex by Pc peptide (■). [35S]Methionine-labelled CK2α was incubated with immobilized MBP–CK2β, followed by the addition of increasing concentrations of Pc peptide. After washing, bound CK2α was determined by radioactivity counting. (C) Inhibition of CK2α–CK2β complex formation by different Pc peptide derivatives. Increasing concentrations of the different peptides were tested as in (B). Pc (▲), linear peptide GAGGRLYGFKIHGGP (■), inverted linear peptide PGGHIKFGYLRGGAG (Δ), head-to-tail peptide analogues: PcL1 RLYGFKIHPGAGG (□), PcL2 RLYGFKIHGPGAGG (●) and PcL3 RLYGFKIHGGPGAGG (○). Results are the means±S.D. (n=3).
Figure 8. Antagonist effects of peptide Pc variants carrying single-site mutations.
(A) Various Pc peptide variants generated by alanine scanning were tested at 1.5 (□) or 15 (■) μM for their antagonist effect on the association of CK2 subunits as described in Figure 7(C). Results are the means±S.D. (n=3) of a representative experiment. WT, wild-type. (B) IC50 values determined for each peptide variant.
Figure 9. Antagonist effect of Pc peptide on CK2-mediated phosphorylation of a CK2β-dependent substrate.
Pre-formed CK2 complexes were incubated for 15 min at 4 °C with increasing concentrations of Pc peptide, followed by the addition of GST–Olig2 (5 μg) and 25 μM [γ-32P]ATP/MgCl2 for 5 min at room temperature. Phosphorylated proteins were separated by SDS/PAGE and analysed by autoradiography (upper panel) or Coomassie Blue staining (lower panel). The results are representative of two independent experiments. C, control without CK2. The molecular mass of the 45 kDa protein is indicated.
DISCUSSION
The irreversible nature of the CK2 holoenzyme formation has been challenged by both its crystal structure and live-cell imaging studies [13,16]. Free populations of each CK2 subunit have been identified in several organs [17], and differential subcellular localizations have also been reported for CK2α and CK2β. In many cases, CK2β operates as a docking platform affecting the accessibility of the catalytic site for binding substrates [15]. From a structural point of view, the CK2β subunit can provide CK2 specificity by either shielding a substantial surface of the CK2α subunit or, at the same time, providing a new surface near the catalytic active site. Since the free catalytic subunit and the holoenzyme exhibit divergent substrate preferences, it could be predicted that such a balance is crucial in the control of the many cellular processes that are governed by this multifaceted enzyme [19].
In the present study, we have shown that the CK2 holoenzyme complex is subject to disassembly and reassembly in vitro. The rather low rate of dissociation of the holoenzyme complex in vitro might not reflect the situation in intact cells, where interacting partners could induce conformational changes, shifting the balance in favour of the dissociation [19]. A relatively small portion of CK2β encompassing the 33-residue C-terminal region constitutes a minimum CK2α-binding domain which is necessary and sufficient for high-affinity interaction with CK2α. The CK2 holoenzyme crystal structure has revealed that several residues in this domain make significant contacts with CK2α. On the basis of these data, the finding that emerges from our mutagenesis study is that only a small set of primary hydrophobic contacts at the centre of the interface dominates affinity in vitro, allowing the identification of Tyr188 and Phe190 as the most important interaction points of CK2β. Mutations of these residues had strong functional consequences, since the CK2β-YF and CK2β-MYF mutants were (i) unable to sustain the formation of a stable CK2 complex, (ii) defective for its supramolecular organization, and (iii) inefficient for CK2-mediated phosphorylation of a CK2β-dependent substrate such as CDC25B.
BiFC analysis of YN-CK2α and YC-CK2β reproducibly demonstrated a specific interaction, whereas the individual tagged proteins did not yield fluorescence. A characteristic BiFC pattern was detected with CK2β-M, CK2β-Y and CK2β-F single mutants, indicating that these CK2β mutants mediate CK2α recognition. In contrast, BiFC fluorescence could be monitored only at a very low intensity with the CK2β-YF double mutant, demonstrating that these two amino acids are essential for a productive complex formation in the normal cellular environment. These observations, which corroborated our in vitro quantitative binding analysis, were supportive of the ability to design peptides that can affect the CK2 subunit interaction. This led us to a structure-based design of CK2β-derived conformationally constrained peptides that can efficiently antagonize this high-affinity interaction. Among a series of cyclic peptides, the disulfide-bridged Pc peptide was the most active peptide variant. This cyclic peptide was considerably more potent than its identical linear form, indicating that cyclization staples the peptide in a fixed conformation, a strategy that is known to strongly enhance peptide affinity for their target by limiting flexibility and multiple conformational changes [38]. Consistent with the identification of Tyr188 and Phe190 as CK2β hotspots, alanine-scanning analysis has confirmed that change of these two hydrophobic residues was highly detrimental for the inhibitory activity of the Pc peptide.
Potential biological relevance of CK2 subunit antagonists
To our knowledge, the 11-mer peptide Pc represents the first small antagonist that binds to the CK2 interface and inhibits its high-affinity subunit interaction. Functional analysis showed that Pc could act as an antagonist of CK2β-dependent phosphorylation through the inhibition/disruption of the CK2 holoenzyme complex. Structural modifications of this cyclic peptide, such as attachment to cell-permeant adducts, will be required to confer in vivo activity. Future efforts should also focus on the design of peptidomimetics with enhanced activity and selectivity.
Alternatively, the present study indicates further that it should be possible to obtain small molecules that bind to the CK2α hydrophobic groove and inhibit its interaction with CK2β in a manner similar to the Pc peptide. Molecules that selectively inhibit the CK2 subunit interaction would be useful in determining the importance of CK2β in the control of the many cellular processes that are governed by this multifunctional kinase. In particular, such inhibitors would provide more rapid and reversible tools than siRNA (small interfering RNA) or overexpression methods for correctly identifying relevant CK2β-dependent substrates. They will also serve as leads for the rational design of function-specific drugs that disrupt some actions of CK2, but leave others intact, allowing the deregulation of specific intracellular pathways. Furthermore, selective disruption of the CK2α–CK2β interaction could find important applications to test pharmacologically the importance of this interaction in tumour cell growth. With the help of structure-based rational design, the idea of finding small molecules with significantly higher affinities for CK2α using the Pc peptide as a lead molecule does not seem far-fetched.
Online data
Acknowledgments
We thank Thierry Buchou for providing recombinant GST–Olig2, Jean-Baptiste Raiser (Cristallographie des Protéines, Institut de Biologie Structurale, Grenoble, France) for assistance in molecular modelling, Nicole Thielens (Enzymologie Moléculaire, Institut de Biologie Structurale, Grenable, France) for assistance with BIAcore and Bernard Ducommun (CNRS UMR5088, Université Paul Sabatier, Toulouse, France) for providing recombinant CDC25B. This work was supported by the Institut National de la Santé et de la Recherche Médicale (Inserm), the Commissariat à l'Energie Atomique (CEA), the Ligue Nationale contre le Cancer (Equipe labellisée 2007), the Institut National du Cancer (grant number 57) and by the Centre National pour la Recherche Scientifique (CNRS) and the Université Joseph Fourier (UJF). We also acknowledge the Plateau de Synthèse de la Plate-Forme Chimie Nanobio and the Ministère de la Recherche for grant number 15432-2004 to V. D.
References
- 1.Andreeva I. E., Livanova N. B., Eronina T. B., Silonova G. V., Poglazov B. F. Phosphorylase kinase from chicken skeletal muscle: quaternary structure, regulatory properties and partial proteolysis. Eur. J. Biochem. 1986;158:99–106. doi: 10.1111/j.1432-1033.1986.tb09726.x. [DOI] [PubMed] [Google Scholar]
- 2.Gibson R. M., Ji-Buechler Y., Taylor S. S. Interaction of the regulatory and catalytic subunits of cAMP-dependent protein kinase: electrostatic sites on the type Iα regulatory subunit. J. Biol. Chem. 1997;272:16343–16350. doi: 10.1074/jbc.272.26.16343. [DOI] [PubMed] [Google Scholar]
- 3.Nigg E. A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 4.Hsu H. L., Yannone S. M., Chen D. J. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair. 2002;1:225–235. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 5.Dyck J. R., Gao G., Widmer J., Stapleton D., Fernandez C. S., Kemp B. E., Witters L. A. Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic β and γ subunits. J. Biol. Chem. 1996;271:17798–17803. doi: 10.1074/jbc.271.30.17798. [DOI] [PubMed] [Google Scholar]
- 6.Rothwarf D. M., Zandi E., Natoli G., Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 7.Gibson R. M., Taylor S. S. Dissecting the cooperative reassociation of the regulatory and catalytic subunits of cAMP-dependent protein kinase: role of Trp-196 in the catalytic subunit. J. Biol. Chem. 1997;272:31998–32005. doi: 10.1074/jbc.272.51.31998. [DOI] [PubMed] [Google Scholar]
- 8.Pinna L. A. Protein kinase CK2: a challenge to canons. J. Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 9.Allende J. E., Allende C. C. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 10.Litchfield D. W. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu E., Rubin C. S. Expression of wild-type and mutated forms of the catalytic (α) subunit of Caenorhabditis elegans casein kinase II in Escherichia coli. J. Biol. Chem. 1990;265:20609–20615. [PubMed] [Google Scholar]
- 12.Grankowski N., Boldyreff B., Issinger O. G. Isolation and characterization of recombinant human casein kinase II subunits α and β from bacteria. Eur. J. Biochem. 1991;198:25–30. doi: 10.1111/j.1432-1033.1991.tb15982.x. [DOI] [PubMed] [Google Scholar]
- 13.Niefind K., Guerra B., Ermakowa I., Issinger O. G. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 2001;20:5320–5331. doi: 10.1093/emboj/20.19.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chantalat L., Leroy D., Filhol O., Nueda A., Benitez M. J., Chambaz E. M., Cochet C., Dideberg O. Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J. 1999;18:2930–2940. doi: 10.1093/emboj/18.11.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibby A. C., Litchfield D. W. The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2β. Int. J. Biol. Sci. 2005;1:67–79. doi: 10.7150/ijbs.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filhol O., Nueda A., Martel V., Gerber-Scokaert D., Benitez M. J., Souchier C., Saoudi Y., Cochet C. Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 2003;23:975–987. doi: 10.1128/MCB.23.3.975-987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerra B., Siemer S., Boldyreff B., Issinger O. G. Protein kinase CK2: evidence for a protein kinase CK2β subunit fraction, devoid of the catalytic CK2α subunit, in mouse brain and testicles. FEBS Lett. 1999;462:353–357. doi: 10.1016/s0014-5793(99)01553-7. [DOI] [PubMed] [Google Scholar]
- 18.Li X., Guan B., Maghami S., Bieberich C. J. NKX3.1 is regulated by protein kinase CK2 in prostate tumor cells. Mol. Cell. Biol. 2006;26:3008–3017. doi: 10.1128/MCB.26.8.3008-3017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filhol O., Martiel J. L., Cochet C. Protein kinase CK2: a new view of an old molecular complex. EMBO Rep. 2004;5:351–355. doi: 10.1038/sj.embor.7400115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allende C. C., Allende J. E. Promiscuous subunit interactions: a possible mechanism for the regulation of protein kinase CK2. J. Cell. Biochem. Suppl. 1998;30–31:129–136. [PubMed] [Google Scholar]
- 21.Berg T. Modulation of protein–protein interactions with small organic molecules. Angew. Chem. Int. Ed. Engl. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 22.Chene P. Drugs targeting protein–protein interactions. ChemMedChem. 2006;1:400–411. doi: 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- 23.Renaudet O., Dumy P. Chemoselectively template-assembled glycoconjugates as mimics for multivalent presentation of carbohydrates. Org. Lett. 2003;5:243–246. doi: 10.1021/ol0270935. [DOI] [PubMed] [Google Scholar]
- 24.Heriche J. K., Lebrin F., Rabilloud T., Leroy D., Chambaz E. M., Goldberg Y. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science. 1997;276:952–955. doi: 10.1126/science.276.5314.952. [DOI] [PubMed] [Google Scholar]
- 25.Leroy D., Filhol O., Quintaine N., Sarrouilhe D., Loue-Mackenbach P., Chambaz E. M., Cochet C. Dissecting subdomains involved in multiple functions of the CK2β subunit. Mol. Cell. Biochem. 1999;191:43–50. [PubMed] [Google Scholar]
- 26.Filhol O., Cochet C., Wedegaertner P., Gill G. N., Chambaz E. M. Coexpression of both α and β subunits is required for assembly of regulated casein kinase II. Biochemistry. 1991;30:11133–11140. doi: 10.1021/bi00110a016. [DOI] [PubMed] [Google Scholar]
- 27.Hu C. D., Kerppola T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerppola T. K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell. Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martel V., Filhol O., Colas P., Cochet C. p53-dependent inhibition of mammalian cell survival by a genetically selected peptide aptamer that targets the regulatory subunit of protein kinase CK2. Oncogene. 2006;25:7343–7353. doi: 10.1038/sj.onc.1209722. [DOI] [PubMed] [Google Scholar]
- 30.Martel V., Filhol O., Nueda A., Cochet C. Dynamic localization/association of protein kinase CK2 subunits in living cells: a role in its cellular regulation? Ann. N.Y. Acad. Sci. 2002;973:272–277. doi: 10.1111/j.1749-6632.2002.tb04648.x. [DOI] [PubMed] [Google Scholar]
- 31.Canton D. A., Zhang C., Litchfield D. W. Assembly of protein kinase CK2: investigation of complex formation between catalytic and regulatory subunits using a zinc-finger-deficient mutant of CK2β. Biochem. J. 2001;358:87–94. doi: 10.1042/0264-6021:3580087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filhol O., Benitez M. J., Cochet C. A zinc ribbon motif is essential for the formation of functional tetrameric protein kinase CK2. In: Iuchi S., Kuldell N., editors. Zinc Finger Proteins: from Atomic Contact to Cellular Function. Austin: Landes Biosciences; 2005. pp. 124–129. [Google Scholar]
- 33.Marin O., Meggio F., Sarno S., Pinna L. A. Physical dissection of the structural elements responsible for regulatory properties and intersubunit interactions of protein kinase CK2 β-subunit. Biochemistry. 1997;36:7192–7198. doi: 10.1021/bi962885q. [DOI] [PubMed] [Google Scholar]
- 34.Theis-Febvre N., Filhol O., Froment C., Cazales M., Cochet C., Monsarrat B., Ducommun B., Baldin V. Protein kinase CK2 regulates CDC25B phosphatase activity. Oncogene. 2003;22:220–232. doi: 10.1038/sj.onc.1206107. [DOI] [PubMed] [Google Scholar]
- 35.Pagano M. A., Sarno S., Poletto G., Cozza G., Pinna L. A., Meggio F. Autophosphorylation at the regulatory β subunit reflects the supramolecular organization of protein kinase CK2. Mol. Cell. Biochem. 2005;274:23–29. doi: 10.1007/s11010-005-3116-y. [DOI] [PubMed] [Google Scholar]
- 36.Theis-Febvre N., Martel V., Laudet B., Souchier C., Grunwald D., Cochet C., Filhol O. Highlighting protein kinase CK2 movement in living cells. Mol. Cell. Biochem. 2005;274:15–22. doi: 10.1007/s11010-005-3115-z. [DOI] [PubMed] [Google Scholar]
- 37.Ermakova I., Boldyreff B., Issinger O. G., Niefind K. Crystal structure of a C-terminal deletion mutant of human protein kinase CK2 catalytic subunit. J. Mol. Biol. 2003;330:925–934. doi: 10.1016/s0022-2836(03)00638-7. [DOI] [PubMed] [Google Scholar]
- 38.Leduc A. M., Trent J. O., Wittliff J. L., Bramlett K. S., Briggs S. L., Chirgadze N. Y., Wang Y., Burris T. P., Spatola A. F. Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.