Abstract
The ubiquitin-like SUMO system functions by a cyclic process of modification and demodification, and recent data suggest that the nucleolus is a site of sumoylation–desumoylation cycles. For example, the tumour suppressor ARF stimulates sumoylation of nucleolar proteins. Here, we show that the nucleolar SUMO-specific protease SENP3 is associated with nucleophosmin (NPM1), a crucial factor in ribosome biogenesis. SENP3 catalyses desumoylation of NPM1–SUMO2 conjugates in vitro and counteracts ARF-induced modification of NPM1 by SUMO2 in vivo. Intriguingly, depletion of SENP3 by short interfering RNA interferes with nucleolar ribosomal RNA processing and inhibits the conversion of the 32S rRNA species to the 28S form, thus phenocopying the processing defect observed on depletion of NPM1. Moreover, mimicking constitutive modification of NPM1 by SUMO2 interferes with 28S rRNA maturation. These results define SENP3 as an essential factor for ribosome biogenesis and suggest that deconjugation of SUMO2 from NPM1 by SENP3 is critically involved in 28S rRNA maturation.
Keywords: ARF tumour suppressor, NPM1/B23, desumoylation, ribosome biogenesis
Introduction
The post-translational modification of proteins with the ubiquitin-like SUMO modifier has an important regulatory function in many cellular processes, including DNA repair, cell-cycle progression and transcription (Muller et al, 2004; Hay, 2005). Human cells express three forms of SUMO—SUMO1, SUMO2 and SUMO3. SUMO2 and SUMO3 are highly similar and share an identity of 97%, whereas SUMO1 and SUMO2/3 are about 50% identical. Conjugation of SUMO to a target protein proceeds by a multistep enzymatic pathway, which requires the E1 activating enzyme Aos1/Uba2, the E2 conjugating enzyme Ubc9 and, in some cases, involves E3 ligases, such as members of the PIAS family or RanBP2 (Pichler et al, 2002; Schmidt & Muller, 2003). Importantly, SUMO modification is a highly dynamic and reversible process and, in consequence, typically only a small fraction of a substrate is sumoylated at a given time (Hay, 2005). The demodification process is catalysed by SUMO-specific cysteine proteases of the SENP family (Mukhopadhyay & Dasso, 2007). In humans, six members of this family, known as SENP1–3 and SENP5–7, have been identified so far. Interestingly, in mammalian cells, SUMO paralogues (Ayaydin & Dasso, 2004; Fu et al, 2005) and the enzymatic components of the SUMO system are compartmentalized in specific subcellular regions, suggesting that sumoylation–desumoylation cycles are primarily active at these sites (Saitoh et al, 2006).
Recent data also implicate the nucleolus in dynamic cycles of sumoylation and desumoylation. For example, two members of the SENP family, SENP3 and SENP5, are specifically concentrated in the nucleolus (Nishida et al, 2000; Di Bacco et al, 2006; Gong & Yeh, 2006). Furthermore, enforced expression of the tumour suppressor ARF (human p14ARF or mouse p19Arf), which also localizes in the nucleolus, induces sumoylation of several nucleolar proteins (Xirodimas et al, 2002; Chen & Chen, 2003; Woods et al, 2004; Rizos et al, 2005; Tago et al, 2005). ARF inhibits cell-cycle progression through both p53-dependent and p53-independent mechanisms, and it has been proposed that the latter mechanisms are related to ARF-dependent regulation of ribosome biogenesis in the nucleolus (Sherr, 2006). In the nucleolus, ribosomal RNA is initially transcribed as a precursor that is subsequently cleaved in multiple steps into the mature 28S and 5.8S rRNA of the 60S ribosome and the 18S rRNA of the 40S ribosomal subunit (Dez & Tollervey, 2004). ARF inhibits the maturation of the 28S rRNA by interfering with the function of the nucleophosmin (NPM1) protein (Itahana et al, 2003; Sugimoto et al, 2003). NPM1 has been implicated in several cellular processes, including centrosome duplication, DNA-damage response and ribosome biogenesis (Grisendi et al, 2006). In ribosome biogenesis, NPM1 is specifically required for the processing of the 32S rRNA species to the mature 28S rRNA form (Savkur & Olson, 1998; Itahana et al, 2003). It has been proposed that ARF antagonizes this function by inducing the ubiquitin-dependent degradation of NPM1 (Itahana et al, 2003). Interestingly, NPM1 is also a target for ARF-mediated sumoylation (Tago et al, 2005); however, the functional significance of this process has remained unclear.
Results
NPM1 is a major binding partner of SENP3
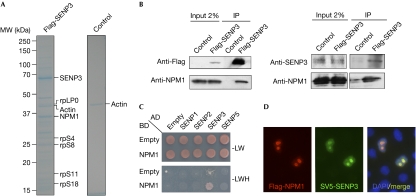
To identify interaction partners of SENP3, a Flag-tagged version of the enzymatically inactive SENP3 protein—in which the catalytic cysteine residue at position 532 was changed to serine—was expressed in human embryonic kidney (HEK)293T cells and proteins associated with SENP3C532S were affinity purified on anti-Flag beads, separated by SDS–polyacrylamide gel electrophoresis and analysed using mass spectrometry (Fig 1A). In addition to the bait protein SENP3 and ribosomal subunit proteins, a major band migrating at 37 kDa was identified as NPM1 (supplementary Fig 1 online). To verify this finding, anti-Flag immunoprecipitates from cells expressing wild-type Flag-SENP3 were probed with an NPM1 antibody. Consistent with the result from mass spectrometry, a 37 kDa band co-precipitating with Flag-SENP3 was detected by the NPM1 antibody (Fig 1B, left). The interaction of both proteins was confirmed at their endogenous levels of expression, when SENP3 was immunoprecipitated from HeLa cells with an SENP3 antibody followed by anti-NPM1 western blotting (Fig 1B, right). A physical interaction of NPM1 with SENP3, but not other members of the human SENP family (SENP1, SENP2 or SENP5), was also detected in the yeast two-hybrid system (Fig 1C). Subsequent experiments with fragments of NPM1, which corresponded to functional domains of NPM1, delineated a region spanning amino acids 1–186 as being sufficient for binding to SENP3 (supplementary Fig 2 online). In line with data on the physical interaction of NPM1 and SENP3, immunofluorescence experiments showed colocalization of both proteins in the nucleolus (Fig 1D).
Figure 1.
Nucleophosmin is a major binding partner of SENP3. (A) Flag-tagged SENP3C352S was expressed in human embryonic kidney (HEK)293T cells and affinity purification was performed using an anti-Flag agarose column. Bound proteins were eluted with Flag peptide, separated by SDS–polyacrylamide gel electrophoresis and analysed using mass spectrometry. (B) Left: Flag-SENP3 was expressed in HEK293T cells and immunoprecipitated (IP) with anti-Flag agarose beads. The bound material was separated by SDS–polyacrylamide gel electrophoresis and probed using western blotting with the indicated antibodies. Right: endogenous SENP3 was immunoprecipitated from HeLa cells with a rabbit polyclonal antibody and probed using western blotting with the indicated antibodies. (C) NPM1 was tested in a directed yeast two-hybrid assay for interaction with SUMO proteases as indicated. (D) HeLa cells were transfected with SV5-SENP3 and Flag-NPM1, and localization was determined by immunostaining with SV5 and Flag antibodies. Nuclei were visualized using 4,6-diamidino-2-phenylindole (DAPI) staining. MW, molecular weight; NPM1, nucleophosmin.
SENP3 catalyses desumoylation of NPM1
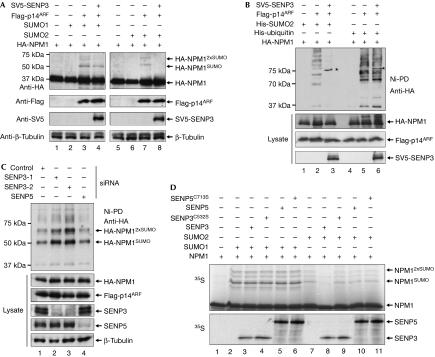
Recent work has reported that NPM1 is modified by SUMO (Tago et al, 2005). Accordingly, a SUMO modified form of endogenous NPM1 was recovered on Ni-NTA beads from HEK293T cells expressing His-SUMO1 or His-SUMO2 (supplementary Fig 3 online). ARF has been shown to stimulate the SUMO modification of NPM1 and, in particular, to induce its multisumoylation. Consistently, on expression of p14ARF with haemagglutinin (HA)-NPM1 and either SUMO1 or SUMO2, two high-molecular-weight anti-HA-reactive NPM1 forms migrating at 50 kDa and 65 kDa were detected in addition to the major 37 kDa NPM1 form (Fig 2A, lanes 3,7). Expression of SENP3 slightly reduced the level of NPM1–SUMO1 conjugates, but induced a complete loss of NPM1–SUMO2 forms (Fig 2A, lanes 4,8). As ARF can also induce ubiquitination of NPM1 (Itahana et al, 2003), we wanted to formally distinguish whether SENP3 acts on ARF-induced SUMO or ubiquitin conjugates. Therefore, HA-NPM1 and p14ARF were coexpressed in the absence or presence of SENP3 with His-tagged SUMO2 or His-ubiquitin, which allowed for affinity purification of the respective conjugates on Ni-NTA beads. On expression of His-SUMO2, the 50 kDa and 65 kDa NPM1 conjugates, as well as high-molecular-weight material, presumably corresponding to chains of SUMO2 on NPM1, were enriched on Ni-NTA beads (Fig 2B, lane 2). Mono- and diubiquitinated forms of NPM1 were found at 45 kDa and 55 kDa, respectively, and polymeric His-ubiquitin–NPM1 conjugates were detected as a typical high-molecular-weight ladder of bands (Fig 2B, lane 5). Importantly, on expression of SENP3, the SUMO2–NPM1 conjugates completely disappeared, whereas the ubiquitin–NPM1 conjugates remained unaltered, indicating that SENP3 counteracts ARF-mediated sumoylation—but not ubiquitination—of NPM1 (Fig 2B, lanes 3,6). Notably, expression of p14ARF did not affect the association of NPM1 with SENP3, making it unlikely that ARF-mediated sumoylation acts by displacing SENP3 from NPM1 (supplementary Fig 4A online). Interestingly, however, the expression of p14ARF recruits SUMO2 into the nucleolus (supplementary Fig 4B online). To study further the involvement of endogenous SENP3 in desumoylation of NPM1, SENP3 was depleted from cells by using two independent short interfering RNA (siRNA) duplexes and efficient downregulation of the protein was verified by immunoblotting with an SENP3 antibody (Fig 2C, lanes 2,3). Importantly, depletion of SENP3 by each of the siRNA duplexes led to a strong increase in His-SUMO2–NPM1 conjugates that were recovered on Ni-NTA beads, indicating that at physiological levels of expression SENP3 reverses modification of NPM1 by SUMO2 (Fig 2C, lanes 2,3). It is noteworthy that depletion of SENP5—the closest homologue of SENP3 in humans—does not affect the sumoylation of NPM1 (Fig 2C, lane 4).
Figure 2.
SENP3 catalyses the desumoylation of nucleophosmin. (A) HA-NPM1 was expressed alone or together with Flag-p14ARF and SUMO1 or SUMO2 in the presence or absence of SV5-tagged SENP3 in human embryonic kidney (HEK)293T cells. Expression of the respective proteins was verified by immunoblotting. Western blotting with anti-tubulin shows equal loading. (B) HA-NPM1 and His-tagged versions of SUMO2 or ubiquitin were expressed in the presence of Flag-p14ARF and SV5-SENP3 in HEK293T cells. His–SUMO2 or His–ubiquitin conjugates were recovered on Ni-NTA beads and subjected to western blotting using an HA antibody. Expression of the respective proteins was monitored by western blotting of cell lysates. Asterisks indicate a crossreactivity of the HA antibody with overexpressed SV5-tagged SENP3. (C) HEK293T cells were depleted from SENP3 or SENP5 by siRNA duplexes and transfected with the indicated plasmids. His–SUMO2 conjugates were recovered on Ni-NTA beads and subjected to western blotting using an HA antibody. Expression of the respective proteins was monitored by western blotting of cell lysates. (D) 35S-labelled NPM1, generated by in vitro transcription/translation, was first incubated with recombinant E1 enzyme, E2 enzyme and either SUMO1 (lanes 2–6) or SUMO2 (lanes 7–11) in the presence of ATP. In the control reaction, SUMO was omitted (lane 1). Following the modification reaction, SENP3 (lanes 3,8) and SENP5 (lanes 5,10) or the catalytically inactive mutants SENP3C532S (lanes 4,9) and SENP5C713S (lanes 6,11), generated by in vitro translation/transcription, were added. The amounts of SENP3, SENP5 and the respective catalytically inactive mutants are shown below. HA, haemagglutinin; NPM1, nucleophosmin; siRNA, short interfering RNA.
To analyse whether SENP3 directly exerts protease activity on NPM1–SUMO conjugates, we set up an in vitro sumoylation–desumoylation assay. To modify NPM1, the 35S-labelled protein, generated by in vitro transcription/translation, was incubated with recombinant components of the sumoylation machinery. In the control reaction, which lacked SUMO, a major band of NPM1 migrating at 37 kDa was detected (Fig 2D, lane 1), whereas several high-molecular-weight forms, which corresponded to either NPM1–SUMO1 or NPM1–SUMO2 conjugates, became apparent in the presence of the E1 enzyme, the E2 enzyme and either SUMO1 or SUMO2 (Fig 2D, lanes 2,7). To test whether SENP3 catalyses the cleavage of NPM1–SUMO conjugates, the reactions were subsequently incubated with wild-type or catalytically inactive forms of either SENP3 or SENP5. NPM1–SUMO1 conjugates were not influenced by the addition of either form of SENP (Fig 2D, lanes 3–6), whereas NPM1–SUMO2 conjugates were lost on incubation with the wild-type SENP3 protein (Fig 2D, lane 8), but remained stable in the presence of the catalytically inactive protein (Fig 2D, lane 9). In comparison, SENP5 did not significantly reduce the amount of NPM1–SUMO2 conjugates (Fig 2D, lane 10).
Together, these data show that SENP3 catalyses desumoylation of NPM1 and, in particular, cleaves SUMO2/3–NPM1 conjugates.
Downregulation of SENP3 inhibits rRNA processing
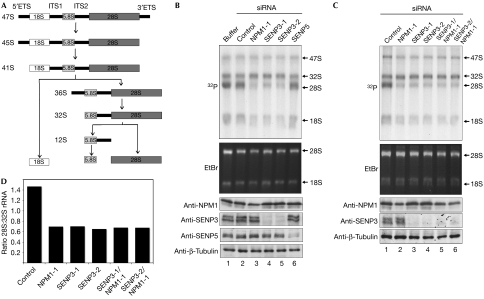
NPM1 is an essential factor in the nucleolar rRNA processing pathway. The main steps in rRNA processing are summarized in Fig 3A. NPM1 controls the maturation of the 28S rRNA species, possibly by inducing the cleavage of the 32S rRNA intermediate within the ITS2 region (Savkur & Olson, 1998; Itahana et al, 2003). To investigate a potential involvement of SENP3 in rRNA processing, we depleted the protein from HeLa cells by using two independent siRNA duplexes (Fig 2C), and the effect on rRNA processing was studied by in vivo labelling of nascent rRNA using 32P-orthophosphate (Fig 3B). For comparison, rRNA processing was also monitored in cells that were depleted of NPM1; depletion of the respective proteins was verified by immunoblotting (Fig 3B, bottom panel). To study pre-rRNA processing, RNA was prepared, separated by denaturing agarose gel electrophoresis and metabolically labelled rRNA species were visualized by autoradiography. The 47S and the 32S precursor rRNAs, as well as the 28S and 18S mature rRNA forms, were detected in all samples. In untransfected cells or cells transfected with a control siRNA, the mature 28S rRNA species was the most prominent form and was typically more abundant than its 32S rRNA precursor. By contrast, and consistent with the role of NPM1 as a 32S processing factor, knockdown of NPM1 caused a marked decrease in the amount of mature 28S species (Fig 3B, lane 3). Intriguingly, depletion of SENP3 phenocopies this defect and severely compromises the production of the mature 28S rRNA form (Fig 3B, lanes 4,5). Similar results were also obtained in U2OS cells on depletion of SENP3 with the same siRNA duplexes or two other independent siRNA sequences (supplementary Fig 5A,B online). Importantly, depletion of SENP5 in either HeLa or U2OS cells did not affect the amount of 28S rRNA. To establish whether SENP3 and NPM1 are part of a common pathway in rRNA processing, we performed a siRNA-based epistasis analysis by co-depletion of both proteins from HeLa or U2OS cells (Fig 3C; supplementary Fig 5C online). Quantification of the 32S and 28S rRNA species showed that co-depletion did not enhance the 32S processing defect observed on the depletion of SENP3 or NPM1 individually. This indicates that SENP3 and NPM1 act in a common and not a parallel pathway of 32S rRNA processing.
Figure 3.
Downregulation of SENP3 interferes with pre-ribosomal RNA processing. (A) Diagram summarizing the main steps of rRNA processing. (B,C) HeLa cells were transfected with siRNA duplexes targeting NPM1, SENP5 or SENP3, as indicated. Transfection of a scrambled oligonucleotide served as a control. At 72 h after transfection, cells were pulse labelled with 32P-orthophosphate for 1 h and chased for 2 h. An equal amount of RNA was loaded into each lane. Ethidium bromide (EtBr) staining of 28S and 18S rRNA is shown. Downregulation of the respective proteins was analysed by western blotting. (D) The signal intensities of the 28S and 32S rRNA forms were quantified by phosphoimager analysis and the 28S:32S ratio was calculated. ETS, external transcribed spacer; ITS, internal transcribed spacer; NPM1, nucleophosmin; siRNA, short interfering RNA.
Sumoylation of NPM1 prevents 28S rRNA maturation
To investigate whether an alteration in SUMO modification of NPM1 affects rRNA processing directly, we tested NPM1K263R, which has been reported to be devoid of SUMO modification, for its ability to mediate 28S rRNA processing (Liu et al, 2007). By using a knockdown/knock-in strategy (Holzel et al, 2007), we depleted cells from endogenous NPM1, reintroduced siRNA-resistant versions of Flag-NPM1WT or Flag-NPM1K263R and performed an in vivo labelling experiment as described above. Both Flag-NPM1WT and Flag-NPM1K263R fully complemented the depletion of endogenous NPM1 (supplementary Fig 6A online). However, subsequent experiments showed that NPM1K263R is still efficiently modified by SUMO in vitro and undergoes ARF-mediated sumoylation in vivo (supplementary Fig 6B online), indicating that mutation of Lys 263 does not abrogate sumoylation of NPM1.
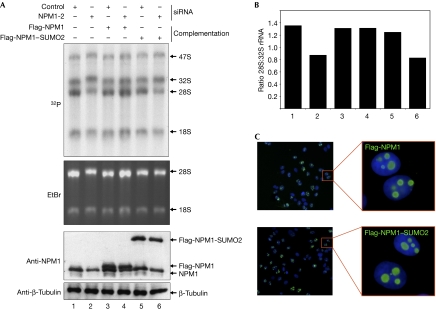
As a more direct approach to determine whether a failure in deconjugation of SUMO2 from NPM1 affects its function in rRNA processing, we mimicked constitutive sumoylation by linearly fusing SUMO2 to NPM1. Cells depleted of endogenous NPM1 were complemented with either Flag-NPM1WT or Flag-NPM1–SUMO2 (Fig 4A). Intriguingly, in contrast to Flag-NPM1WT, re-expression of Flag-NPM1–SUMO2 could not rescue the defect in 28S maturation caused by the depletion of endogenous NPM1 (Fig 4B), although Flag-NPM1–SUMO2 was expressed homogenously at a level comparable to Flag-NPM1 and showed normal localization to the nucleolus (Fig 4C). It is noteworthy that Flag-NPM1–SUMO2 retained the ability to interact with endogenous NPM1 (supplementary Fig 7 online, upper panel, compare lanes 7 and 8) and associated normally with known binding partners of NPM1, such as p14ARF, retinoblastoma protein, poly(ADP-ribose) polymerase-1 or histone H3 (supplementary Fig 7 online, lower panel, compare lanes 7 and 8). These observations indicate that tethering of SUMO2 to NPM1 does not generally affect the structure and functions of NPM1, and thus validate the fusion protein as a suitable system to mimic persistent modification by SUMO2. In summary, the data are consistent with the idea that a defect in desumoylation of NPM1 directly interferes with the correct processing of the 32S rRNA species.
Figure 4.
Constitutive SUMO2 conjugation of NPM1 interferes with pre-ribosomal RNA processing. (A) HeLa cells expressing Flag-NPM1 or Flag-NPM1–SUMO2 from a tetracycline-inducible promoter were transfected with an siRNA duplex targeting NPM1 or a control siRNA. At 72 h after transfection, in vivo labelling and processing of the samples was carried out as described in Fig 3. (B) The signal intensities were quantified as described in Fig 3D. (C) Homogenous expression and localization of Flag-NPM1 or Flag-NPM1–SUMO2 was monitored by indirect immunofluorescence. EtBr, ethidium bromide; NPM1, nucleophosmin; siRNA, short interfering RNA.
Discussion
Eukaryotic ribosome synthesis is a tightly controlled multistep process that requires the coordinated action of a series of cellular components. Here, we have shown that SENP3 is crucial for 32S rRNA processing. Furthermore, we have shown that SENP3 catalyses the deconjugation of SUMO2 from NPM1 and provided evidence that this process is a critical step for 28S maturation. The related nucleolar SUMO protease SENP5 is unable to catalyse demodification of NPM1 and does not affect 32S processing. However, the loss of SENP5 reduces the amount of the primary 47S rRNA transcript (supplementary Fig 5B online; M.H. and S.M., unpublished data), indicating that both SENP3 and SENP5 function in ribosome biogenesis, but control distinct steps in this process by acting on specific substrates. Importantly, the involvement of the SUMO system in ribosome biogenesis seems to be an evolutionarily conserved mechanism, as genetic studies in the yeast Saccharomyces cerevisiae provide evidence that both sumoylation and desumoylation are required for the efficient formation and nuclear export of pre-ribosomal particles (Panse et al, 2006).
SUMO modification of nucleolar proteins has been proposed to control their distribution between the nucleolus and the nucleoplasm. For example, a SUMO-dependent re-localization from the nucleolus to the nucleus has been postulated for topoisomerase I (Mo et al, 2002). Although it has been reported that a presumed non-sumoylatable mutant of NPM1 (NPM1K263R) shows an altered nuclear/nucleolar distribution (Liu et al, 2007), we were unable to reproduce these data (supplementary Fig 6D online). Furthermore, we could not confirm the reported loss of sumoylation with NPM1K263R. Although SUMO-dependent partitioning of nucleolar regulators is an attractive concept, we did not observe a change in NPM1 localization on depletion or overexpression of SENP3 (data not shown), indicating that enhanced modification of NPM1 by SUMO2/3 affects NPM1 function within the nucleolus. In line with this idea, expression of p14ARF triggers accumulation of SUMO paralogues in the nucleolus (Tago et al, 2005; supplementary Fig 4B online). Therefore, we propose a nucleolar modification–demodification cycle of NPM1, which involves p14ARF in the conjugation process and requires SENP3 for deconjugation. Furthermore, we suggest that the balanced conjugation/deconjugation of NPM1 and possibly other nucleolar proteins is part of the regulatory network controlling ribosome synthesis and cell proliferation.
Accumulating evidence indicates that ribosome synthesis is coupled with cell-cycle progression (Dez & Tollervey, 2004). Interestingly, SENP3 was recently identified in an siRNA screen as a potential component of the spindle assembly checkpoint, indicating that it is also involved in the control of mitotic processes (Stegmeier et al, 2007). Similarly, SENP5 is essential for the proper progression of mitosis and cytokinesis (Di Bacco et al, 2006). It will be exciting to study whether SENP3 and SENP5 might indeed represent cellular factors that coordinate ribosome biogenesis with the cell division cycle.
Methods
Immunoprecipitation, Ni-NTA pulldown and western blotting. For purification of SENP3-associated proteins, 4 × 108 HEK293T cells were transfected with Flag-SENP3C532S or empty control vector. At 48 h after transfection, cells were lysed in buffer A (50 mM HEPES pH 7.2, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40). Cell lysates were incubated with anti-Flag agarose beads (Sigma-Aldrich, St Louis, MO, USA) for 3 h, washed three times with buffer A and bound proteins were eluted with Flag peptide (100 μg/ml; Sigma-Aldrich). For the immunoprecipitation of transfected or endogenous SENP3, 1.5 × 106 HEK293T or HeLa cells were used. Western blotting and Ni-NTA pulldown experiments were carried out as described previously (Muller et al, 2000; Ledl et al, 2005).
In vitro sumoylation and desumoylation. 35S-labelled NPM1 was generated by in vitro transcription/translation using the TNT Quick Coupled T7 kit (Promega, Madison, WI, USA) and sumoylation was carried out as described previously (Schmidt & Muller, 2002). For demodification, SENP3 and SENP5, generated by in vitro transcription/translation, were added to the in vitro modification reactions and samples were incubated for an additional 90 min at 30°C. Proteins were separated on SDS gels and detected by autoradiography.
Knockdown/knock-in experiments, RNA analysis and 32P in vivo labelling. Knockdown/knock-in experiments and metabolic labelling of rRNA were carried out as described previously (Holzel et al, 2005, 2007) with some minor modifications (see supplementary information online). Sequences of siRNA duplexes are listed in the supplementary information online.
Immunofluorescence. For immunofluorescence, cells were fixed in 3.4% paraformaldehyde and processed using standard protocols. Images were acquired with an AX10 microscope (Zeiss, Jena, Germany). Antibodies are listed in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary information
Acknowledgments
We thank E. Yeh and M. Oren for reagents, J. Rech for help with experiments, and A. Buchberger, S. Jentsch, P. Stehmeier and E. Finkbeiner for discussions and comments on the manuscript. S.M. was supported by the ‘Deutsche Forschungsgemeinschaft' and the ‘German-Israeli Foundation for Scientific Research and Development'. D.E. is supported by the ‘Deutsche Forschungsgemeinschaft', SFB 684. S.M. is indebted to S. Jentsch for generous and continuous support.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ayaydin F, Dasso M (2004) Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15: 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen J (2003) MDM2–ARF complex regulates p53 sumoylation. Oncogene 22: 5348–5357 [DOI] [PubMed] [Google Scholar]
- Dez C, Tollervey D (2004) Ribosome synthesis meets the cell cycle. Curr Opin Microbiol 7: 631–637 [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G (2006) The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol 26: 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C et al. (2005) Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3. Oncogene 24: 5401–5413 [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET (2006) Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem 281: 15869–15877 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP (2006) Nucleophosmin and cancer. Nat Rev Cancer 6: 493–505 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Holzel M et al. (2005) Mammalian WDR12 is a novel member of the Pes1–Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Grimm T, Rohrmoser M, Malamoussi A, Harasim T, Gruber-Eber A, Kremmer E, Eick D (2007) The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res 35: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y (2003) Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell 12: 1151–1164 [DOI] [PubMed] [Google Scholar]
- Ledl A, Schmidt D, Muller S (2005) Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24: 3810–3818 [DOI] [PubMed] [Google Scholar]
- Liu X, Liu Z, Jang SW, Ma Z, Shinmura K, Kang S, Dong S, Chen J, Fukasawa K, Ye K (2007) Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. Proc Natl Acad Sci USA 104: 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo YY, Yu Y, Shen Z, Beck WT (2002) Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem 277: 2958–2964 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32: 286–295 [DOI] [PubMed] [Google Scholar]
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 275: 13321–13329 [DOI] [PubMed] [Google Scholar]
- Muller S, Ledl A, Schmidt D (2004) SUMO: a regulator of gene expression and genome integrity. Oncogene 23: 1998–2008 [DOI] [PubMed] [Google Scholar]
- Nishida T, Tanaka H, Yasuda H (2000) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem 267: 6423–6427 [DOI] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E (2006) Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Rizos H, Woodruff S, Kefford RF (2005) p14ARF interacts with the SUMO-conjugating enzyme Ubc9 and promotes the sumoylation of its binding partners. Cell Cycle 4: 597–603 [DOI] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M (2006) In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res 312: 1418–1430 [DOI] [PubMed] [Google Scholar]
- Savkur RS, Olson MO (1998) Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res 26: 4508–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci 60: 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ (2006) Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Stegmeier F et al. (2007) Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446: 876–881 [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Kuo ML, Roussel MF, Sherr CJ (2003) Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell 11: 415–424 [DOI] [PubMed] [Google Scholar]
- Tago K, Chiocca S, Sherr CJ (2005) Sumoylation induced by the Arf tumor suppressor: a p53-independent function. Proc Natl Acad Sci USA 102: 7689–7694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK (2004) p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem 279: 50157–50166 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Chisholm J, Desterro JM, Lane DP, Hay RT (2002) P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett 528: 207–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information