Mitochondria are known to be semi-autonomous organelles that are responsible for energy production and cellular respiration. Mitochondria break down glucose to release energy in the form of ATP through oxidative phosphorylation. However, as a by-product, a steady stream of reactive oxygen species (ROS) is released from these cellular powerhouses. ROS can potentially cause damage to cellular components, and are therefore closely linked to diseases such as Alzheimer disease and cancer.
Recently, mitochondria were also found to have a crucial role in innate immunity. Innate immune responses against invading viruses rely on the detection of viral pathogen-associated molecular patterns (PAMPs) and the subsequent production of antiviral cytokines such as type I interferons (IFNs). One prototypical viral PAMP is double-stranded (ds)RNA, which can be detected by Toll-like receptor 3 (TLR3) in endosomes (Akira & Takeda, 2004). TLR3 was the first reported dsRNA receptor able to signal to interferon regulatory factor (IRF) and NF-κB, which are essential transcription factors that regulate type I IFN production (Fig 1). Since then, two homologous RNA helicases—retinoic-acid-inducible gene I (RIG-I) and melanoma-differentiation-associated protein 5 (MDA5)—have been identified as cytoplasmic sensors of viral-derived RNA (Yoneyama et al, 2004).
Figure 1.
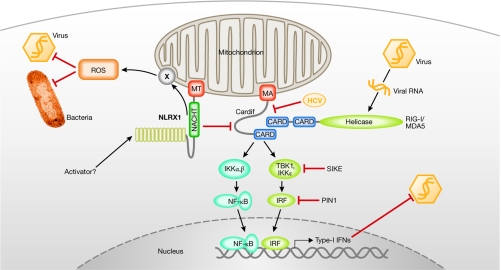
Mitochondria as anti-pathogen platforms. Through its mitochondrial anchor (MA) sequence, Cardif is targeted to the outer membrane of mitochondria, where it orchestrates RLH-dependent antiviral responses through the recruitment of both viral RNA sensors (RIG-I or MDA5) and effector proteins (IKK). Several cellular (SIKE, PIN1) but also viral (for example, the protease of HCV) proteins tightly regulate the antiviral response. NLRX1, another mitochondria-targeted protein (MT), might act as a negative regulator of Cardif signalling, diminishing virally induced Cardif–RLH interactions. However, NLRX1 also promotes ROS production at the mitochondria, which consequently helps to fight bacteria and viruses. How NLRX1 performs these functions, and how it becomes activated, remain unanswered questions. CARD, caspase recruitment domain; HCV, hepatitis C virus; IFN, interferon; IKK, IκB kinase; IRF, interferon regulatory factor; MDA5, melanoma-differentiation-associated protein 5; NLRX1, NOD-like receptor X1; PIN1, peptidylpropyl isomerase 1; RIG-I, retinoic-acid-inducible gene I; ROS, reactive oxygen species; RLH, RIG-like helicase; SIKE, supressor of IKKɛ; TBK, TANK-binding kinase.
The RIG-like helicases (RLHs), RIG-I and MDA5, both contain two amino-terminal caspase recruitment domains (CARDs) and an RNA helicase domain (Fig 1; Meylan et al, 2005). During viral infection, RIG-I and MDA5 trigger the activation of NF-κB, IRF3 and IRF7, which cooperate in the induction of antiviral type I IFN (Fig 1). RIG-I—but not MDA5—mounts an antiviral response against 5'pppRNA molecules present during the replication of certain single-stranded (ss)RNA viruses, such as Sendai virus and influenza virus. By contrast, MDA5 detects the presence of synthetic poly I:C—a synthetic dsRNA analogue—and the positive-strand ssRNA picornavirus encephalomyocarditis virus (Takeuchi & Akira, 2007). Mechanistically, RIG-I and MDA5 use their CARDs to connect with Cardif (also known as MAVS, IPS1 or VISA) and initiate downstream events (Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005). Cardif contains an N-terminal CARD that interacts with both RIG-I and MDA5, whereas its carboxyl terminus contains a membrane-anchor region—similar to the anti-apoptotic BCL2—which targets Cardif to the outer mitochondrial membrane (Fig 1). When Cardif is released from the mitochondria into the cytoplasm, it no longer mediates downstream IRF and NF-κB activation (Meylan & Tschopp, 2006). Once activated by viral RNA, Cardif recruits appropriate signalling intermediates, such as IκB kinases (IKKs, namely IKKα, β, ɛ and TANK-binding kinase 1 (TBK1)), to activate NF-κB and IRF transcription factors (Fig 1).
The importance of antiviral systems are often highlighted by the number of viruses that target the signalling pathway; for example, Cardif is cleaved and inactivated by the hepatitis C virus (HCV; Meylan et al, 2005). Owing to the importance of this antiviral mitochondrial pathway, it therefore seems reasonable that it could be tightly controlled by other cellular proteins. This would ensure the inhibition of unwarranted—and possibly deleterious—antiviral responses. Recently, further insights have been gained into the mechanism by which cells control these potentially harmful signalling events (Fig 1). First, suppressor of IKKɛ (SIKE), which interacts with the kinases TBK1 and IKKɛ, inhibits both RLH and TLR-dependent IRF activation by preventing interactions between various proteins and these two kinases (Huang et al, 2005). Second, PIN1, a peptidyl-prolyl isomerase, interacts with phosphorylated IRF3 and promotes its polyubiquitination and proteasome-dependent degradation (Saitoh et al, 2006). In two recent reports—one by Jenny Ting' group in Nature (Moore et al, 2008) and the other by Stephen Girardin and colleagues in this issue of EMBO reports (Tattoli et al, 2008)—an additional mitochondrial protein, NLRX1, was found to modulate innate immune responses.
NLRX1 is a member of the cytoplasmic NOD-like receptors (NLRs; Fritz et al, 2006; Martinon et al, 2007). The most understood NLR members are NALP3 (also known as cryopyrin) and NOD2. NALP3 detects exogenous and host ligands, such as bacterial peptidoglycan, ATP or uric acid (Petrilli et al, 2007), and recruits the inflammatory caspase 1 through the adaptor protein ASC. This forms an interleukin-1β-processing molecular complex termed the ‘inflammasome” (Agostini et al, 2004; Petrilli et al, 2007). NOD2 is a NF-κB-activating NLR, which detects the bacterial PAMP muramyl-dipeptide (Franchi et al, 2008). NOD2 has attracted much attention as mutations in NOD2 are associated with Crohn disease (Le Bourhis et al, 2007). By contrast, almost nothing is known about the biological function of NLRX1. This NLR protein is highly conserved between species and is expressed ubiquitously. NLRX1 is a close sequence homologue of NOD3—one of six members of the NOD subfamily of NLRs—and is also called NOD5 or NOD9; unfortunately, the nomenclature of NLRs is confusing, particularly since the recent introduction of additional names by the Human Genome Organization. NOD subfamily members commonly have a CARD at their N-terminus, followed by a NACHT domain and several leucine-rich repeat domains (Martinon et al, 2007). NLRX1 (NOD5) is distinct as it lacks the N-terminal CARD, which is instead replaced by a putative mitochondrial targeting sequence located within the first 20 amino acids. Indeed, NLRX1 was found to localize to mitochondria (Tattoli et al, 2008)—more precisely to their outer membrane (Moore et al, 2008)—provided that this crucial N-terminal sequence is present. On the basis of this subcellular localization, the authors of both studies searched for a function of NLRX1 that is connected to the known roles of mitochondria.
Interestingly, although the two functions of NLRX1 identified by the different groups are indeed both related to mitochondria, they are not identical and, at first glance, seem to be mutually exclusive. Ting' group proposes that the NACHT domain of NLRX1 interacts physically with the CARD of Cardif (Moore et al, 2008). As this domain is required for the interaction of Cardif with RLHs, competition between NLRX1 and RLH is predicted. Indeed, overexpression of NLRX1 disrupts the RLH–Cardif interaction induced by Sendai virus and consequently interferes with IFN-β production. Similarly, RLH-induced IRF3 and NF-κB activation (triggered by transfection of poly I:C) is inhibited, resulting in impaired IFN-β production (Fig 1). Importantly, replication of Sindbis virus is highly attenuated in the presence of NLRX1 small interfering RNA. By contrast, the TLR3 signalling pathway is not affected as both IFN-β and NF-κB reporter assays remain normal when poly I:C is added to the cellular exterior, showing the specificity of NLRX1 for the cytoplasmic antiviral effector system.
In contrast to Ting' results, no inhibitory function of NLRX1 on the IFN-β and NF-κB signalling pathways is reported by Girardin' group (Tattoli et al, 2008). Instead, amplified activity of NF-κB, and c-Jun N-terminal kinase (JNK), is observed on overexpression of NLRX1 following activation of these pathways by tumour necrosis factor (TNF) or Shigella. Mechanistically, this is explained by the observation that overexpression of NLRX1 leads to the generation of ROS. Furthermore, NLRX1 further increases ROS production induced by poly I:C, TNF and Shigella. It is well documented that ROS potentiates pro-inflammatory signalling pathways (Gloire et al, 2006) and as several pro-inflammatory stimuli, such as bacterial and viral infections, are also known to trigger ROS (Gloire et al, 2006), the observed synergism of NLRX1 and poly I:C, TNF or Shigella seems possible. NLRX1 lacking the mitochondrial anchor sequence is inactive, which suggests that the increased ROS production is of mitochondrial origin and not attributable to, for example, increased activity of one of the NADPH oxidases. However, the precise amplification mechanism remains elusive.
The data published by the two groups have important biological consequences. NLRX1 is the first NLR member that is constitutively localized to the mitochondria—NALP1 binds only transiently to BCL2 and BCL-XL. In addition to the mitochondrial targeting signal, NLRX1 uses its NACHT domain to interact with Cardif and therefore mitochondrial association seems to be enforced. Nevertheless, this direct NLRX1–Cardif interaction must be weak as deletion of the mitochondrial targeting sequence abolishes the mitochondrial localization of NLRX1 and also its function. Furthermore, proteins with mitochondrial targeting sequences are usually imported into the mitochondria by crossing at least the outer membrane, making an interaction with Cardif difficult to explain.
Both reports agree on the mitochondrial localization of NLRX1, but the proposed functions of NLRX1 are different. Ting' group identifies NLRX1 as an inhibitor of antiviral mitochondrial signalling. The evidence for such a role seems convincing as markedly increased IFN-β responses are clearly observed in cells with downregulated NLRX1 expression. However, a pronounced effect on the replication activity of a non-modified virus such as vesicular stomatitis virus or Sendai virus has not been shown. How can we reconcile Ting' data with the idea of Girardin', which identifies NLRX1 as an amplifier of ROS generation? ROS are required for IRF3 (Chiang et al, 2006) and STAT activation (Liu et al, 2004), and are therefore clearly implicated in the antiviral innate immune response. It is counterintuitive that NLRX1 activates an antiviral response while at the same time inhibits this response. One explanation for this dilemma is to postulate that NLRX1 first suppresses spontaneous activation of the RLH–Cardif pathway but, on activation of the RLH by viruses, NLRX1 dissociates from Cardif, converting it into an antiviral molecule by increasing ROS production at the mitochondria. This latter function might be more in line with the general function of NLRs—that is, the detection and elimination of danger. Alternatively, putative antiviral functions of NLRX1 might dominate over its proviral role in certain cell types, or in the context of infections with particular viruses, and vice versa. We will have to wait for the generation of mice that are deficient in NLRX1 to determine fully the role of this new mitochondrial protein. Nevertheless, the two studies do confirm the emerging role of mitochondria not only as powerhouses of the cell but also as crucial regulators of immunity.


Acknowledgments
We thank M. Eckert for discussions and critical reading of the manuscript. The authors are supported by grants from the Swiss National Science Foundation and the Human Frontier Science Program.
References
- Agostini L, Martinon F, Burns K, McDermott EM, Hawkins PN, Tschopp J (2004) NALP3 forms an IL-1β processing inflammasome with increased activity in Muckle–Wells auto-inflammatory disorder. Immunity 20: 319–325 [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K (2004) Toll-like receptor signalling. Nat Rev Immunol 4: 499–511 [DOI] [PubMed] [Google Scholar]
- Chiang E, Dang O, Anderson K, Matsuzawa A, Ichijo H, David M (2006) Cutting edge: apoptosis-regulating signal kinase 1 is required for reactive oxygen species-mediated activation of IFN regulatory factor 3 by lipopolysaccharide. J Immunol 176: 5720–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, Kim YG, Nunez G (2008) Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE (2006) Nod-like proteins in immunity, inflammation and disease. Nat Immunol 7: 1250–1257 [DOI] [PubMed] [Google Scholar]
- Gloire G, Legrand-Poels S, Piette J (2006) NF-κB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol 72: 1493–1505 [DOI] [PubMed] [Google Scholar]
- Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB (2005) SIKE is an IKKɛ/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J 24: 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Le Bourhis L, Benko S, Girardin SE (2007) Nod1 and Nod2 in innate immunity and human inflammatory disorders. Biochem Soc Trans 35: 1479–1484 [DOI] [PubMed] [Google Scholar]
- Liu T, Castro S, Brasier AR, Jamaluddin M, Garofalo RP, Casola A (2004) Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J Biol Chem 279: 2461–2469 [DOI] [PubMed] [Google Scholar]
- Martinon F, Gaide O, Petrilli V, Mayor A, Tschopp J (2007) NALP inflammasomes: a central role in innate immunity. Semin Immunopathol 29: 213–229 [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J (2006) Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell 22: 561–569 [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Moore CB et al. (2008) NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451: 573–577 [DOI] [PubMed] [Google Scholar]
- Petrilli V, Dostert C, Muruve DA, Tschopp J (2007) The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol 19: 615–622 [DOI] [PubMed] [Google Scholar]
- Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S (2006) Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol 7: 598–605 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signalling protein that activates NF-κB and IRF3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S (2007) Recognition of viruses by innate immunity. Immunol Rev 220: 214–224 [DOI] [PubMed] [Google Scholar]
- Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, Arnoult D, Girardin S (2008) NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep 9: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-β signalling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]