Abstract
The mammalian heterochromatin protein 1 (HP1) family of proteins was recently shown to be involved in transient repression of inducible promoters. One of these promoters is the HIV1 long terminal repeat, which, during viral latency, recruits a non-processive RNA polymerase II (RNAPII) that synthesizes a short regulatory transcript. Here, we have used this promoter to examine the interplay of HP1α, HP1β and HP1γ with RNAPII. We find that, in the absence of stimulation, HP1β is present on the promoter together with the non-processive RNAPII and functions as a negative regulator. On activation, HP1β bound to methylated H3K9 is rapidly released concurrent with histone H3 phospho-acetylation, and is replaced by HP1γ. This isoform localizes to the promoter but also inside the coding region, together with the processive RNAPII. Our data show that HP1 recruitment–release is a sequential mechanism that is precisely regulated and highly dependent on transcription.
Keywords: histone code, RNA polymerase II, HIV1, mitogen-activated protein kinases
Introduction
The heterochromatin protein 1 (HP1) proteins are transcriptional regulators conserved from Schizosaccharomyces pombe to mammals. These proteins bind histone H3 methylated on lysine 9 (H3K9), but they also interact with DNA, a wide range of regulatory and structural proteins and a yet uncharacterized RNA component. HP1 proteins are mainly considered to be silencers involved in the spreading of heterochromatin (reviewed by Lomberk et al, 2006). In mammalian cells, the HP1 family is composed of HP1α, HP1β and HP1γ. These three isoforms are concentrated in foci of dense pericentromeric heterochromatin but are also present in the rest of the nucleus. Consistent with their general distribution, the mammalian HP1 proteins are detected not only in dense heterochromatic regions but also on active euchromatic genes (reviewed by Hediger & Gasser, 2006). For example, HP1β is present on the repressed cyclin E promoter (Nielsen et al, 2001). Similarly, HP1γ participates in the repression of the mouse mammary tumour virus promoter in the absence of hormonal stimulation (Vicent et al, 2006). Interestingly, HP1γ is also recruited to the coding region of a subset of actively transcribed erythroid-specific genes in a transcription-dependent manner (Vakoc et al, 2005).
Recently, HP1 proteins were also shown to be present on the long terminal repeat (LTR) of HIV1 during the phases of viral latency (Chéné et al, 2007; Marban et al, 2007). This promoter is stimulated by the nuclear factor-κB and the mitogen-activated protein kinase signal-transduction pathways that are both known from other promoters to induce phosphorylation on serine 10 of histone H3 (H3S10; Nabel & Baltimore, 1987; Yang et al, 1999; Saccani et al, 2002; Anest et al, 2003; Clayton & Mahadevan, 2003; Yamamoto et al, 2003). This is noteworthy because phosphorylation of H3S10 is part of the mechanism that causes delocalization of HP1 proteins from the condensing chromosomes during mitosis (Mateescu et al, 2004; Fischle et al, 2005; Hirota et al, 2005). Another interesting aspect of the HIV1 LTR is its constant activity. Even during latency, the promoter recruits non-phosphorylated RNA polymerase II (RNAPII) that produces a short RNA known as TAR (Kim et al, 2006, and references therein). It is this RNA that allows the recruitment of the strong viral transactivator Tat on reactivation of the virus (for a recent review, see Zhou & Yik, 2006).
The coincidence of transcriptional activity and HP1 recruitment on the HIV1 LTR provides an opportunity to study how RNAPII and HP1 proteins affect each other. We therefore carefully examined the movements and the distribution of the HP1 proteins and the RNAPII on integrated single-copy LTR-derived reporter constructs during transcriptional activation. We observed a well-localized sequential recruitment of HP1β and HP1γ that is orchestrated by histone H3 phosphorylation and RNAPII distribution.
Results
Decreased levels of HP1β reactivate the HIV1 LTR
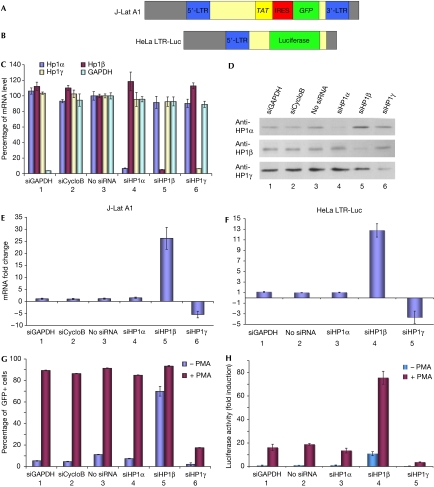
To examine HP1-mediated repression of the HIV1 LTR, we used two cell lines with different reporter constructs integrated at different loci, but which both showed low levels of basal LTR transcription while being efficiently stimulated in the presence of the protein kinase C activator phorbol-myristate acetate (PMA). The T-cell-derived J-Lat A1 cells harbour one copy of a construct containing a tat and a green fluorescent protein (GFP) gene framed by the 5′ and 3′ HIV1 LTRs (Fig 1A; Jordan et al, 2003), whereas the HeLa LTR-Luc cells have integrated a single copy of an HIV1 LTR upstream of a luciferase reporter gene (Fig 1B; Treand et al, 2006). To determine whether the three HP1 isoforms have similar roles in the transcriptional control of the LTRs, we performed short interfering RNA (siRNA) knockdown of each of the three proteins in J-Lat A1 cells (Fig 1C–E) and in HeLa LTR-Luc cells (Fig 1F). In both cell lines, inactivation of HP1β resulted in increased production of long HIV1 transcripts, as measured by reverse transcription quantitative PCR (RT-qPCR; Fig 1E, bar 5, and Fig 1F, bar 4). This effect was sensitive to the efficiency of the siRNAs (supplementary Fig S1 online). Unexpectedly, knockdown of HP1γ reduced rather than increased transcription, indicating that this HP1 isoform is involved in reactivation rather than repression of the promoters (Fig 1E, bar 6, and Fig 1F, bar 5). Finally, we noted that knockdown of HP1α had no effect on the transcriptional activity of integrated HIV1 LTRs (Fig 1E, bar 4, and Fig 1F, bar 3). Owing to the presence of tat in the HIV1 construct of the J-Lat A1 cells, a transient stimulation with PMA initiated continuous transcriptional activity of the LTR and, consequently, production of GFP. Knockdown of HP1β launched permanent transcriptional activity with an efficiency comparable with PMA stimulation (Fig 1G, bar 5). By contrast, siRNAs directed against HP1γ reduced the number of GFP-expressing cells and prevented efficient stimulation with PMA (Fig 1G, bar 6). In HeLa LTR-Luc cells that express luciferase but not Tat and GFP, luciferase activity was increased by HP1β knockdown and was further stimulated by PMA (Fig 1H, bar 4), whereas it was reduced by HP1γ knockdown (Fig 1H, bar 5). These observations are consistent with HP1β functioning as a transcriptional repressor of the HIV1 LTR, whereas HP1γ has a positive effect on transcription when the RNAPII becomes processive.
Figure 1.
Heterochromatin protein 1 inactivation modifies HIV1 transcription. (A,B) Schematic representation of HIV1 constructions. (C,D) J-Lat A1 cells were transfected with the indicated siRNAs. After 2 days, messenger RNA and protein levels were determined by RT-qPCR (C) or western blot (D), respectively. (E,F) At 48 h after siRNA transfection in the indicated cell lines, transcript levels were quantified by RT-qPCR. (G,H) Cells were transfected with the indicated siRNAs and treated or not treated with 10 nM PMA. For J-Lat A1 cells, percentage of GFP-positive cells was scored by fluorescence-activated cell sorting (G). For HeLa LTR-Luc cells, luciferase activity was measured (H). CycloB, cyclophilin B; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HP1, heterochromatin protein 1; LTR, long terminal repeat; PMA, phorbol-myristate acetate; RT-qPCR, real-time quantitative PCR; siRNA, short interfering RNA.
A switch from HP1β to HP1γ during activation
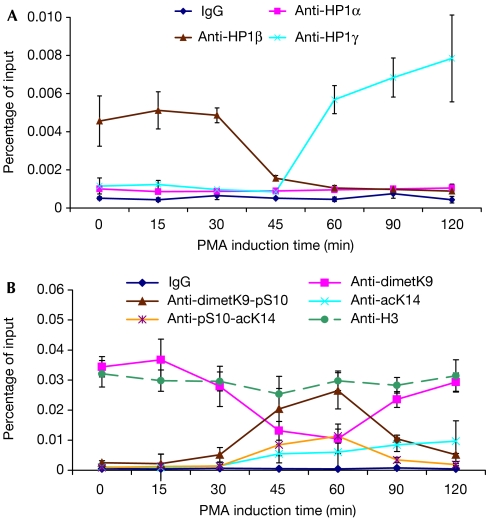
We next investigated the presence of the HP1 isoforms on the HIV1 LTR using chromatin immunoprecipitation (ChIP) assays and J-Lat A1 cells. HP1β was detected on the promoter before addition of PMA but disappeared after 45 min of stimulation (Fig 2A, brown curve). This shows that HP1β is present on the HIV1 LTR during phases of repression, but is then released by the machinery of transcriptional activation. Interestingly, HP1γ absent initially from the promoter before stimulation was recruited shortly after the removal of HP1β (Fig 2A, blue curve). HP1α was not detected on the LTR before or after the stimulation (Fig 2A, pink curve). Control ChIP experiments, however, showed the presence of HP1α on the α-satellites (supplementary Fig S3 online). To gain an insight into the mechanisms leading to the release of HP1β during activation, we followed several modifications of histone H3 (Fig 2B). Earlier studies have established that HP1 proteins bind to methylated H3K9 and that this binding can be disrupted by either phosphorylation of H3S10 or phosphorylation of H3S10 associated with acetylation of H3K14 (Mateescu et al, 2004; Fischle et al, 2005; Hirota et al, 2005). Our experiments showed that, after stimulation, the release of HP1β could be correlated with peaks of phospho-H3S10 dimethyl-H3K9 and phospho-H3S10 acetyl-H3K14 double modifications. For HP1γ, recruitment started during these peaks and continued thereafter. Dimethyl-H3K9 was detected before and after but not during the peaks because phosphorylation of H3S10 masks the epitope (Mateescu et al, 2004). Altogether, these data show that on the LTR, the shift from HP1β to HP1γ is correlated with a short burst of H3S10 phosphorylation on the methylated tails of histone H3.
Figure 2.
Activation-dependent recruitment and release of heterochromatin 1 proteins on the HIV1 long terminal repeat. After 2 h of PMA treatment of J-Lat A1 cells, chromatin immunoprecipitation experiments were performed with antibodies specific for HP1α, HP1β and HP1γ (A), or histone H3 tail modifications (B). Enrichment in HP1 LTR chromatin was measured by quantitative PCR using primers spanning the TS region (see Fig 3C). Values are averages from three independent experiments. ac, acetylated; dimet, dimethylated; H, histone; HP1, heterochromatin protein 1; K, lysine; LTR, long terminal repeat; PMA, phorbol-myristate acetate; pS, phospho-Serine; TS, transcription start.
HP1β co-distributes with RNA polymerase II
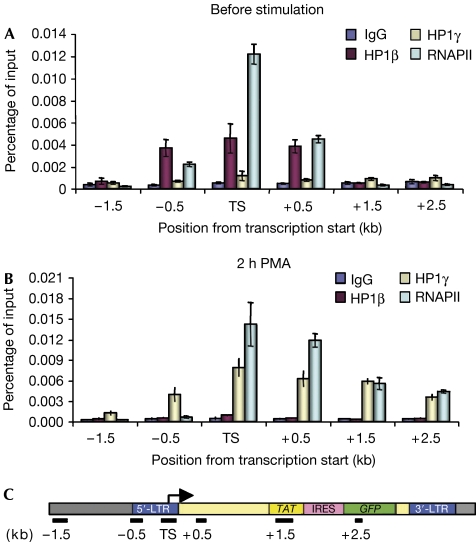
It has frequently been suggested that repression by HP1 proteins can spread from the site of recruitment to neighbouring chromatin. We therefore used ChIP assays and J-Lat A1 cells to investigate the distribution of HP1β and HP1γ over a 4 kb region centred on the +1 site of HIV1 transcription (TS). Unexpectedly, we found that HP1β was present only within a narrow area spanning from −0.5 to +0.5 kb (Fig 3A, red bars). Association of the protein with the promoter was not detected after stimulation with PMA (Fig 3B, red bars). By contrast, HP1γ was detectable only after PMA addition and showed a distribution wider than that of HP1β, spreading more downstream than upstream from the TS site (Fig 3B, yellow bars). This observation was consistent with an earlier study showing that HP1γ is present on the coding region of several erythroid-specific genes (Vakoc et al, 2005). The study also indicated that HP1γ required elongating RNAPII for its recruitment. We therefore used ChIP assays to probe for the presence of RNAPII on the HIV1 LTR. As this promoter is largely regulated at the level of elongation, RNAPII was present both before and after stimulation but with differing distributions. Before addition of PMA, the polymerase was colocalized with HP1β inside a 1 kb region framing the +1 site (Fig 3A, light blue bars). After stimulation, the RNAPII was distributed with the newly recruited HP1γ inside the coding region, although some HP1γ was also present upstream from the +1 site where no polymerase was detected (Fig 3B, light blue and yellow bars).
Figure 3.
Heterochromatin protein 1 factors colocalize with RNA polymerase II on HIV1 chromatin. After 2 h of PMA treatment of J-Lat A1 cells, chromatin immunoprecipitation experiments were performed with antibodies specific for HP1β, HP1γ or RNAPII amino terminus. Enrichments after immunoprecipitation from (A) unstimulated or (B) PMA-treated cells were measured using quantitative PCR. (C) Schematic representation of PCR amplicons on the HIV1 construct. HP1, heterochromatin protein 1; LTR, long terminal repeat; PMA, phorbol-myristate acetate; RNAP, RNA polymerase II.
Recruitment of HP1β requires active RNA polymerase II
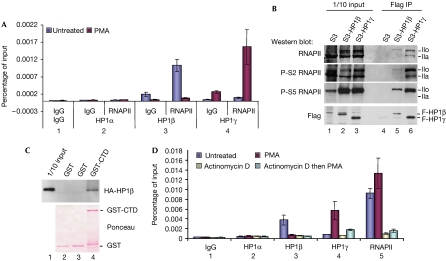
To establish that the colocalization of the HP1 proteins with RNAPII was not just due to heterogeneity of the cell population, we performed ChIP–reChIP experiments using either HP1β or HP1γ antibodies first and then RNAPII antibodies. These experiments showed that, on a given LTR, the RNAPII was associated with HP1β when transcription was repressed and with HP1γ after stimulation with PMA (Fig 4A). We next investigated whether HP1β would interact with the RNAPII. To this end, HP1β and HP1γ were immunoprecipitated from extracts of HeLa cells expressing Flag-tagged versions of these proteins, and western blots were carried out with different phospho-specific RNAPII antibodies. HP1β and HP1γ interacted with both hypo- and hyperphosphorylated RNAPII (Fig 4B, lanes 5,6,IIa,IIo). For HP1γ, the hyperphosphorylated species (IIo) were detected with antibodies specific for carboxy-terminal domain (CTD) phosphorylation on both Ser 2 and Ser 5 (Fig 4B, lane 6). By contrast, only Ser 5 phosphorylation was well detected in the HP1β co-immunoprecipitate, consistent with a preference of HP1β for non-elongating RNAPII (Fig 4B, lane 5). Also, haemagglutinin (HA)-tagged HP1β produced in Escherichia coli interacted in vitro with a glutathione S-transferase (GST)-RNAPII CTD construct, showing that the interaction was direct and not strictly dependent on phosphorylation (Fig 4C). These observations prompted us to test the role of RNAPII in the recruitment of HP1β. Using ChIP assays, we identified the intercalating agent actinomycin D as a good inhibitor of RNAPII recruitment on the LTR integrated in the J-Lat A1 cells (Fig 4D, bar 5). Additional ChIP assays using HP1β or HP1γ antibodies before and after stimulation with PMA showed that actinomycin D also decreased recruitment of the two HP1 proteins to the LTR (Fig 4D, bars 2–4). These experiments indicate that for HP1β, similar to that for HP1γ, ongoing transcription is required for recruitment.
Figure 4.
HP1β and HP1γ recruitment to the HIV1 long terminal repeat is dependent on the RNA polymerase II. (A) ChIP–reChIP was performed on J-Lat A1 cells treated with PMA for 2 h or untreated. Chromatin was immunoprecipitated first with HP1 or control antibodies and then with anti-RNAPII. (B) Flag-HP1β and Flag-HP1γ were immunoprecipitated (IP) from HeLa S3 cells stably expressing these proteins. Hypo- and hyperphosphorylated forms of RNAPII (IIa and IIo, respectively) were then detected by western bolt with antibodies against RNAPII CTD either non-phosphorylated (8WG16) or phosphorylated on serine 2 or on serine 5 (H5 or H14, respectively), or with an anti-Flag antibody. (C) Pulldown of HA-HP1β with GST-RNAPII CTD expressed in Escherichia coli. (D) J-Lat A1 cells were treated with PMA and actinomycin D as indicated. ChIP experiments were performed using the indicated antibodies. In (A) and (D), chromatin enrichment after immunoprecipitation was measured by using quantitative PCR with primers spanning the transcription start region. Data shown are averaged from two independent experiments. ChIP, chromatin immunoprecipitation; CTD, carboxy-terminal domain; GST, glutathione S-transferase; HA, haemagglutinin; HP1, heterochromatin protein 1; PMA, phorbol-myristate acetate; RNAPII, RNA polymerase II.
Discussion
Our data show that the HIV1 LTR can recruit and release HP1 proteins and that these movements are linked to phosphorylation of H3S10 and positioning of the RNAPII. The regulatory effect of H3S10 phosphorylation is frequently referred to as a phospho-switch. In this mechanism, which was initially observed during mitosis, the release of HP1 binding to methylated histones H3 or H1 is initiated by phosphorylation of the serine neighbouring the methylated lysine and does not require demethylation of this lysine (Mateescu et al, 2004; Daujat et al, 2005; Fischle et al, 2005; Hirota et al, 2005; Vicent et al, 2006). The H3S10 phosphorylation on the HIV1 LTR could be mediated by either the nuclear factor-κB or the mitogen-activated protein kinase pathways via Iκ-B kinase-α or MSK1/2, respectively (Thomson et al, 1999; Anest et al, 2003).
On our two LTR constructs that are both integrated in euchromatin, the three HP1 isoforms seem to have distinct roles, with HP1β present at the idle promoter, HP1γ localizing to the transcribed coding region after the induction and HP1α apparently not participating in the regulation. This selective recruitment diverges from what has been observed on latent integrated intact HIV1 virus where all three HP1 isoforms were detected on the LTR (Marban et al, 2007). This difference might be explained by the heterogeneity of the cell population after an infection in which all the cells are likely to have integrated the virus in various loci that might be associated with different HP1 isoforms depending on their heterochromatic or euchromatic position. Consistent with this, we have so far been unable to detect HP1α on any cellular euchromatic gene. We also note that another study shows a reactivation of the HIV1 LTR with an anti-HP1γ siRNA (Chéné et al, 2007). We found that this reactivation could be visualized only with this particular siRNA, which seems to induce a stress response (supplementary Figs S2,S5 online).
Our data show that recruitment of HP1β and HP1γ to the HIV1 LTR requires transcription. This does not seem to be a rule for all genes (Vicent et al, 2006), but it is consistent with the implication of the S. pombe RNAPII subunit Rpb2 in the recruitment of the HP1 homologue SWI6 to pericentromeric heterochromatin (Djupedal et al, 2005; Kato et al, 2005). Similarly, in Drosophila, HP1 proteins have been associated with transcriptionally active regions (Piacentini et al, 2003; de Wit et al, 2007). We found that HP1β interacts with the RNAPII with an apparent counter-selection of CTD phosphorylation on Ser 2. By contrast, HP1γ associates with both Ser 5- and Ser 2-phosphorylated CTDs (Fig 4B; Vakoc et al, 2005). It is therefore possible that the RNAPII recruits HP1 proteins as a function of the CTD phosphorylation status. Alternatively, as HP1 proteins rely on RNA-binding activity for association with chromatin (Muchardt et al, 2002), the HP1β–HP1γ switch could be regulated by the length of the RNA transcript.
From our data, HP1β seems to be an atypical repressor that does not antagonize RNAPII recruitment but rather functions as a chaperone that accompanies the polymerase, possibly to prevent inappropriate elongation activity. A recent study has shown that in Drosophila, hundreds of promoters carry a stalled RNAPII, indicating that pre-recruitment of the polymerase is a general phenomenon (Muse et al, 2007). We have identified several cellular genes, including the actin-like ACTA2 and ACTL8, the myomesin MYOM1 and the dehydrogenase–reductase DHRS2, that are stimulated by knockdown of HP1β but not HP1γ (supplementary Fig S4 online), and we anticipate a role for HP1β in the transient repression of many promoters beyond the HIV1 LTR.
Methods
Cell culture. Jurkat J-Lat tat-IRES-gfp clone A1 (NIH AIDS Research & Reference Reagent Program) was maintained in RMPI 1640 Glutamax medium (Invitrogen, Cergy-Pontoise, France) supplemented with 10% fetal bovine serum and 100 U ml−1 penicillin and streptomycin. HeLa LTR-Luc cells were cultured in DMEM. When indicated, the medium was supplemented with 10 nM phorbol-myristate acetate (PMA) and 0.2 μM actinomycin D (Sigma-Aldrich, Lyon, France) or DMSO.
RNA interference and Amaxa nucleofection. HP1 siRNAs are listed in the supplementary information online. Glyceraldehyde-3-phosphate dehydrogenase and cyclophilin B control siRNAs were purchased from Dharmacon (Perbio Science, Brebières, France). siRNAs (50 nM) were delivered by nucleofection (Amaxa, Cologne, Germany) using the Nucleofector kit V and program I-10. HP1α (2G9), HP1β (1A9) and HP1γ (1G6) antibodies from Euromedex (Souffelweyersheim, France) were used for western blots.
Messenger RNA and protein quantification. Total RNA was isolated using Trizol (Invitrogen). After DNase treatment (Turbo DNA-free kit; Ambion, Courtaboeuf, France), reverse transcription was performed using SuperScript II (Invitrogen) and random hexanucleotides following the manufacturer's instructions. Complementary DNAs were quantified by RT-qPCR (Mx3005P; Stratagene, Amsterdam, The Netherlands) using SYBR Green PCR master mix (Applied Biosystems, Courtaboeuf, France).
GFP analysis by flow cytometry. Samples were analysed on a COULTER Epics XL flow cytometer after propidium iodide labelling. Live cells (propidium-iodide-negative) were further gated using forward scatter compared with FL1 to discriminate GFP-positive from GFP-negative cells.
Chromatin immunoprecipitation. Anti-histone-modification ChIPs were performed as described (Batsché et al, 2006). The protocol for ChIP with HP1 and RNAPII antibodies is provided as the supplementary information online. The following antibodies were used: H3K9me2 (07-441, Upstate, Saint-Quentin en Yvelines, France), H3K9me2S10ph (Mateescu et al, 2004), H3K14ac (07-353, Upstate) or H3S10phK14ac (07-081, Upstate), HP1α (1H5), HP1β (1A9), HP1γ (1G6), RNAPII (N20, Santa Cruz, Tebu Bio, Le Perray en Yvelines, France). HIV1 primers are listed in the supplementary information online.
Protein interactions. Co-immunoprecipitation and GST pull-down assays are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
We thank S. Emiliani and O. Bensaude for the gift of plasmids and cell lines, D. Oficjalska for sharing unpublished observations, Saliha Azebi for technical assistance and M. Yaniv and J. Seeler for critical reading of the manuscript. B.M. received a fellowship from Agence Nationale de Recherche sur le Sida (ANRS). The experimental work was supported by a grant from the ANRS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS (2003) A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423: 659–663 [DOI] [PubMed] [Google Scholar]
- Batsché E, Yaniv M, Muchardt C (2006) The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13: 22–29 [DOI] [PubMed] [Google Scholar]
- Chéné I et al. (2007) Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, Mahadevan LC (2003) MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett 546: 51–58 [DOI] [PubMed] [Google Scholar]
- Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R (2005) HP1 binds specifically to Lys 26-methylated histone H1.4, whereas simultaneous Ser 27 phosphorylation blocks HP1 binding. J Biol Chem 280: 38090–38095 [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B (2007) High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: 346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Hediger F, Gasser SM (2006) Heterochromatin protein 1: don't judge the book by its cover!. Curr Opin Genet Dev 16: 143–150 [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM (2005) Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E (2003) HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (2005) RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J (2006) Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J 25: 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G, Wallrath L, Urrutia R (2006) The heterochromatin protein 1 family. Genome Biol 7: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O (2007) Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 26: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, England P, Halgand F, Yaniv M, Muchardt C (2004) Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep 5: 490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326: 711–713 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S (2003) Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 161: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G (2002) p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat Immunol 3: 69–75 [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC (1999) The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J 18: 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S (2006) Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25: 1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M (2006) Induction of progesterone target genes requires activation of erk and msk kinases and phosphorylation of histone H3. Mol Cell 24: 367–381 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB (2003) Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423: 655–659 [DOI] [PubMed] [Google Scholar]
- Yang X, Chen Y, Gabuzda D (1999) ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-κB. J Biol Chem 274: 27981–27988 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH (2006) The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information