Abstract
Werner Syndrome (WS) is a human genetic disorder with many features of premature aging. The gene defective in WS (WRN) has been cloned and encodes a protein homologous to several helicases, including Escherichia coli RecQ, the human Bloom syndrome protein (BLM), and Saccharomyces cerevisiae Sgs1p. To better define the function of WRN protein we have determined its subcellular localization. Indirect immunofluorescence using polyclonal anti-human WRN shows a predominant nucleolar localization. Studies of WRN mutant cells lines confirmed the specificity of antibody recognition. No difference was seen in the subcellular localization of the WRN protein in a variety of normal and transformed human cell lines, including both carcinomas and sarcomas. The nucleolar localization of human WRN protein was supported by the finding that upon biochemical subcellular fractionation, WRN protein is present in an increased concentration in a subnuclear fraction enriched for nucleolar proteins. We have also determined the subcellular localization of the mouse WRN homologue (mWRN). In contrast to human WRN protein, mWRN protein is present diffusely throughout the nucleus. Understanding the function of WRN in these organisms of vastly differing lifespan may yield new insights into the mechanisms of lifespan determination.
Werner syndrome (WS) is a rare autosomal recessive disorder (1). Affected individuals appear normal during the first decade of life; the first manifestation of the disorder is lack of the adolescent growth spurt. This symptom is classically followed by the development of bilateral ocular cataracts and premature graying of the hair in the twenties. Osteoporosis, type II diabetes mellitus, accelerated atherosclerosis, and cancer occur in the thirties and forties. Eventually, WS patients succumb to cardiovascular disease or cancer in the fourth or fifth decade (2).
Cells explanted from individuals with WS show a greatly shortened division potential in vitro (3–5). Genomic instability has been observed in WS cells, including chromosomal rearrangements (6), as well as an increased frequency of deletion mutations within the HPRT gene (7). In vivo, an increased frequency of HPRT− lymphocytes has also been observed in patients with WS (8).
The gene defective in WS, WRN, encodes a predicted protein of 1,432 amino acids with the seven signature motifs characteristic of DNA and RNA helicases, enzymes that unwind nucleic acids in an ATP-dependent and directionally specific manner. WRN demonstrates greatest similarity to the RecQ subfamily of DNA helicases, including Escherichia coli RecQ (9, 10), Saccharomyces cerevisiae Sgs1p (11, 12), Schizosaccharomyces pombe rqh1p (13), and the human proteins BLM [defective in the human disease Bloom’s syndrome (14)] and RecQL (15). Purified WRN protein has been demonstrated to be a DNA helicase, apparently functioning in a 3′-to-5′ direction (16, 17).
Studies of S. cerevisiae Sgs1p have revealed a mechanism by which loss of a RecQ family DNA helicase may cause premature aging. SGS1 was identified genetically as a suppressor of the slow growth phenotype of yeast bearing mutations in the TOP3 topoisomerase gene (11). Yeast mutant for SGS1 show increased rates of recombination throughout the genome, and particularly at the ribosomal DNA (rDNA) array (11, 12). In a striking parallel to WS in humans, the sgs1 mutation shortens yeast lifespan by approximately 60% (18). Immunolocalization of Sgs1p demonstrates that the protein is concentrated in the nucleolus, where it acts to suppress the formation of extrachromosomal rDNA circles (ERCs) (19). In the absence of Sgs1p, these circles readily form and subsequently replicate to high levels as yeast age. The sgs1 mutant appears to provide an accurate model of accelerated aging in yeast, because many of the changes that are observed in aging sgs1 cells [accumulation of ERCs, nucleolar fragmentation, silent information regulator (SIR) relocalization, increase in cell size] occur in aging wild-type cells as well (18, 19). Indeed, the presence of high levels of ERCs is a major determinant of yeast lifespan, as the premature introduction of an ERC markedly diminishes lifespan (19). Thus, in yeast it appears that Sgs1p, a WRN homologue, promotes longevity by regulating recombination at the rDNA array.
Here we demonstrate by immunofluorescence and biochemical fractionation the subcellular localization of endogenous WRN protein in wild-type human cells, WS mutant cell lines, and mouse cells. Like Sgs1p in yeast, the human WRN protein is concentrated in the nucleolus, suggesting that these proteins may function in a similar fashion to determine longevity in organisms as diverse as humans and yeast. A closely conserved mouse WRN homologue, mWRN, however, is not concentrated in the nucleolus but is distributed throughout the nucleoplasm.
MATERIALS AND METHODS
Cell Lines and Tissue Culture.
WI38 (75.1-CCL), GCT (TIB-223), PC-3 (CRL-1435), and HeLa S3 (CCL-2.2) cells were obtained from the American Type Culture Collection. All WRN mutant cell lines were obtained from the National Institute on Aging, Aging Cell Culture Repository, Coriell Institute for Medical Research (Camden, NJ). Cell lines were grown by using standard techniques, in the medium recommended by the supplier. X3 cells, a mouse fibroblastoid cell line derived from mammary tissue, were provided by Barbara Panning (Massachusetts Institute of Technology).
Antibodies.
Two μg of poly(A)+ RNA, prepared from HeLa cells by using the Micro-Fast Track kit (Invitrogen), was used as a template for a first-strand cDNA system using SuperScript II reverse transcriptase (SuperScript preamplification system, GIBCO/BRL). The resultant single-stranded DNA was used as a template in a PCR with Pfu DNA polymerase (Stratagene), using primers 5′-CGGCCATATGGATGACTCAGAGGATACATCCT-3′ and 5′-GCGCGTCGACCTATTAACTAAAAAGACCTCCCCTTTTCG-3′. The 1,475-bp PCR product, coding for amino acids 949-1432 of human WRN, was cloned into the NdeI and SalI sites of pET-28a(+) (Novagen). Recombinant WRN protein produced by using the pET system (Novagen) was purified from total cellular lysates by using Ni-nitrilotriacetate-agarose (Qiagen, Chatsworth, CA). The protein so purified was used to immunize rabbits by following standard protocols (20). Antibodies were affinity purified by using 200 μg of the immunizing antigen immobilized on nitrocellulose as described (21).
A polyclonal antiserum against a His6-tagged carboxyl-terminal fragment of the mWRN protein corresponding to residues 1191–1390 was produced in chickens (Covance, Denver, PA). IgY was isolated from eggs by using the EGGstract IgY purification system (Promega), and antibodies were purified with 5 mg of immunizing antigen covalently coupled to a diaminopropylamine column (Pierce).
Mouse monoclonal antibodies to human nucleolar protein B23 (22, 23), nucleolin (24, 25), and nucleolar protein P120 (26) were gifts of Harris Busch (Baylor Univ., Houston, TX). Antibody 8WG16 to the human RNA polymerase II was from Berkeley Antibody, Richmond, CA. Affinity-purified fluorochrome-conjugated secondary antibodies were obtained from Vector Laboratories.
Indirect Immunofluorescence.
Cells were grown on multiwell slides (CellPoint Scientific, Rockville, MD) and fixed in 50% methanol/50% acetone for 2 min at room temperature. Slides were washed three times for 2 min in 50 ml of PBS (8.1 mM Na2HPO4/1.5 mM KH2PO4/2.7 mM KCl/140 mM NaCl) and incubated with primary antibody diluted in binding buffer (PBS/1% BSA/0.2% Tween-20) for 45 min at 37°C in a humid chamber. Slides were washed in PBS as above and incubated with secondary antibody for 45 min at 37°C. After washing in PBS, slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) at 50 μg/ml in PBS for 1 min at room temperature and mounted in VectaShield (Vector Laboratories). Affinity-purified antibody was used at a dilution of 1:5; all secondary antibodies were used at a dilution of 1:100.
Mouse cells grown on multiwell slides (CellPoint) were washed once in PBS. Subsequent treatments were at 4°C, unless otherwise indicated. Slides were then washed for 30 sec in CSK (100 mM NaCl/300 mM sucrose/3 mM MgCl2/10 mM Pipes, pH 7.5), treated 2 min in CSK/0.5% Triton X-100, washed 30 sec in CSK, and fixed for 10 min in PBS/4% paraformaldehyde. Slides were then treated for 30 min at 37°C in blocking solution [PBS/0.2% cold water fish skin gelatin (Sigma)/5% goat serum/0.2% Tween-20]. Primary and secondary antibody incubations were performed as described above with blocking solution used in place of binding buffer.
Reverse Transcriptase–PCR Sequencing of WRN Mutants.
cDNA was prepared from WRN mutant cell lines as described above and used as a template in a nested PCR using Pfu DNA polymerase. For the amino-terminal half of the WRN sequence, the primers used were 5′-GTTGGACTGGATCTTCTCGG-3′ and 5′-GGAATTTGAAGGTCCAACAATC-3′; and 5′-TCGGGTTTTCTTTCAGATATTG-3′ and 5′-GCAGCCATTTCTTGTCAAAAC-3′. For the carboxyl-terminal half of the sequence, the primers used were 5′-TCACCTGTACTGGTTTTGATCG-3′ and 5′-AAGAACTGGCATCTTAAATC-3′; and 5′-GAAGTTAGGCGAAAAACAGGG-3′ and 5′-GGCATCTTAAATCAGCCTTCC-3′. Each nested reaction consisted of 25 cycles (94°C for 45 sec, 56°C for 1 min, 72°C for 5 min) under standard conditions (Stratagene). PCR products purified from 0.6% agarose gels by using Qiaex II resin (Qiagen) were sequenced by automated fluorescence sequencing (Massachusetts Institute of Technology Biopolymers Lab).
Subcellular Fractionation of HeLa Extracts and Preparation of Immunoblots.
Logarithmically growing HeLa S3 cells were harvested and washed once with PBS. Whole cell extracts were prepared by resuspending cells in S buffer (2% SDS/65 mM Tris⋅HCl, pH 6.8). Nuclear extracts were prepared from 2 × 107 cells as described (27). Nucleoli were prepared as described (28), with the following modifications. Cytoplasmic material was removed from the sedimented nuclei, which were then resuspended in 2 ml of N buffer (10 mM Hepes, pH 7.6/0.05 mM MgCl2/880 mM sucrose/0.05 mM spermidine/0.015 mM spermine/0.5 mM phenylmethanesulfonyl fluoride/2 mM benzamidine/5 ng/ml leupeptin/5 ng/ml pepstatin/2 ng/ml aprotinin). A portion (10%) of the nuclei were immediately centrifuged and resuspended in S buffer. The remaining nuclei were sonicated until >95% of nuclei were disrupted and intact nucleoli, stained with 0.1% azure C, were visible by microscopy. The sonicate was centrifuged at 1,000 × g for 20 min at 4°C, and nonsedimented material was removed. The nucleolar pellet was resuspended in N buffer and resedimented as before, followed by resuspension in S buffer.
For Western blots, 10 μg (protein) of whole cell extract or subcellular fraction was separated by SDS/PAGE, and specific proteins were visualized by immunoblotting and ECL (Amersham) detection, using standard techniques (20). Immunoblot signal intensities were measured by transmission scanning.
RESULTS
Nucleolar Localization of WRN Protein.
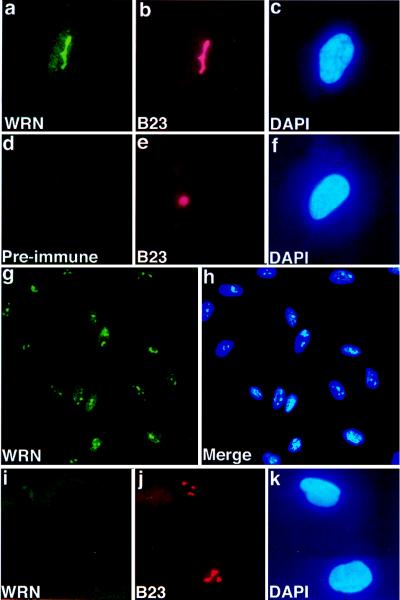
When antibody raised in rabbits against a carboxyl-terminal WRN fragment is used, indirect immunofluorescence of fibroblasts reveals a predominant subnuclear structure (Fig. 1a). This structure is present one to several times per nucleus, varying in different cells (Fig. 1g). Costaining the same cell with antibody to human nucleolar protein B23 (22, 23) identifies this structure as the nucleolus. Similar consecutive staining with anti-WRN and mouse monoclonal antibody to human nucleolin (24, 25), mouse monoclonal antibody to human nucleolar protein P120 (26), or human autoimmune anti-nucleolar antiserum (Sigma) shows identical patterns of fluorescence. A similar pattern of WRN indirect immunofluorescence was seen with cells that had been fixed with paraformaldehyde, although diffuse nuclear staining was increased slightly (data not shown).
Figure 1.
Nucleolar localization of WRN in human cell lines. Immunofluorescence images of human fetal lung fibroblasts (WI38, a–h) or adult skin fibroblasts derived from a patient with WS (cell line AG0780, i–k). Indirect immunofluorescence was performed on methanol/acetone-fixed cells, using anti-WRN antibody (a, d, g) or antibody to nucleolar protein B23 (b, e, j) or by DAPI, which binds to nuclear DNA (c, f, h, k). The bound antibody is visualized with a fluorescein-conjugated secondary antibody (green fluorescence), or Texas red-conjugated secondary antibody (red fluorescence). WI38 cells were treated consecutively with anti-WRN (a), anti-nucleolar protein B23 (b), or DAPI (c). No fluorescence was seen when preimmune serum was used in place of anti-WRN antibody (d) and cells were treated consecutively with anti-B23 (e) and DAPI (f). A low-power view of WI38 cells treated with anti-WRN shows little cell-to-cell variation in the immunofluorescence signal (g), as also shown in the composite photomicrograph of WI38 cells stained with anti-WRN and DAPI (h). Fibroblasts from a patient with WS show no staining when treated with anti-WRN (i), although no difference in gross nucleolar (j) or nuclear (k) morphology is observed. (g and h, × 200 ; all others, × 500.)
No specific fluorescence was seen with preimmune serum (Fig. 1d). These results, and the results from immunofluorescence of WRN mutants (discussed below), show that the pattern seen on immunofluorescence is specific for the endogenous WRN protein.
The pattern of staining varied little from cell to cell (Fig. 1 g). No change in subnuclear localization or level of WRN protein expression was seen when cells were growth arrested by serum starvation (data not shown). Likewise, no cell cycle variation was seen when cells were synchronized by serum starvation and aphidicolin block and cell cycle stage was assayed after release by flow cytometry and bromodeoxyuridine incorporation (data not shown).
WRN Immunofluorescence in WRN Mutants.
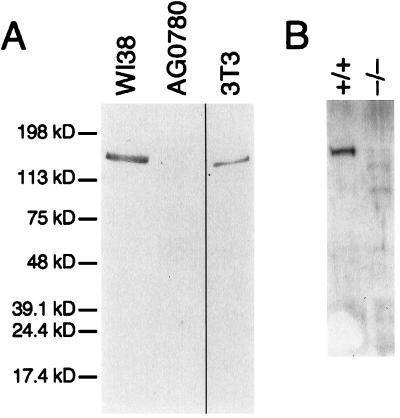
To further assess the specificity of WRN localization, we examined the staining pattern of multiple WRN mutant cell lines. In a skin fibroblast cell line containing a nonsense mutation at codon 369 (AG0780, (29)), no signal is detected when anti-WRN antibody is used (Fig. 1i). There is no abnormality in overall nucleolar structure (Fig. 1j). A single protein of approximate molecular mass 170 kDa is seen when the same antibody is used to probe immunoblots of whole cell extract of normal WI38 fibroblasts, but no reactive protein is seen in extracts of this WRN mutant cell line (Fig. 2). These results unequivocally demonstrate the specificity of the immunofluorescence pattern of anti-WRN antibody.
Figure 2.
Immunoblot of human and mouse WRN proteins. Whole cell extracts of the indicated cell lines were prepared, and 10 μg of each was analyzed by Western blotting. (A) Immunoblot of 8% polyacrylamide gel, using anti-WRN antibody. Anti-human WRN recognizes a single protein of apparent mobility corresponding to 170 kDa in fibroblast cell line WI38. In a WRN mutant fibroblast cell line, AG0780, no reactive protein is seen. A single protein of slightly increased mobility is seen in an extract prepared from mouse 3T3 cells, in agreement with the previous identification of a single, highly conserved WRN homologue in mouse (38). (B) Immunoblot of a 7.5% polyacrylamide gel, using anti-mWRN antibody. Anti-mWRN recognizes a single protein of apparent mobility corresponding to 165 kDa in normal mouse ear fibroblasts (+/+). No cross-reacting protein is seen in ear fibroblasts prepared from a mouse in which mWRN has been disrupted (−/−).
We screened a total of nine WS cell lines by immunofluorescence for WRN protein (Table 1). Five of these cells lines showed no detectable immunofluorescence when analyzed for WRN subcellular localization. Four of these also showed no detectable WRN protein on immunoblot; one (AG05229) showed a faint band of apparent molecular mass 60 kDa not seen in wild-type fibroblasts. No missense mutations have so far been detected at the WRN locus in affected individuals (29–32), and it has been observed that WRN mutant RNAs are present at decreased concentration (33). All published mutations cause premature termination producing truncated WRN proteins lacking the nuclear localization signal located at its carboxyl terminus (34).
Table 1.
Summary of indirect immunofluorescence, immunoblot, and sequencing of WRN in mutant cell lines
| Cell line | Type | Immunofluorescence | Immunoblot | Predicted protein sequence |
|---|---|---|---|---|
| AG0780 | PF | − | − | Arg369 → Stop (29) |
| AG05229 | PF | − | − | ND |
| AG12798 | PF | − | ND | ND |
| AG11395 | TF | − | − | Arg369 → Stop (29) |
| AG04103 | LCL | − | − | Arg369 → Stop (29) |
| AG04110 | PF | WT | WT | WT |
| AG06300 | PF | WT | WT | Phe1074 → Leu |
| AG07066 | TF | WT | WT | Phe1074 → Leu |
| AG07896 | LCL | WT | WT | Phe1074 → Leu, Cys1367 → Arg |
Cell types were PF, primary fibroblast; TF, simian virus 40-transformed fibroblast; and LCL, Epstein–Barr virus-transformed lymphoblastoid cell line. A − pattern of WRN immunofluorescence indicates no fluorescent signal with anti-WRN antibody. A − WRN immunoblot indicates no band of appropriate mobility on immunoblot with anti-WRN. In cell lines showing WT WRN immunofluorescence, there was no change in the fluorescence signal with anti-WRN as compared to appropriate wild-type WRN control cell lines. Likewise, WT immunoblot indicates a band of the same mobility and intensity as an extract from normal control cells when probed with anti-WRN antiserum. Sequences were determined from reverse transcriptase–PCR products prepared from total cellular RNA. ND, not determined.
Four WRN mutant cell lines both showed a wild-type pattern of WRN immunofluorescence and demonstrated a protein of wild-type mobility and abundance on immunoblots of cellular extracts (summarized in Table 1). We determined the nucleotide sequence of WRN RNA in these cell lines by direct sequencing of reverse transcriptase–PCR (RT-PCR) products (see Materials and Methods). In three cell lines, the conservative amino acid replacement leucine for phenylalanine at amino acid 1074 was present. This allele is most likely a normal variant, as identical substitutions in the WRN sequence in cells from patients with Hutchinson–Gilford progeria were also found (data not shown). No WRN mutations are present in this disorder (35). The arginine for cysteine substitution at amino acid 1367 is a normal variant in the Japanese population and may be associated with a protective effect against myocardial infarction (36). WRN in HeLa cells was found to be heterozygous for this allelic variant (data not shown). It is likely all four of these WRN mutant cell lines produce a functional WRN protein.
The simplest explanation for this result is phenotypic misclassification. WS has a complex phenotype with variable presentation. Alternatively, a wild-type WRN locus might be generated during tissue culture in vitro by mitotic recombination within the WRN locus in a compound heterozygote. A third possibility is that mutations outside the WRN locus may produce a syndrome that is clinically indistinguishable from WS.
Nucleolar Localization of WRN Is Independent of Tissue Type.
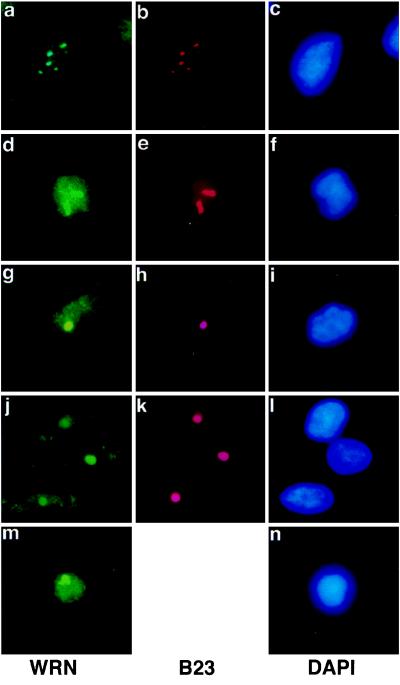
The subcellular localization of WRN in cell lines derived from a variety of tissues was determined (Fig. 3). Predominant nucleolar localization was observed in a transformed fibroblast cell line derived from WI38 (a–c), in HeLa cells (d–f), in a prostatic carcinoma (g–i), in a malignant fibrous histiocytoma (j–l), and in lymphoblastoid cells (m and n). In HeLa cells, diffuse nuclear staining was increased (d–f).
Figure 3.
WRN nucleolar localization in multiple cell lines. Cells were subjected to double immunolabeling with anti-WRN and anti-nucleolar protein B23, as described in the legend to Fig. 1. In each row, Left shows the anti-WRN signal, Center shows the anti-B23 (anti-nucleolar signal), and Right shows the DAPI (nuclear) signal. (a–c) WI38 75.1, simian virus 40-transformed human fetal lung fibroblast cell line. (d–f) HeLa, human cervical carcinoma cell line. (g–i) PC-3, prostatic adenocarcinoma. (j–l) GCT, fibrous histiocytoma. (m and n) AG07877, lymphoblastoid cell line. (×500.)
Individuals with WS are prone to developing a number of malignancies, primarily sarcomas (37). No difference in localization was seen in the sarcoma cell line analyzed (a malignant fibrous histiocytoma) and the two carcinoma cell lines (a cervical carcinoma and a prostatic carcinoma).
Nucleolar Localization of WRN upon Biochemical Fractionation.
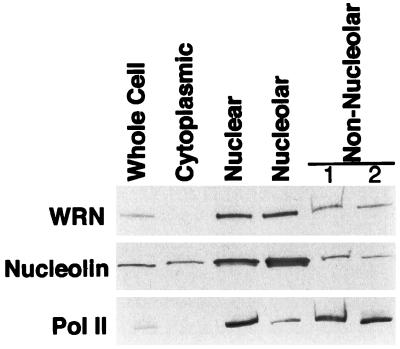
To confirm the nucleolar localization of WRN protein observed by immunofluorescence, HeLa extract was fractionated into cytoplasmic, nuclear, nucleolar, and nonnucleolar nuclear components (Fig. 4). Little WRN protein was found in the cytoplasm. WRN protein was found in a 3-fold higher concentration in the nucleolar fraction, compared with the nonnuclear fraction (containing nuclear proteins left after sedimentation of nucleoli).
Figure 4.
Distribution of WRN protein among HeLa subcellular fractions. Extracts of HeLa cells were prepared and fractionated into cytoplasmic, nuclear, nucleolar, and nonnucleolar fractions. A 10-μg sample of each fraction was separated by electrophoresis on an SDS/4–15% polyacrylamide gradient gel and then analyzed by Western blotting. Blots, prepared in parallel, were probed with the antibodies indicated to the left. To control for the fidelity of the fractionation, blots were probed with antibodies to proteins of preestablished subnuclear localization: nucleolin, which is present in increased concentration in the nucleolus, and RNA polymerase II (Pol II), which is excluded from the nucleolus. Nonnucleolar lanes 1 and 2 are the nonsedimented material from the first and second cycle of sedimentation through sucrose (see Materials and Methods). An obvious decrease in concentration of WRN or nucleolin in the total nuclear compared with the nucleolar fractions was not observed. There are two possible explanations for this finding. The nucleolar fraction is contaminated with some amount of adherent chromatin, and some nucleolar proteins diffuse out of nucleoli during fractionation. Both of these artifacts will decrease the apparent concentration of a protein in the nucleolar fraction. The diffusion of nucleolar protein during fractionation will have a lesser effect on its concentration in the nonnucleolar fraction, as the nucleolar fraction contains 20% of the total nuclear protein, whereas the nonnucleolar fraction contains 80%.
To control for the fidelity of the fractionation, we probed duplicate immunoblots for nucleolin, a protein known to be concentrated in the nucleolus (24, 25), and for RNA polymerase II (Pol II), a protein that has no known function in the nucleolus. As expected, nucleolin was found 8-fold more concentrated in the nucleolar fraction compared with the nonnucleolar nuclear fraction, and Pol II was 5-fold more concentrated in the nonnucleolar nuclear fraction than in the nucleolus.
WRN in Mouse Cells.
The mouse genome contains a single locus of high homology to the WRN gene located on mouse chromosome 8A4 (38), in a region that is syntenic with human 8p12 containing the human WRN locus (39). The mouse WRN cDNA can code for a protein of 1,401 amino acids that is highly homologous to the human WRN protein (38).
Additional evidence that this locus is indeed the single mouse homologue of the human WRN gene is that antibodies raised against human WRN recognize a single protein in extracts of mouse 3T3 cells (Fig. 2A). When antibodies raised against amino acids 1191–1390 of the mWRN protein were used to probe mouse fibroblast extracts, a single protein of the same mobility as that recognized by antibody to human WRN in 3T3 cells is seen (Fig. 2B). Affinity-purified antibody to mWRN recognizes a single protein in extracts of human fibroblasts, of mobility identical to that of human WRN (data not shown). The high degree of protein sequence similarity, syntenic location, and specific immunologic bidirectional crossreactivity all support that this is the single WRN homologue in mice.
A mouse containing a disruption in the genomic WRN locus has been constructed (D.B.L., C. Beard, J. Dausman, R. Jaenisch, and L.G., unpublished results). The targeted mutation is predicted to eliminate expression of a portion of the helicase domain and all of the carboxyl terminus of the mWRN protein. Extracts of fibroblasts from mice homozygous for the targeted mutation at the mWRN locus show no mWRN protein on immunoblots (Fig. 2B).
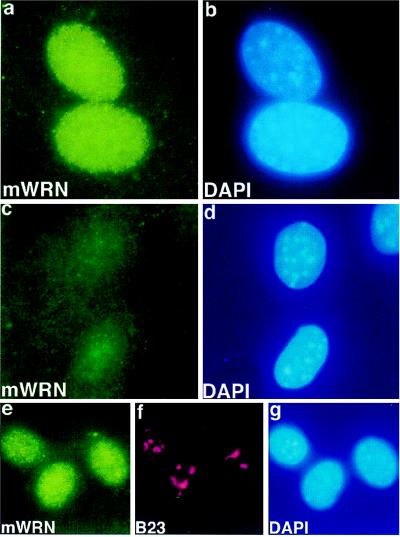
Immunofluorescence in skin fibroblasts of normal mice with antibody prepared against recombinant mWRN protein shows a diffuse nuclear pattern (Fig. 5a). These cells, stained at approximate population doubling level 5, were normal morphologically and exhibited normal contact inhibition of growth. There is no appreciable increase in fluorescence intensity in nucleoli in mouse cells (compare Fig. 5 e and f), in marked contrast to that observed with human WRN immunofluorescence in human fibroblasts (compare Fig. 1 a and b). When antibody to human WRN was used for indirect immunofluorescence of mouse cells, no specific fluorescent signal was seen (data not shown). No specific signal was seen when ear fibroblasts homozygous for disruption of the mouse WRN locus were stained in parallel (Fig. 5c).
Figure 5.
WRN localization in mouse cell lines. Immunofluorescence images of ear fibroblasts derived from wild type (a and b, e–g) or from a mouse homozygous for a targeted mutation in the mouse homologue of WRN (c and d). Indirect immunofluorescence was performed on fixed cells with anti-mWRN antibody (a, c, e), with anti-nucleolar protein B23 (f), or with DAPI (b, d, g). Fixation in this experiment was with 4% paraformaldehyde. Indirect immunofluorescence of mWRN in wild-type ear fibroblasts shows a diffuse nuclear pattern (a). No mWRN fluorescence is seen in ear fibroblasts derived from a mouse homozygous for a mWRN disruption (c). No increase in mWRN immunofluorescence is seen in nucleolar regions (compare e, mWRN, and f, nucleolar protein B23), in a normal mouse fibroblastoid cell line (X3). (×750.)
DISCUSSION
In this study, we demonstrate nucleolar localization of the human WRN gene product by both indirect immunofluorescence and biochemical fractionation. Both techniques yield the identical result, that WRN protein is present in higher concentration in the nucleolus than in the nucleoplasm. No change in the pattern of WRN fluorescence was observed in a number of cell lines, including both sarcomas and carcinomas. Our results examining the localization of endogenous WRN protein differ from previous reports, which showed diffuse nuclear staining of the products of tagged WRN transgenes (16, 34).
What is the significance of the predominant nucleolar localization of WRN? Some insight into the possible function of WRN in the nucleolus might be gained by examining the function of a yeast WRN homologue, SGS1.
In yeast, sgs1 mutants were found by several criteria to age prematurely (18). Sgs1p immunolocalizes to the nucleus, with an increased concentration in the nucleolus (18), similar to WRN localization in human fibroblasts. In old sgs1 mother cells, the nucleolus was enlarged and fragmented. These changes caused the nucleolus to occupy at least 50% of the nuclear volume in most old cells. Significantly, similar changes were found in nucleoli of old wild-type cells, indicating that the changes were not the consequence of the sgs1 mutation per se.
Two striking changes were found to be present in the rDNA of old yeast mother cells: the rDNA copy number increased greatly, and this increase in rDNA sequences was due the presence of large numbers of ERCs. The number of ERCs was estimated to be between 500 and 1,000 copies per cell in aged yeast (19). The accumulation of rDNA circles appears capable of causing yeast senescence, as premature introduction of a exogenous rDNA circle by using the Cre-lox recombination system markedly diminished lifespan. Thus, in S. cerevisiae, replicative capacity is limited by the accumulation of ERCs in old cells, and SGS1, the yeast WRN homologue, functions to extend lifespan by regulating recombination at the rDNA array.
An identical process does not appear to occur in normal human fibroblasts undergoing replicative senescence. The rDNA copy number in senescent WI38 cells remains constant (40), and the number of nucleoli per cell was found to be constant (41) or to decrease (42, 43) with successive population doublings in vitro. In none of these studies was the topological state of rDNA determined. If ERCs do occur in senescent human fibroblasts, they do not accumulate and do not cause nucleolar fragmentation.
One requirement for the amplification of ERCs in yeast is asymmetric segregation of the ERCs to the mother, rather than the newly formed daughter. It is unlikely that such asymmetric segregation occurs in fibroblasts. A more promising human cell type in which ERCs might be amplified are those cells undergoing more pronounced asymmetric division—possibly stem cells—as in intestinal crypts or in the basal layer of the epidermis. It remains to be determined whether rDNA copy number or rDNA topology of such stem cell populations varies as a function of donor age or the number of doublings.
Changes in rDNA have been observed in postmitotic tissues. In 1979, Strehler et al. (44, 45) observed a decrease in rDNA in neuronal and cardiac tissues. When liquid phase hybridization to 18S and 28S ribosomal RNA was used, the amount of rDNA was found to decrease by 0.5% per year in human myocardium (44), and a model was proposed in which deletion of repeats within the rDNA arrays would occur by intrachromosomal homologous recombination and ERC formation (45). In these postmitotic cells, rDNA sequence loss was suggested to occur by ERC formation and subsequent degradation of the rDNA episome. Clearly, these initial results using less accurate solution hybridization need to be verified by using current membrane-hybridization-based techniques with single-copy gene controls.
The mouse homologue of human WRN, mWRN, does not show nucleolar localization. It remains to be determined whether this apparent difference in subcellular localization implies a difference in the function of WRN in these two organisms. A comparison of the human (32) and mouse (38) sequences shows a single structural difference—the mouse sequence lacks the acidic direct repeat found just amino-terminal to the helicase domain in the human protein (38). Such sequence variation in the two proteins may be responsible for their different subcellular localization. Alternatively, factors may be expressed in human cells and not in mouse that targets WRN to the nucleolus.
Appropriate analysis of expression of human WRN in mWRN knockout cells, and the expression of mWRN in human WRN-negative cells should be able to distinguish among these possibilities. Such a cross-species analysis may aid in our understanding of the importance of nucleolar structure and function in mammalian aging and yield new insights into mechanisms of lifespan determination.
Acknowledgments
We thank David McNabb, Shin Imai, Barbara Panning, Dale Talbot, Rudolph Jaenisch, and members of the Guarente laboratory for many stimulating discussions, and Harris Busch for providing monoclonal antibodies to nucleolar proteins. This work was supported by National Institutes of Health Grant K08AG00790-01 to R.A.M., Grant K08AG00775-01 to F.B.J., a Medical Scientist Training Program Training Grant to D.L., and Grant R01AG11119 to L.G.
ABBREVIATIONS
- WS
Werner syndrome
- rDNA
DNA encoding ribosomal RNA
- ERC
extrachromosomal rDNA circle
- mWRN
mouse homologue of WRN protein
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Salk D. Hum Genet. 1982;62:1–5. doi: 10.1007/BF00295598. [DOI] [PubMed] [Google Scholar]
- 2.Epstein C J, Martin G M, Schultz A L, Motulsky A G. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Martin G M, Sprague C A, Epstein C J. Lab Invest. 1970;23:86–91. [PubMed] [Google Scholar]
- 4.Holliday R, Thompson K V A, Huschtscha L I, Rattan S I S, Sedgwick S G, Spanos A. In: Werner’s Syndrome and Human Aging. Salk D, Fujiwara Y, Martin G M, editors. Vol. 190. New York: Plenum; 1982. pp. 331–339. [Google Scholar]
- 5.Salk D, Bryant E, Hoehn H, Johnston P, Martin G M. In: Werner’s Syndrome and Human Aging. Salk D, Fujiwara Y, Martin G M, editors. Vol. 190. New York: Plenum; 1982. pp. 305–312. [Google Scholar]
- 6.Salk D, H, A, Hoehn H, Martin G M. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 7.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuchi K, Tanaka K, Kumahara Y, Marumo K, Pride M B, Martin G M, Monnat R J., Jr Hum Genet. 1990;84:249–252. doi: 10.1007/BF00200569. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 10.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangloff S, McDonald J P, Bendixen C, Lane A, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watt P M, Louis E J, Borta R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 13.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis N A, Groden J, Ye T, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 15.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 16.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray M D, Shen J C, Kamath-Loeb A S, Blank A, Sopher B L, Martin G M, Oshima J, Loeb L A. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 18.Sinclair D A, Mills K, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 19.Sinclair D, Guarente L. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 21.Tang W-J. In: Antibodies in Cell Biology. Asai D, editor. Vol. 37. San Diego: Academic; 1993. pp. 95–117. [Google Scholar]
- 22.Spector D, Ochs R, Busch H. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- 23.Ochs R, Lischwe M, O’Leary P, Busch H. Exp Cell Res. 1983;146:139–149. doi: 10.1016/0014-4827(83)90332-4. [DOI] [PubMed] [Google Scholar]
- 24.Olson M O, Guetzow K, Busch H. Exp Cell Res. 1981;135:259–265. doi: 10.1016/0014-4827(81)90161-0. [DOI] [PubMed] [Google Scholar]
- 25.Freeman J W, Chatterjee A, Ross B E, Busch H. Mol Cell Biochem. 1985;68:87–96. doi: 10.1007/BF00219392. [DOI] [PubMed] [Google Scholar]
- 26.Freeman J, Busch R, Gyorkey F, Gyorkey P, Ross B, Busch H. Cancer Res. 1988;48:1244–1251. [PubMed] [Google Scholar]
- 27.Descombes P, Schibler U. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 28.Kistler J, Duncombe Y, Laemmli U. J Cell Biol. 1984;99:1981–1988. doi: 10.1083/jcb.99.6.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C-E, Oshima J, Wijsim E, Nakura J, Miki T, Piussan C, Matthews S, Fu Y-H, Mulligan J, Martin G, Schellenberg G. Am J Hum Genet. 1997;60:330–341. [PMC free article] [PubMed] [Google Scholar]
- 30.Miki T, Nakura J, Ye L, Mitsuda N, Morishima A, Sato N, Kamino K, Ogihara T. Mech Ageing Dev. 1997;98:255–265. doi: 10.1016/s0047-6374(97)00112-7. [DOI] [PubMed] [Google Scholar]
- 31.Oshima J, Yu C-E, Piussan C, Klein G, Jabkowski J, Balci S, Miki T, Nakura J, Ogihara T, Ells J, et al. Hum Mol Genet. 1996;5:1909–1913. doi: 10.1093/hmg/5.12.1909. [DOI] [PubMed] [Google Scholar]
- 32.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 33.Yamabe Y, Sugimoto M, Satoh M, Suzuki N, Sugarawa M, Goto M, Furuichi Y. Biochem Biophys Res Commun. 1997;236:151–154. doi: 10.1006/bbrc.1997.6919. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Nat Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- 35.Oshima J, Brown W T, Martin G M. Lancet. 1996;348:1106. doi: 10.1016/S0140-6736(05)64456-X. [DOI] [PubMed] [Google Scholar]
- 36.Ye L, Miki T, Nakura J, Oshima J, Kamino K, Rakugi H, Ikegami H, Higaki J, Edland S, Martin G, Ogihara T. Am J Med Gen. 1997;68:494–498. doi: 10.1002/(sici)1096-8628(19970211)68:4<494::aid-ajmg30>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 37.Goto M, Miller R W, Ishikawa Y, Sugano H. Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 38.Imamura O, Ichikawa K, Yamabe Y, Goto M, Sugawara M, Furuichi Y. Genomics. 1997;41:298–300. doi: 10.1006/geno.1997.4661. [DOI] [PubMed] [Google Scholar]
- 39.Mouse Genome Database (MGD), Mouse Genome Informatics (1998) (The Jackson Laboratory, Bar Harbor, ME).
- 40.Peterson C R D, Cryar J R, Gaubatz J W. Arch Gerontol Geriatr. 1984;3:115–125. doi: 10.1016/0167-4943(84)90004-9. [DOI] [PubMed] [Google Scholar]
- 41.Weinstein M E, Mukherjee A B. Mech Ageing Dev. 1988;42:215–227. doi: 10.1016/0047-6374(88)90048-6. [DOI] [PubMed] [Google Scholar]
- 42.Bemiller P M, Lee L H. Mech Ageing Dev. 1978;8:417–427. doi: 10.1016/0047-6374(78)90041-6. [DOI] [PubMed] [Google Scholar]
- 43.Bemiller P, Krauss E. Mech Ageing Dev. 1983;22:79–87. doi: 10.1016/0047-6374(83)90008-8. [DOI] [PubMed] [Google Scholar]
- 44.Strehler B L, Chang M P, Johnson L K. Mech Ageing Dev. 1979;11:371–378. doi: 10.1016/0047-6374(79)90012-5. [DOI] [PubMed] [Google Scholar]
- 45.Strehler B L, Chang M P. Mech Ageing Dev. 1979;11:379–382. doi: 10.1016/0047-6374(79)90013-7. [DOI] [PubMed] [Google Scholar]