Abstract
We have previously cloned the human Na+/H+ exchanger NHE2 gene and its promoter region. In the present study, the regulatory elements responsible for the constitutive expression of NHE2 were studied. Transient transfection assays revealed that the −40/+150 promoter region contains the core promoter responsible for the optimal promoter activity. A smaller fragment, −10/+40, containing the TIS (transcription initiation site) showed minimal activity. We identified a palindrome that overlaps the TIS and binds to the transcription factors Sp1 and Sp3. Mutations in the 5′ flank of the palindrome abolished the Sp1/Sp3 interaction and reduced promoter activity by approx. 45%. In addition, a conserved GC-box centered at −25 was found to play a critical role in basal promoter activity and also interacted with Sp1 and Sp3. An internal deletion in the GC-box severely reduced the promoter activity. Sp1/Sp3 binding to these elements was established using gel-mobility shift assays, confirmed by chromatin immunoprecipitation and co-transfections in Drosophila SL2 cells. Furthermore, we identified two positive regulatory elements in the DNA region corresponding to the 5′-UTR (5′-untranslated region). The results in the present study indicate that Sp1 and Sp3 are required for constitutive NHE2 expression and that the positive regulatory elements of the 5′-UTR may co-operate with the 5′-flanking region to achieve the optimal promoter activity.
Keywords: C2BBe1 cell line, Na+/H+ exchanger (NHE), stimulating protein 1/3 (Sp1/Sp3), transcriptional regulation, 5′-untranslated region
Abbreviations: AP2, activator protein 2; ChIP, chromatin immunoprecipitation; EGR-1, early growth response gene product-1; GMSA, gel mobility- shift assay; Inr, initiator; NF-κB, nuclear factor κB; NHE, Na+/H+ exchanger; PANDER, pancreatic-derived factor; Sp, stimulating protein; TBP, TATA-binding protein; TIS, transcription initiation site; USF, upstream stimulatory factor; UTR, untranslated region
INTRODUCTION
Human NHE2 (Na+/H+ exchanger 2) is one of nine members of the family of Na+/H+ exchangers. NHEs act as plasma membrane transporters, ultimately functioning to regulate pHi (intracellular pH), cell volume and Na+ absorption (for a review see [1–4]). Protein and mRNA of NHE isoforms 1, 2, 3 and 8 have been reported to be expressed in intestinal and colonic tissue [3]. NHE1 is localized to the basolateral membrane of intestinal cells and is thought to serve a housekeeping function. NHE2 and NHE3 are localized to the apical membrane in polarized cells and play potentially similar roles in intestinal Na+ absorption with differential activities in various segments of the gastrointestinal tract; however, in knockout mice of either isoform, the other isoform did not show any compensatory effect [5,6]. In rats, NHE8 has been suggested to be important for apical Na+ absorption early in life [7].
There is a wealth of data on short-term (post-translational) regulation of the NHE isoforms by various stimuli such as growth factors, hormones and protein kinases [8,9] and their interactions with the cytoplasmic regulatory proteins [10–12]. However, little is known about the long-term (transcriptional) regulation of these genes and data in this regard have previously been emerging, mostly on the NHE1 isoform [13,14]. Among the other isoforms the human and rat promoter sequences for the NHE2 and NHE3 genes have been cloned [15–19] and their specific characteristics are beginning to be unraveled. For example, the effect of hyperosmolarity, the transcription factor Sp1 (stimulating protein 1) and EGF (epidermal growth factor) on rat NHE2 expression has been described [20–22].
Our previous studies demonstrated that the human NHE2 gene is located on chromosome 2q11.2 and is composed of 12 exons and 11 introns [15]. The promoter that governs the NHE2 gene expression is highly GC-rich, lacks both TATA and CCAAT-boxes and contains multiple Sp1 sites [15]. It also contains other potential transcription factor binding sites, including AP2 (activator protein 2), NF-κB (nuclear factor κB), Oct1 (ocatmer-binding protein 1), CdxA and Cdx-2. The rat NHE2 promoter has also been shown to lack both TATA and CCAAT consensus sequences [16]. A number of potential transcription factor binding sites are conserved between the human and the rat NHE2 promoters. These include recognition sequences for Sp1, AP-2, CACCC and NF-κB; however, variations in some other cis-elements exist that may suggest the existence of differences in regulation of NHE2 in different species [15].
We have recently conducted several studies demonstrating the stimulation of promoter activities of both NHE2 and NHE3 genes in response to PMA. Both genes exhibited up-regulation by PMA through an EGR-1 (early growth response gene product-1)-dependent mechanism [23,24]. An enhanced protein–DNA interaction between the PMA-induced EGR-1 and a distal GC-box at −330, in respect to the TIS (transcription initiation site), appears to be responsible for the increased NHE2 mRNA and promoter expression. Binding of PMA-induced EGR-1 to the distal GC-box was coupled with displacement of transcription factors Sp1 and Sp3 from this element [23].
In the present study, we characterize further the human NHE2 promoter with a focus on the proximal promoter and the 5′-UTR (5′-untranslated region) and report that a GC-box at the −25 position is critical for transcriptional activity and binds to the transcription factors Sp1 and Sp3. Furthermore, we report that transcription factors Sp1 and Sp3 also bind to an inverted repeat sequence overlapping the TIS. In co-transfection experiments using Drosophila SL2 cells both Sp1 and Sp3 transactivated the NHE2 promoter individually, whereas combined expression of these factors led to attenuation of the stimulatory effect of Sp3 by Sp1. Finally, our functional analyses detected two positive regulatory elements in the DNA sequences corresponding to the 5′-UTR. These elements seem to be required for optimal activity of the NHE2 core promoter.
EXPERIMENTAL
Chemicals
Chemicals were purchased from Fisher Scientific or Sigma. Restriction endonucleases or other modifying enzymes were purchased from New England BioLabs, Invitrogen or Promega. All radioisotope nucleotides were obtained from Amersham-Pharmacia Biotechnologies. Molecular biology techniques including restrictions, ligations, plasmid isolations and transformations were performed using standard procedures as described previously [25]. Anti-human Sp1, Sp3, EGR-1, USF (upstream stimulatory factor)-1 and USF-2 antibodies were purchased from Santa Cruz Biotechnology.
Oligonucleotides and plasmid construction
All oligonucleotides were synthesized by Life Technologies, Invitrogen. To clone the various fragments of the NHE2 proximal promoter region, we employed PCR amplification. For cloning purposes, XhoI or HindIII restriction enzyme recognition sequences were introduced into the 5′-end of the sense or antisense oligonucleotides respectively. Fragments of the NHE2 promoter were amplified using construct p−85/+150 as a template along with primers specific for the desired truncation end-points. The nucleotide sequences of the sense strand of the oligonucleotides used to generate these clones are shown in Table 1, and were based on the human NHE2 promoter sequence (GenBank® accession no. AF273748). Subsequent to PCR amplifications, the amplicons were digested with XhoI and HindIII, gel purified and cloned into pGL2-basic, which contains a promoter-less luciferase reporter gene. Mutant constructs containing base substitutions were created from p−85/+150 using the QuikChange® site-specific mutagenesis kit from Stratagene according to the manufacturer's protocol with complementary primers containing the desired mutation. Plasmid p−85/+150ΔGC, containing a deletion at bp −23 to −11 of p−85/+150, was an artifact of PCR amplification randomly isolated during mutagenesis of an unrelated site, which was subsequently corrected by site-specific mutagenesis with wild-type primers to obtain the p−85/+150ΔGC. To create the 3′-deletion construct p−85/+113, p−85/+150 was double- digested first with the restriction enzyme HindIII, ends were filled in using the Klenow fragment of DNA polymerase I, digested again with restriction enzyme PmlI, purified and ligated. These digestions released the DNA region between +113 and +150. All constructs were verified by sequence analysis.
Table 1. Primers used for PCR amplification of the truncated fragments of the NHE2 proximal promoter region.
The direction of the primers is indicated by F (forward) or R (reverse). Oligonucleotides are named by the first nucleotide corresponding to the NHE2 promoter sequence with respect to the TIS. Restriction enzyme recognition sites are shown in bold.
| Direction/position | Sequence |
|---|---|
| F/−85 | 5′-TATCTCGAGTGTGCCGGCCAGGTCCC-3′ |
| F/−40 | 5′-TATCTCGAGCCGCGCCCGCCCCGCCC-3′ |
| F/−20 | 5′-TATCTCGAGCCCGCGGGCCGCCC-3′ |
| F/−10 | 5′-TATCTCGAGCGCCCTGGGCAGGGC-3′ |
| F/+4 | 5′-AGCTCGAGCCTGCCGCTGCGGCTGGAGAGC-3′ |
| F/+11 | 5′-AGCTCGAGCTGCGGCTGGAGAGCAGC-3′ |
| R/+4 | 5′-TATAAGCTTCCCTGCCCAGGGCGGCCCGC-3′ |
| R/+11 | 5′-TATAAGCTTCGGCAGGCCCCTGCCCAG-3′ |
| R/+150 | 5′-TATAAGCTTCCTCGCTGCGAGCAGGCGCAC-3′ |
| R/+71 | 5′-TATAAGCTTCTCAGCCGCCCTCCGCCGCCG-3′ |
| R/+41 | 5′-TATAAGCTTCCATGCCGGTGCGCTGCTCTCCAGC-3′ |
Cell culture and transfection
The C2BBe1 cell line, a sub-clone of Caco-2 human colonic epithelial cells, was cultured according to standard techniques as previously described [15]. Transient transfection was performed using Lipofectamine™ 2000 reagent (Life Technologies, Invitrogen) according to the manufacturer's protocol. Briefly, C2BBe1 cells were seeded at 1.5×105 cells/well on 12-well culture dishes and were transfected after 24 h at 80–90% confluency. A total of 2 μg of DNA, 1.6 μg of test plasmid DNA plus 0.4 μg of pSV-βgal plasmid DNA (a control for transfection efficiency) was used for each transfection. At 4 h after transfection, the DNA/Lipofectamine™ mixture was removed and replaced by regular growth medium (Dulbecco's modified Eagle's medium supplemented with 10% foetal bovine serum, 10 μg/ml transferrin, 50 units/ml penicillin G sodium and 50 μg/ml streptomycin sulfate). Cells were washed with PBS and lysed using a kit from Promega 48 h post-transfection. Luciferase activity was normalized to β-galactosidase activity in the same lysate and expressed either as a relative increase over the normalized activity of the vector or as a percentage of the control. Co-transfection experiments were performed as described above, except that 1.5 μg of test plasmid was co-transfected with 0.5 μg of either the expression vector or the empty vector. In co-transfection experiments luciferase activity was normalized to total protein concentration as determined by the Bradford protein assay system (Bio-Rad) and are presented as the fold-increase over that obtained with the control empty vector. Transfections in Drosophila SL2 cells were as described previously [24]. All transfection experiments were performed at least three times and each time in triplicate. Values are presented as means±S.E.M.
Nuclear extract preparation, GMSA (gel mobility-shift assay) and ChIP (chromatin immunoprecipitation)
Nuclear proteins were prepared from C2BBe1 cells, and DNA–protein interactions were assessed by GMSA as previously described [25]. ChIP assays were performed using the ChIP-It Express kit from Active Motifs according to the manufacturer's instructions. Briefly, C2BBe1 cells were grown to confluency and protein/chromatin was cross-linked with 1% formaldehyde for 10 min at room temperature (21–23 °C). The cross-linking was quenched by adding Glycine Stop-Fix solution for 5 min. The cells were rinsed twice with 1×PBS, collected and lysed in lysis buffer supplemented with protease inhibitor cocktail (both provided in the ChIP-It kit), on ice for 30 min. Subsequently, cells were homogenized in an ice-cold dounce homogenizer and nuclei were collected by gentle centrifugation (2400 g) for 10 min. The nuclei were suspended in 1.0 ml of shearing buffer and sheared to fragments of 200–800 bp using the enzymatic shearing cocktail provided in the kit for 8 min. Shearing efficiency was confirmed by gel electrophoresis prior to immunoprecipitation of the cross-linked chromatin with the Sp1 and Sp3 antibodies. A negative control antibody and a no antibody control were also used along with the experimental samples. A 50 μl aliquot of the sheared chromatin was used for each immunoprecipitation with 5 μg of antibody and 20 μl of Protein G magnetic beads. Immunoprecipitation was carried out overnight on a rotator at 4 °C. Subsequently the magnetic beads along with the co-precipitated immuno-complexes were collected, washed and resuspended in 100 μl of elution buffer (provided in the ChIP-It kit). Cross-linking of DNA proteins was reversed by incubation at 65 °C in the presence of NaCl and proteins were removed by proteinase K treatment at 42 °C. Immunoprecipitated DNA was purified and used for PCR amplification. As a positive control, a 10 μl aliquot of the sheared chromatin was used for input DNA preparation, and was subjected to reversal of the cross-linking, purification with phenol/chloroform extraction and ethanol precipitation and dissolved in 50 μl of elution buffer. For PCR, 2 μl of each ChIP DNA was examined using the forward 5′-GTGTGCCGGCCAGGTCCCCTTG-3′ and reverse 5′-TGCGAGCAGGCGCACTGCCTGGCG-3′ primers. The input DNA was diluted 10-fold prior to PCR amplification. The amplification products were resolved on a 1.5% agarose gel, stained with ethidium bromide and photographed.
Statistical analysis
All graphs and statistics were calculated in Microsoft Excel X (Microsoft Office X). Student's t tests were used to compare samples. P<0.05 was considered significantly different from control.
RESULTS
The NHE2 core promoter region
In a previous study using primer extension experiments we mapped the NHE2 TIS to an adenine residue 320 nucleotides upstream from the ATG translation start codon [15]. Additionally, by transient transfection assays, we showed that maximal promoter activity was retained in the shortest promoter construct containing only 85 bp upstream from the TIS. These observations suggested that the elements required for basal transcription activity are located in a DNA region downstream from the −85 position [23].
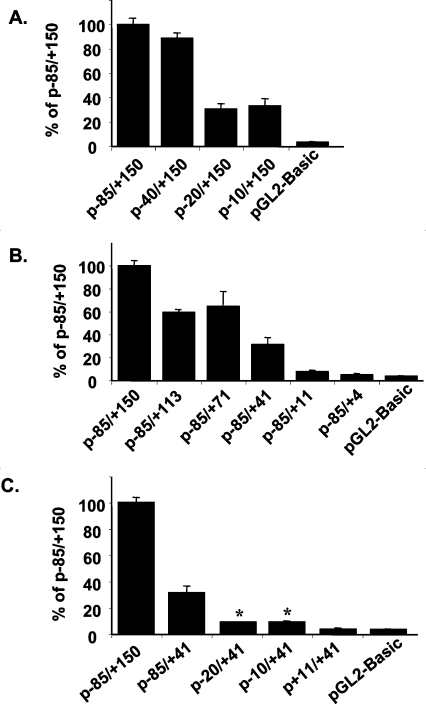
To characterize the functional regulatory elements of the proximal promoter region, we evaluated a series of promoter–reporter constructs containing various portions of the −85 to +150 fragment of the NHE2 promoter, which had the highest promoter activity. These plasmids were transiently transfected into C2BBe1 cells and reporter gene activity as a determinant of the NHE2 promoter activity was measured. The results in Figure 1 are presented as a percentage of the activity of the most active construct (p−85/+150), which was set arbitrarily at 100%. A 5′-deletion to position −40 revealed a minimal effect on promoter activity, indicating that all cis-acting elements necessary for the basal NHE2 promoter activity were located in the −40/+150 region. Further 5′-truncation of the promoter fragment to positions −20 and −10 exhibited an approx. 70% reduction in reporter gene activity compared with p−85/+150, indicating that a major positive regulatory element was harboured between nucleotides −40 to −20. The promoter-less pGL2-basic vector exhibited a 3% luciferase activity (Figure 1A).
Figure 1. Deletion and mutational analysis of the human NHE2 promoter.
Functional analyses were carried out with 5′- (A), 3′- (B) and internal- (C) deletion constructs. Luciferase reporter constructs containing the indicated promoter fragments of the NHE2 promoter along with pSV-βgal were transiently transfected into C2BBe1 cells. At 48 h post-transfection the cells were lysed and reporter gene activity was measured and normalized to β-galactosidase activity as indicated in the Experimental section. The luciferase activity of each deletion mutant is shown as a percentage of the activity of the parental plasmid p−85/+150, which was set at 100%. Values are the means±S.E.M., n≥4. *Significantly different from pGL2-basic.
Since p−10/+150 was able to promote approx. 30% reporter gene activity, we sought to obtain further information about the contribution of the DNA sequences corresponding to the 5′-UTR to the NHE2 promoter activity. We performed functional studies with a series of 3′-deletion constructs of p−85/+150 (Figure 1B). Removal of nucleotide sequences from positions +150 to +113 (p−85/+113), +150 to +71 (p−85/+71) and +150 to +41 (p−85/+41) exhibited 40%, 30% and 70% lower luciferase activity than p−85/+150 respectively, suggesting the presence of positive regulatory elements within the 5′-UTR. Further 3′-deletions to positions +11 and +4 led to the total loss of promoter activity.
To delineate further a functional minimal promoter region, we tested the constructs p−85/+41, p−20/+41, p−10/+41 and p+11/+41 for promoter activity (Figure 1C). The construct containing the −85 to +41 region, p−85/+41, which has lost the positive regulatory elements present in the 5′-UTR had substantially lower (31%) promoter activity compared with p−85/+150. A weak but reproducible reporter activity was observed in constructs p−20/+41 and p−10/+41 (Figure 1C). An approx. 10% promoter activity was exhibited by these constructs and, although this was much less than that of the parental construct, it was significantly higher (3-fold) than the background levels produced by pGL2-basic (3%). A further 5′-deletion to position +11 (p+11/+41) abolished the minimal promoter activity showing an activity similar to the background level (Figure 1C). The minimally active construct contained only 10 bp upstream from the transcription start site, suggesting that the nucleotide sequences surrounding the TIS are responsible for mediating transcription initiation as well as low-level promoter activity. Thus these results reveal that this small promoter region is sufficient to direct low-level promoter activity, and this activity is stimulated in the presence of upstream (p−85/+41) and downstream (p−10/+150) positive regulatory elements.
Identification of the potential positive regulatory elements in the NHE2 5′-UTR
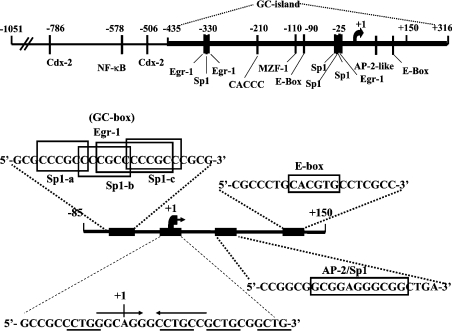
To correlate the results of our functional studies (Figure 1) with the cis-elements present in the proximal NHE2 promoter and its 5′-UTR, we compared the nucleotide sequences of the NHE2 promoter from bp −85 to +150 with the databases for DNA-binding transcription factors using MatInspector application (http://www.genomatix.de). A GC-box located between nucleotide −33 and −13 upstream from the TIS [15], and two sequence motifs corresponding to an AP2/Sp1 overlapping sequence and an E-box were identified in the 5′-UTR DNA. A schematic drawing of the NHE2 promoter region spanning the nucleotides −1051 to +150 (with respect to the TIS, at +1) and the location of the potential regulatory elements including those of the −85 to +150 region is shown in Figure 2. The results from our functional analyses (Figure 1) suggested that 5′-UTR DNA regions corresponding to bp +41 to +71 and +70 to +150 function as positive regulators of the NHE2 basal promoter activity.
Figure 2. Schematic representation of the human NHE2 promoter and core promoter region.
The promoter region between nucleotide −1051 to the ATG codon at +316 (with respect to the TIS shown by a bent arrow) including the location of the potential regulatory elements is shown. The region corresponding to bp −85 to +150 is amplified and the sequences of the GC-box centred at position −25 (containing potential overlapping binding sites for three Sp1 and one EGR-1), the TIS, an overlapping AP2/Sp1 binding site at +60 and an E-box at +113 are shown. The arrows in the TIS oligonucleotide represent the palindromic sequence. The TIS is shown as +1. C/GCTGC/G repeats are underlined. The boxes drawn around nucleotide sequences mark the core sequence of the protein binding sites as indicated.
Identification of a CpG-island spanning the NHE2 promoter
One of the features of the NHE2 proximal promoter region is the unusually high frequency of CpG-dinucleotides. CpG-dinucleotides normally are under-represented in the mammalian genome [26]. A greater than expected rate of CpG-dinucleotides is one of the criteria for the presence of CpG-islands. Thus the high incidence of these doublets prompted us to search the DNA surrounding the NHE2 proximal promoter for the presence of a CpG-island. Using the EMBOSS CpGplot, it was shown that the proximal promoter region was embedded in a CpG-island. To identify the 5′- and 3′-boundaries of this potential CpG-island, we used DNA sequences of a 1.6 kb genomic DNA from our previous studies that contained sequences corresponding to nucleotide sequences 1.0 kb upstream and 0.6 kb downstream from the TIS of NHE2 (GenBank® accession no. AF273748 and AF073299). Using CpGplot, a potential CpG-island was detected that spanned from bp −435 to +501 extending beyond the translation start site at +316 (Figure 2). The NHE2 putative CpG-island conforms well to the criteria set for the CpG-islands [27] by having a GC content of 76%, a length of 936 bp, and a ratio of observed compared with expected CpG-dinucleotides of 0.73.
Transcription factors Sp1 and Sp3 interact with the TIS of the human NHE2 gene
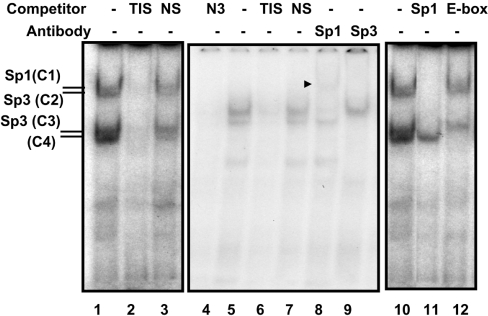
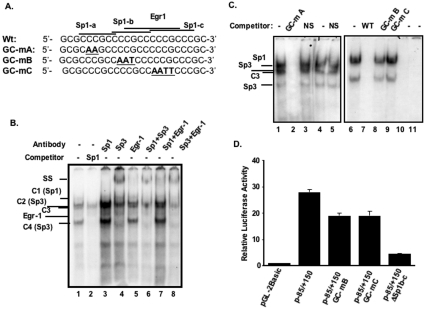
The results obtained from the functional analysis of the p−85/+4 and p−85/+11 (Figure 1B) were intriguing, as these plasmids, which contain the TIS and the upstream GC-box, had complete loss of promoter activity, whereas sequences corresponding to −10 to +41 exhibited low promoter activity. Therefore we hypothesized that either factors binding to the TIS extend beyond the sequences immediately surrounding the transcription start site, which may be disrupted in p−85/+4 and p−85/+11, or the transcription is initiated in a region between +11 to +41, as p−85/+41 exhibited a moderate (∼30%) promoter activity (Figure 1C). We ruled out the latter possibility since p+11/+41 had luciferase activity similar to the background level determined from the promoter-less vector and our previous primer extension experiments with RNA from Caco-2, T84 and C2BBe1 cells had displayed only one extension product mapping to the +1 position as in Figure 2 [15]. To test our first hypothesis and identify the potential proteins that might bind to the TIS region, we performed GMSAs. In these studies, incubation of nuclear proteins from C2BBe1 cells with an end-labelled probe (−12/+20) produced four DNA–protein complexes (Figure 3, lane 1). These nucleoproteins were shown to be specific by competition assays with an unlabelled probe, which eliminated all DNA–protein interactions, whereas an unrelated oligonucleotide used as a non-specific competitor did not alter DNA–protein complex formation (Figure 3, lanes 1 and 2). A careful visual examination of the nucleotide sequences surrounding the TIS revealed the presence of a palindrome sequence (GGCA+1GGGCCTGCC), which overlaps with the TIS indicated as +1 and extends to position +10. The inverted repeat elements have sequence homology with the Sp1 consensus sequence and we noted that the pattern of migration of these nucleoproteins (Figure 3) was similar to those of the Sp1 and Sp3 interactions with the Sp1-binding site that we have observed previously with the NHE3 and NHE2 promoters [23,24] (and see below). To verify the possibility of the presence of Sp1 family members in these complexes we used an unlabelled Sp1 oligonucleotide containing the Sp1 consensus sequence in competition experiments (Figure 3, lane 11). This competitor removed nucleoproteins C1–C3, but not C4, indicating that the protein components of these complexes were Sp1 related. In addition, an oligonucleotide from the NHE3 proximal promoter region, which binds Sp1 and Sp3 [24], also eliminated all of these complexes (Figure 3, lane 4). These findings were confirmed further by supershift assays where an antibody specific for Sp1 (Figure 3, lane 8) supershifted C1, and an anti-Sp3 antibody eliminated C2 and C3 (Figure 3, lane 9) revealing that the protein involved in the formation of C1 is Sp1 and that the protein component involved in the formation of C2 and C3 is Sp3.
Figure 3. Sp1 and Sp3 bind to the NHE2 TIS.
Gel-shift analysis was carried out using 5 μg of C2BBe1 cell nuclear proteins. The end-labelled probe used in these assays was the double-stranded oligonucleotide from −12 to +20, shown as TIS in Table 2. Binding reactions were performed in the presence or absence of competitors (50-fold molar excess) or in the presence of anti-Sp1 and -Sp3 antibodies (2 μg/lane) as indicated at the top of the Figure. All oligonucleotides used in the experiments are shown in Table 2. Four bands showed specific binding to the −12/+20 probe and are indicated by C1–C4. To separate the DNA–protein complexes the gel represented by numbers 4–9 was run for a longer time than the others that resulted in weaker interactions of C4. NS, non-specific nucloetide; N3, an oligonucleotide from the NHE3 promoter that we have shown previously binds Sp1 and Sp3. An arrowhead indicates the Sp1-supershifted band.
In the competition assays C4 was removed with the unlabelled probe, but not with the non-specific oligonucleotide or Sp1 oligonucleotide, suggesting that its interaction with the probe may be specific and unrelated to Sp1 family members. The TIS probe (−13 to +20) contains a sequence CCGCTG, which shows a weak similarity to the consensus E-box motif CAGCTG. To determine whether the C4 complex could be related to E-box-binding proteins, we performed competition assays with the consensus E-box oligonucleotide. Interestingly, the C4 complex was eliminated by the E-box-consensus-binding site (Figure 3, lane 9), however, antibodies to USF1 or USF2 did not affect the C4 complex in supershift assays (results not shown), indicating that the protein component of this complex may share homology with USF1/USF2 for the DNA-binding site but it is serologically different from these transcription factors. We also noticed that a longer electrophoresis time resulted in the elimination of the majority of the C4 complex, implying that it might form weak binding with the probe, perhaps due to low binding affinity of the protein to a non-canonical cis-element or a suboptimal ionic strength of the buffers used in the binding reaction or running buffer. The identity of the protein in nucleoprotein C4 is not clear at this point. In addition, there are several C/GCTGC/G repeats in the TIS region, the significance of which is not known. These sequences are underlined in Figure 2.
Mutational analysis of the TIS
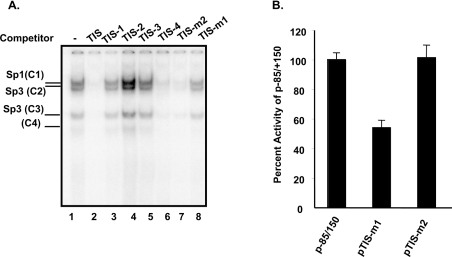
To determine whether sequences contained within the transcription initiation region participate in the transcriptional activity, specific mutations were designed to disrupt the potential protein-binding sites (Table 2). When the unlabelled TIS probe was divided into two portions at nucleotide +4 (TIS-1 and TIS-2) and used as competitors neither one was able to compete with the formation of any of the DNA–protein complexes (Figure 4A, lanes 3 and 4). Oligonucleotide TIS-3, containing nucleotides −3 to +12, was also unable to compete with interactions of these proteins with the probe (Figure 4A, lane 5), whereas TIS-4, which contained five additional base pairs in it 5′-end compared with TIS-3, removed all complexes. These results suggested that sequences around the +4 position are required for complex formation. Next, we introduced base substitutions in the 3′- and 5′- flanking regions of the palindrome. In competition assays, TIS-m1 (which contains base substitutions in the 5′ inverted sequence) was unable to compete (Figure 4A, lane 8), whereas mutations in the 3′ inverted repeat sequence competed out all complexes (Figure 4A, lane 7). The TFIID consensus sequence was also ineffective for competition with any of the nucleoproteins (results not shown). When TIS-m1 was used as an end-labelled probe in GMSAs, these complexes were no longer detectable, indicating that the altered bases in TIS-m1 resulted in loss of factor recognition sequences. In contrast, mutations in TIS-m2 had no effect on complex formation (results not shown).
Table 2. Primers used for GMSA and site-specific mutagenesis of the TIS.
Name, sequence and end-points of the oligonucleotides are indicated. Base substitutions are shown in bold. The nucleotide sequences of the sense strands are given.
| Name | Sequence | Position |
|---|---|---|
| TIS | 5′-GCCGCCCTGGGCAGGGCCTGCCGCTGCGGCTG-3′ | −12 to +20 |
| TIS-1 | 5′-GCCGCCCTGGGCAGG-3′ | −12 to +3 |
| TIS-2 | 5′-GCCTGCCGCTGCGGCTG-3′ | +4 to +20 |
| TIS-3 | 5′-GGCAGGGCCTGCCGC-3′ | −3 to +12 |
| TIS-4 | 5′-CCCTGGGCAGGGCCTGCCGC-3′ | −8 to +12 |
| TIS-m1 | 5′-CCCTGGGTTCGGCCTGCCGC-3′ | −10 to +12 |
| TIS-m2 | 5′-CCCTGGGCAGGGCTTTCCGC-3′ | −6 to +18 |
| TIS-m3 | 5′-CTTTGGGCAGGGCCTGCCGC-3′ | −8 to +12 |
| m-TIS #1 | 5′-GCCGCCCTGGGTTCGGCCTGCCGC-3′ | −12 to +12 |
| m-TIS #2 | 5′-CTGGGCAGGGCTTTCCGCTGCGGC-3′ | −6 to +18 |
Figure 4. Mapping of the Sp1-binding site and functional analysis of the mutated TIS element.
(A) GMSAs were performed with nuclear proteins from C2BBe1 cells and with the wild-type TIS oligonucleotide as the end-labelled probe. As expected, four nucleoproteins were formed and are indicated on the left-hand side. Nuclear proteins (5 μg) and a 50-fold molar excess of competitor oligonucleotides were used in each reaction with 50000 c.p.m. of the end-labelled probe. The oligonucleotides that were truncated or altered at specific nucleotides and used for competition assays or construction of mutated plasmids are shown in Table 2. (B) Mutated constructs were generated in the context of p−85/+150 by site-specific mutagenesis using primers m-TIS#1 and m-TIS#2 shown in Table 2 and as described in the Experimental section. C2BBe1 cells were transfected with the wild-type and mutated constructs along with pSV-βgal. Normalized luciferase activities were determined and compared with the wild-type promoter activity, which was set arbitrarily at 100%. All transfections were repeated three times each in triplicate.
To analyse further the contribution of the palindromic sequence to the NHE2 promoter activity, TIS-m1 and TIS-m2 mutations were introduced into the construct p−85/+150 and the luciferase activity of the mutant constructs was determined after transient transfection into C2BBe1 cells (Figure 4B). Transfection of pTIS-m1, in which the TIS along with the overlapping 5′ half of the palindrome were mutated, led to a 43% reduction in reporter gene activity compared with the wild-type construct. In contrast, transfection of pTIS-m2, containing a mutation in the 3′ half of palindrome, did not alter the reporter gene activity.
The transcription initiation region is trans-activated by the transcription factor Sp1
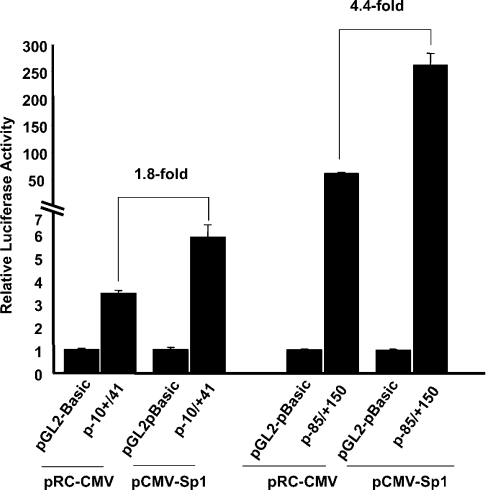
We have shown that transcription factors Sp1 and Sp3 interact with the transcription initiation region (Figures 3 and 4). These findings imply that these proteins may trans-activate the promoter construct p−10/+41. To determine this directly, we performed co-transfection assays with a pRC-CMV derivative Sp1 expression vector and p−10/+41 in C2BBe1 cells. In these experiments 1.5 μg of test plasmid was co-transfected with 0.5 μg of pCMV-Sp1 or the empty vector. The results are shown in Figure 5 and are compared with the activity of p−85/+150 in the presence or absence of the Sp1 expression vector. As shown, Sp1 could trans-activate the activity of p−10/+41(1.8-fold), although at the same concentration it had a higher effect on p−85/+150 (4.4-fold).
Figure 5. Co-transfection experiments in C2BBe1 cells.
C2BBe1 cells were co-transfected with 1.0 μg of test plasmid along with either 0.5 μg of pCMV-Sp1 or the empty vector pRC-CMV. Luciferase activity was measured 48 h post-transfection and normalized to total cellular protein. The results are shown as normalized activity relative to the activity of pGL2-basic, and the fold-increase by co-transfected Sp1 expression vector was calculated as the activity in the presence of the expression vector divided by the activity of reporter construct in the presence of the empty vector. The results represent at least three individual experiments performed in triplicate.
Sp1 and Sp3 bind to the GC-box centered at −25 and enhance NHE2 promoter activity
We next sought to characterize the GC-box at the 5′-flanking region and investigate its contribution to the NHE2 promoter activity. For this purpose a combination of DNA–protein binding assays along with site-specific mutagenesis in the factor binding sites and transient transfection experiments were employed. Nuclear proteins binding to the GC-box were examined by GMSA in combination with competition and supershift analyses using nuclear extracts from C2BBe1 cells and a probe spanning the GC-box from bp −37 to −14. This element is composed of three overlapping Sp1- and one EGR-1-binding sites (Figure 6A) and formed four nucleoprotein complexes (C1–C4) with C2BBe1 nuclear proteins in GMSA (Figures 6B or 6C, lane 1). To examine the specificity of these complexes, competition assays were carried out. Competition experiments in the presence of unlabelled Sp1 oligonucleotide containing the consensus Sp1-binding site resulted in removal of C1, C2 and C4 bands (Figure 6B, lane 2), suggesting that the protein components of these complexes belonged to Sp1 family members. All four complexes were eliminated when unlabelled probe was used as a competitor (Figure 6C, lane 7). The identity of the protein component of these DNA–protein complexes was determined by supershift analysis using anti-Sp1, -Sp3 and -EGR-1 antibodies (Figure 6B, lanes 3–8). These results revealed that Sp1formed C1, and C2 and C4 were generated by interaction with Sp3. Moreover a very faint band corresponding to EGR-1–DNA binding was only visible subsequent to the removal of Sp1 or Sp3 by their cognate antibodies or competitor Sp1 oligonucleotide. The identity of the protein forming the C3 band, which is unrelated to Sp1, is not known.
Figure 6. Characterization of the factors binding to the GC-box and their role in NHE2 promoter activity.
(A) The sequences of the oligonucleotide probes or competitors used in DNA-binding studies are shown. (B) GMSA was performed with the wild-type end-labelled oligonucleotide spanning from nucleotide residues −37 to −14 (GC-WT) of the human NHE2 promoter and incubated with 5 μg of C2BBe1 nuclear proteins (lanes 1–8). Lane 1, no competitor; lane 2, 50-fold molar excess of an unlabelled competitor oligonucleotide containing the Sp1 consensus sequence; lanes 3–8 supershift assays, in the presence of 2 μg of antibodies specific for Sp1, Sp3 and EGR-1 (lanes 3–5) or a combination of those antibodies (lanes 6–8). (C) GMSA was performed with nuclear extracts from C2BBe1 cells and end-labelled wild-type probe (lanes 1–3 and 6–9), GC-mA (lanes 4 and 5), GC-mB (lane 10) and GC-mC (lane 11). The competitor oligonucleotides employed in these assays are shown in (A), and are indicated at the top the Figure. (D) Transfection analysis of the mutant constructs containing altered Sp1-binding sites. Transient transfections were performed with constructs harbouring base substitutions (GC-mB and GC-mC) or deletions (ΔSp1b-c) in the GC-box and the control p−85/+150. Each construct (1.6 μg) was co-transfected along with pSV-βgal (0.4 μg) into C2BBe1 cells and processed as described in the legend to Figure 1. The y-axis shows the normalized luciferase activity of each construct relative to the normalized luciferase activity of the promoter-less vector pGL2-basic. The values presented are means±S.E.M. of at least three independent experiments performed in triplicate.
Further competition assays were carried out with mutated oligonucleotides carrying base substitutions in Sp1-binding sites (Figure 6A). Competition with the unlabelled GC-mA oligonucleotide eliminated all nucleoproteins (Figure 6C, lane 2), indicating that this oligonucleotide was competent for binding to all of the proteins that interacted with the probe. This was confirmed further by GMSA, where GC-mA was used as the end-labelled probe (Figure 6C, lanes 4 and 5). In contrast, interaction of nuclear proteins with the labelled probe was not altered in the presence of GC-mC (Figure 6C, lane 9), which retains an unaltered Sp1-a and Sp1-b, whereas competition with mutated Sp1-b oligonucleotide, GC-mB, showed a minor reduction in the intensity of all of these complexes (Figure 6C, lane 8). These results revealed that, although the consensus Sp1-binding sequence CCCGCC at the Sp1-b site is present in GC-mC, it is unable to interact with its respective proteins, indicating that sequences flanking the consensus sequence are important for the DNA–protein interaction at this site. Furthermore, in GC-mB the Sp1-c motif is intact yet it could barely compete with complex formation with the probe, suggesting that the Sp1-c motif also does not bind to Sp1/Sp3 efficiently. As shown in Figure 6(C) (lanes 10 and 11) no DNA–protein interactions were detected in GMSA using GC-mB and GC-mC as end-labelled probes.
Sp1 and Sp3 are essential for NHE2 promoter activity
To evaluate the functional role of Sp1 and Sp3 binding to the GC-box at −25 in NHE2 promoter activity, the same site-specific mutations that were used in the GMSA and prevented the Sp1/Sp3 interaction with the probe were introduced into p−85/+150 to generate p−85/+150 (GC-mB) and p−85/+150 (GC-mC). Another mutant construct was generated by deletion of nucleotide sequences from bp −23 to −11, which removed binding sites Sp1-b and Sp1-c. The wild-type and mutant constructs were transiently transfected into C2BBe1 cells and the reporter gene activity was analysed. The results shown in Figure 6(D), revealed that mutations GC-mB or GC-mC each reduced the reporter gene activity by approx. 30% of the wild-type promoter, whereas the internal deletion mutant in which the nucleotide residues corresponding to both Sp1-b and Sp1-c were deleted had a dramatic decrease in transcriptional activity (80%). Thus these studies reveal that the altered nucleotides in Sp1-b and Sp1-c contribute to the promoter activity and underscore the critical role of the GC-box element at −25 in the basal transcriptional activity of the NHE2 gene.
Sp1 and Sp3 act as positive regulators of the human NHE2 promoter in Drosophila SL2 cells
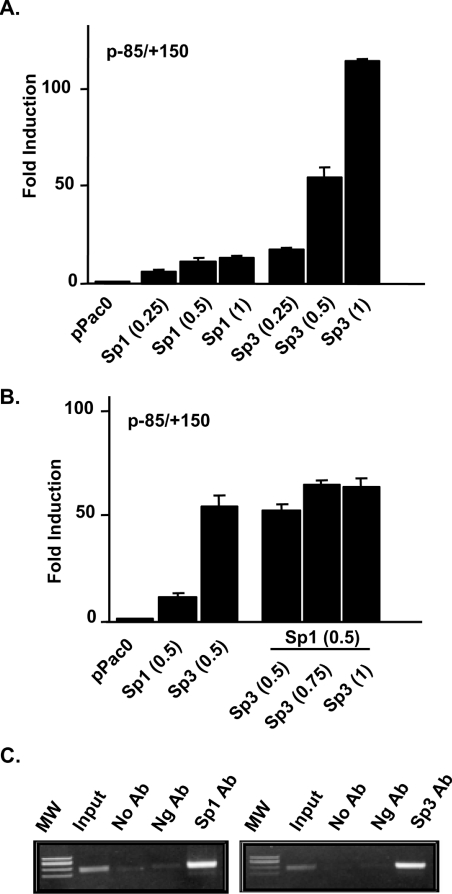
To confirm the involvement of transcription factors Sp1 and Sp3 in transcriptional regulation of the NHE2 promoter, the effect of Sp1 and Sp3 overexpression on NHE2 promoter regulation was examined by co-transfection studies in Drosophila SL2 cells. SL2 cells are devoid of endogenous Sp1 [28,29] and have been used as an invaluable model system to study the role of exogenously introduced factors on transactivation of co-transfected promoters from various genes. The NHE2 promoter–reporter construct p−85/+150 along with pPacSp1, pPacSp3 and pPacSp4 or a combination of these expression vectors were co-transfected into SL2 cells and the effect of these transcription factors on the NHE2 promoter activity was determined relative to the empty vector (Figure 7). In the absence of these transcription factors, the reporter construct p−85/+150 was completely inactive in SL2 cells. Sp1 and Sp3 were both able to dose-dependently transactivate the NHE2 promoter; however, Sp3 trans-activated the basal promoter activity at dramatically higher levels than those of the Sp1 expression vector (Figure 7A). In contrast, Sp4 expression had no effect on NHE2 promoter activity (results not shown). When added together, co-expression of a constant amount of Sp1 with increasing concentrations of Sp3 expression vector led to diminished Sp3-stimulated promoter activity (Figure 7B), suggesting that, at higher concentrations of Sp3, Sp1 might play an inhibitory role in controlling the human NHE2 promoter activity and that the ratio of Sp1 to Sp3 in the cell may be a factor in the level of NHE2 gene expression.
Figure 7. Sp1 and Sp3 trans-activate the proximal promoter of the NHE2 gene.
(A) Drosophila SL2 cells were co-transfected with 1.0 μg of p−85/+150 construct and concentrations of the Sp1 or Sp3 expression vectors as indicated. (B) SL2 cells were co-transfected with a constant concentration of pPacSp1 (0.5 μg) plus 0.5, 0.75 or 1.0 μg of pPacSp3. Transfected cells were harvested after 48 h and luciferase activities were determined using the luciferase assay system from Promega. Luciferase activity was normalized to cellular protein content. The activity indicated represents the fold-induction in luciferase activity relative to that obtained from co-transfection with the empty vector pPac0. These experiments were repeated three times each in duplicate and the results from one such experiment is shown. (C) ChIP assays with anti-Sp1 and -Sp3 antibodies. Cross-linked chromatin was isolated from C2BBe1 cells subsequent to formaldehyde treatment. ChIP assays were performed with anti-Sp1 and -Sp3 antibodies. Primers indicated in the text were used to amplify the NHE2 promoter region using the co-immunoprecipitated DNA as template. The PCR products were resolved on a 1.5% agarose gel. Ab, antibody; Ng Ab, negative control antibody.
Sp1 and Sp3 are associated with the NHE2 promoter in vivo
We have demonstrated that Sp1 and Sp3 can bind to the NHE2 proximal promoter region and the exogenous expression of these transcription factors can trans-activate NHE2 promoter-driven reporter gene activity. To examine the interactions of these transcription factors with the NHE2 promoter in vivo, we performed ChIP assays. C2BBe1 cells were grown to confluence, chromatin was cross-linked using formaldehyde and sheared chromatin was isolated and subjected to immunoprecipitation using anti-Sp1, -Sp3 and negative control antibodies. Following purification and reversal of cross-linking, proteins were digested, immunoprecipitated DNA was purified and subjected to PCR amplification. These results revealed that the NHE2 promoter region was enriched by immunoprecipitation with Sp1 and Sp3 antibodies compared with the negative control antibody (Figure 7C), demonstrating that both Sp1 and Sp3 are associated with the NHE2 promoter on the chromatin in vivo in basal growth conditions.
DISCUSSION
In the present study, we aimed to identify the cis-acting DNA elements and their cognate transcription factors that are required for the expression of the human NHE2 gene in the basal constitutive condition. We have specifically focused on the DNA elements in the proximal promoter region and the 5′-UTR. Our functional analyses of the promoter region show that a GC-box just upstream from the TIS is essential for NHE2 promoter activity. We have identified transcription factors Sp1 and Sp3 as activators of the basal NHE2 promoter activity that not only bind to the upstream GC-box, but also to the TIS.
In an attempt to delineate the cis-elements that mediate the constitutive activity of the NHE2 promoter, we generated a number of 5′- and 3′-deletions in a construct carrying the NHE2 proximal promoter. Our functional analyses revealed that, in addition to a critical GC-box in the 5′-flanking region, the 5′-UTR of the human NHE2 gene carries important cis-elements that are involved in the constitutive expression of the human NHE2 gene. We mapped the positive regulatory elements of the 5′-UTR to an AP2/Sp1 element at +60 and an E-box at +113. We showed that maximal constitutive promoter activity is directed by sequences from −40 to +150, indicating that not only the sequences immediately upstream from and spanning the TIS are critical for the constitutive activity of the NHE2 promoter, but also elements located in the 5′- UTR are essential for the full activity of this promoter. In addition, modulation of these factors under certain stimulatory conditions may serve to mediate the regulatory effects of those particular signals. E-box motifs have been identified within the 5′-UTR of the PANDER (pancreatic-derived factor) promoter [30] and shown to mediate the glucose-responsiveness as well as tissue-specific expression of the PANDER gene. Similarly, multiple AP-2 elements in the first exon of the human vacuolar H+-ATPase B2 subunit gene are required for transcriptional activation of the B2 promoter during monocyte to macrophage differentiation [31].
Promoters such as the NHE2 promoter that lack the TATA- and CCAAT-box often contain multiple Sp1-binding sites and may also contain additional regulatory elements such as the transcription Inr (Initiator). The Inr elements have been shown to act either with a TATA-box or in its absence independently to promote transcription [32]. The sequences immediately surrounding the TIS of the NHE2 gene, GCA+1GGG, exhibit weak homology with the transcription initiator consensus sequence YCA+1NT/AY [33] (Y is a pyrimidine and N is a T or G or C, but rarely an A), where only the core nucleotides CA are conserved. However, as suggested by Smale [32], pyrimidines may not be required in all positions indicated in the consensus sequence, although their presence enhances Inr activity. One of the factors that may bind the Inr element is TFII-I [34,35], which, upon binding through its interactions with the TBP (TATA-binding protein), recruits the transcription pre-initiation complex to the transcription start site and potentiates the transcription. TFII-I is a helix–loop–helix protein, which has been shown to interact with the transcription factor USF and activate transcription [36–38]. Among other proteins, that bind to the Inr element and enhance transcription of the target gene are the E-box-binding protein USF1 [36], Yin Yang protein YYI [39] and Sp1 [40,41]. Analysis of the DNA sequences surrounding the NHE2 TIS revealed that the TIS is overlapped with a palindrome. Using GMSAs and supershift experiments we showed that transcription factors Sp1 and Sp3 interact with the 5′ flank of the palindrome and TIS. A small DNA fragment spanning the TIS (bp −10 to +41) exhibited low levels of reporter gene activity in the absence of the upstream and downstream activator sequences. Activity of this fragment was enhanced approx. 3.5-fold in the presence of either upstream GC-box or downstream activator elements. Mutations in the TIS sequence, which also eliminated Sp1/Sp3 binding, led to a 43% reduction in NHE2 promoter activity. Through these experiments we established that Sp1 and Sp3 interact with the NHE2 transcription initiation region and may be involved in both transcriptional activation as well as transcription. These results are consistent with the notion that Sp1 can recruit the basal transcription machinery to the promoter region in the absence of the TATA element. Sp1 is important both in transcriptional activation and initiation [41] and can be regulated by multiple regulatory mechanisms in a cell- and tissue-specific manner [42,43].
Functional analyses of the NHE2 promoter suggested that the GC-box centered at −25 is essential for the optimal promoter activity. By GMSA and supershift experiments, we identified Sp1 and Sp3 as trans-acting factors that bind to this region at basal growth conditions. Competition and mutational analysis revealed that the Sp1-b-binding site might be the only authentic Sp1 motif, as mutant oligonucleotides with intact Sp1-a or Sp1-c did not bind to nuclear proteins. Despite the lack of protein binding to GC-mB and GC-mC probes, the constructs containing the same mutations had relatively high promoter activity (70% of the control). In contrast, an internal deletion that removed sequences corresponding to the Sp1-b and Sp1-c motifs severely reduced the activity by 80% compared with control. These results suggest that co-operation with the downstream regulatory elements may be responsible for stabilizing DNA–protein interactions in the mutated constructs and their promoter activity.
The contribution of Sp1 and Sp3 to the basal activity of NHE2 was confirmed by co-transfections in SL2 cells. Addition of Sp1 or Sp3 individually resulted in a dose-dependent activation of the reporter gene activity and Sp3 consistently stimulated the NHE2 promoter activity to a greater extent than that of Sp1. Furthermore, co-expression of Sp1 with different concentrations of Sp3 exhibited a negative effect on the Sp3-stimulated trans-activation of the reporter gene. These findings are in contrast with the effects of Sp1 and Sp3 on the rat NHE2 promoter, where a positive regulatory role for Sp1 and an inhibitory role for Sp3 have been reported [21]. In addition, a single DNA–protein complex was formed with the rat GC-box [21], whereas our results show four nucleoprotein complexes with the same region. These discrepancies may reflect cell- or species-specific differences and a differential mechanism of transcriptional control between the human and rat promoters. These findings strongly suggest that transcription factors Sp1 and Sp3 are involved in the constitutive expression of the NHE2 gene and show that these factors may be involved in both transcriptional activation and initiation of transcription. Previously we have shown by Western blot analysis that both Sp1 and Sp3 are expressed in C2BBe1 cells [23]; therefore they may be constantly available for the constitutive expression of the NHE2 gene. Metabolic acidosis in rats has been shown to increase the NHE2 transport activity, as well as mRNA and protein levels [44]. In addition, low pH has been implicated in enhanced DNA-binding activity of Sp1 to Sp1-binding sites and its stable interaction with TBP in the human epidermoid cancer KB cells [45]. Our recent findings also show increased DNA-binding affinity of Sp1/Sp3 to both the GC-box at −25 and the TIS in nuclear proteins from acidified (pH 7.0) C2BBe1 cells (J. Malakooti, Y. Zhu and K. Ramaswamy, unpublished work). Therefore it is possible that the enhanced DNA-binding activity of the Sp1 transcription factor could contribute to up-regulation of the NHE2 gene under conditions of cellular acidosis.
We also found that the NHE2 proximal promoter is embedded in a CpG-island. The CpG-islands often coincide with the promoter region in housekeeping genes where they are unmethylated; they may also be present in the promoter region of genes that are expressed in a tissue-/cell-specific fashion. The CpG-islands in the promoter region of the latter group of genes are usually methylated and the state of DNA methylation of the CpG-dinucleotides may govern the expression of the downstream gene. Although the NHE2 promoter contains a number of attributes of the housekeeping gene such as high GC-content, lack of TATA and CCAAT-boxes and the presence of a putative CpG-island, it is well-known that the NHE2 gene is expressed in a tissue-/cell-specific fashion [46,47]. However, the significance of a CpG-island in the NHE2 promoter has not yet been determined and needs to be established experimentally. In summary, our results revealed that cis-elements for Sp1/Sp3 binding in the NHE2 promoter are mainly responsible for constitutive NHE2 promoter activity and positive regulatory elements in the 5′-UTR may co-operate with the upstream elements to mediate the optimal NHE2 activity. These results also suggest that NHE2 gene expression may be regulated by several mechanisms, including the use of activator elements and their corresponding binding factors and by epigenetic mechanisms involving the potential CpG-island embedded in the promoter region. It is of interest to determine whether these elements affect regulation of the NHE2 gene expression in response to various extracellular stimuli, developmental and tissue-/cell-specific conditions.
Acknowledgments
These studies were supported by the NIDDK (National Institute of Diabetes and Digestive and Kidney diseases) grants RO1-DK33349 (to K. R.), RO1-DK67990 (to K. R.) and PO1-DK067887 (to K. R., P. K. D. and J. M.). We thank Dr R. Tjian (Department of Molecular and Cell Biology, University of California, Berkeley, CA, U.S.A.) and Dr G. Suske (Institut für Molekularbiologie und Tumorforschung, Philipps-Universität Marburg, Marburg, Germany) for their generosity in providing the expression vectors used in the present study.
References
- 1.Counillon L., Pouyssegur J. The expanding family of eucaryotic Na+/H+ exchangers. J. Biol. Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Orlowski J., Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 3.Zachos N. C., Tse M., Donowitz M. Molecular physiology of intestinal Na+/H+ exchange. Annu. Rev. Physiol. 2005;67:411–443. doi: 10.1146/annurev.physiol.67.031103.153004. [DOI] [PubMed] [Google Scholar]
- 4.Kiela P. R., Xu H., Ghishan F. K. Apical Na+/H+ exchangers in the mammalian gastrointestinal tract. J. Physiol. Pharmacol. 2006;57(Suppl. 7):51–79. [PubMed] [Google Scholar]
- 5.Gawenis L. R., Stien X., Shull G. E., Schultheis P. J., Woo A. L., Walker N. M., Clarke L. L. Intestinal NaCl transport in NHE2 and NHE3 knockout mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;282:G776–G784. doi: 10.1152/ajpgi.00297.2001. [DOI] [PubMed] [Google Scholar]
- 6.Schultheis P. J., Clarke L. L., Meneton P., Miller M. L., Soleimani M., Gawenis L. R., Riddle T. M., Duffy J. J., Doetschman T., Wang T., et al. Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat. Genet. 1998;19:282–285. doi: 10.1038/969. [DOI] [PubMed] [Google Scholar]
- 7.Xu H., Chen R., Ghishan F. K. Subcloning, localization, and expression of the rat intestinal sodium-hydrogen exchanger isoform 8. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G36–G41. doi: 10.1152/ajpgi.00552.2004. [DOI] [PubMed] [Google Scholar]
- 8.Lee-Kwon W., Kawano K., Choi J. W., Kim J. H., Donowitz M. Lysophosphatidic acid stimulates brush border Na+/H+ exchanger 3 (NHE3) activity by increasing its exocytosis by an NHE3 kinase A regulatory protein-dependent mechanism. J. Biol. Chem. 2003;278:16494–16501. doi: 10.1074/jbc.M300580200. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Silva N. L., Lucchesi P. A., Haworth R., Wang K., Michalak M., Pelech S., Fliegel L. Phosphorylation and regulation of the Na+/H+ exchanger through mitogen-activated protein kinase. Biochemistry. 1997;36:9151–9158. doi: 10.1021/bi970802f. [DOI] [PubMed] [Google Scholar]
- 10.Wade J. B., Liu J., Coleman R. A., Cunningham R., Steplock D. A., Lee-Kwon W., Pallone T. L., Shenolikar S., Weinman E. J. Localization and interaction of NHERF isoforms in the renal proximal tubule of the mouse. Am. J. Physiol. Cell Physiol. 2003;285:C1494–C1503. doi: 10.1152/ajpcell.00092.2003. [DOI] [PubMed] [Google Scholar]
- 11.Weinman E. J., Shenolikar S. The Na-H exchanger regulatory factor. Exp. Nephrol. 1997;5:449–452. [PubMed] [Google Scholar]
- 12.Weinman E. J., Steplock D., Tate K., Hall R. A., Spurney R. F., Shenolikar S. Structure–function of recombinant Na/H exchanger regulatory factor (NHE- RF) J. Clin. Invest. 1998;101:2199–2206. doi: 10.1172/JCI204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horie S., Moe O., Yamaji Y., Cano A., Miller R. T., Alpern R. J. Role of protein kinase C and transcription factor AP-1 in the acid-induced increase in Na/H antiporter activity. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5236–5240. doi: 10.1073/pnas.89.12.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao G. N., Sardet C., Pouyssegur J., Berk B. C. Na+/H+ antiporter gene expression increases during retinoic acid- induced granulocytic differentiation of HL60 cells. J. Cell. Physiol. 1992;151:361–366. doi: 10.1002/jcp.1041510217. [DOI] [PubMed] [Google Scholar]
- 15.Malakooti J., Dahdal R. Y., Dudeja P. K., Layden T. J., Ramaswamy K. The human Na+/H+ exchanger NHE2 gene: genomic organization and promoter characterization. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G763–G773. doi: 10.1152/ajpgi.2001.280.4.G763. [DOI] [PubMed] [Google Scholar]
- 16.Muller Y. L., Collins J. F., Bai L., Xu H., Ghishan F. K. Molecular cloning and characterization of the rat NHE-2 gene promoter. Biochim. Biophys. Acta. 1998;1442:314–319. doi: 10.1016/s0167-4781(98)00191-2. [DOI] [PubMed] [Google Scholar]
- 17.Malakooti J., Memark V. C., Dudeja P. K., Ramaswamy K. Molecular cloning and functional analysis of the human Na+/H+ exchanger NHE3 promoter. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;282:G491–G500. doi: 10.1152/ajpgi.00273.2001. [DOI] [PubMed] [Google Scholar]
- 18.Cano A. Characterization of the rat NHE3 promoter. Am. J. Physiol. 1996;271:F629–F636. doi: 10.1152/ajprenal.1996.271.3.F629. [DOI] [PubMed] [Google Scholar]
- 19.Kandasamy R. A., Orlowski J. Genomic organization and glucocorticoid transcriptional activation of the rat Na+/H+ exchanger Nhe3 gene. J. Biol. Chem. 1996;271:10551–10559. doi: 10.1074/jbc.271.18.10551. [DOI] [PubMed] [Google Scholar]
- 20.Bai L., Collins J. F., Muller Y. L., Xu H., Kiela P. R., Ghishan F. K. Characterization of cis-elements required for osmotic response of rat Na+/H+ exchanger-2 (NHE-2) gene. Am. J. Physiol. 1999;277:R1112–R1119. doi: 10.1152/ajpregu.1999.277.4.R1112. [DOI] [PubMed] [Google Scholar]
- 21.Bai L., Collins J. F., Xu H., Ghishan F. K. Transcriptional regulation of rat Na+/H+ exchanger isoform-2 (NHE- 2) gene by Sp1 transcription factor. Am. J. Physiol. Cell. Physiol. 2001;280:C1168–C1175. doi: 10.1152/ajpcell.2001.280.5.C1168. [DOI] [PubMed] [Google Scholar]
- 22.Xu H., Collins J. F., Bai L., Kiela P. R., Lynch R. M., Ghishan F. K. Epidermal growth factor regulation of rat NHE2 gene expression. Am. J. Physiol. Cell. Physiol. 2001;281:C504–C513. doi: 10.1152/ajpcell.2001.281.2.C504. [DOI] [PubMed] [Google Scholar]
- 23.Malakooti J., Sandoval R., Memark V. C., Dudeja P. K., Ramaswamy K. Zinc finger transcription factor Egr-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G653–G663. doi: 10.1152/ajpgi.00010.2005. [DOI] [PubMed] [Google Scholar]
- 24.Malakooti J., Sandoval R., Amin M. R., Clark J., Dudeja P. K., Ramaswamy K. Transcriptional stimulation of the human NHE3 promoter activity by PMA: PKC independence and involvement of the transcription factor EGR-1. Biochem. J. 2006;396:327–336. doi: 10.1042/BJ20051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: Wiley & Sons; 1995. Current Protocols in Molecular Biology. [Google Scholar]
- 26.Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 27.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 28.Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 29.Seto E., Shi Y., Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 30.Burkhardt B. R., Yang M. C., Robert C. E., Greene S. R., McFadden K. K., Yang J., Wu J., Gao Z., Wolf B. A. Tissue-specific and glucose-responsive expression of the pancreatic-derived factor (PANDER) promoter. Biochim. Biophys. Acta. 2005;1730:215–225. doi: 10.1016/j.bbaexp.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Lee B. S., Underhill D. M., Crane M. K., Gluck S. L. Transcriptional regulation of the vacuolar H+-ATPase B2 subunit gene in differentiating THP-1 cells. J. Biol. Chem. 1995;270:7320–7329. doi: 10.1074/jbc.270.13.7320. [DOI] [PubMed] [Google Scholar]
- 32.Smale S. T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 33.Javahery R., Khachi A., Lo K., Zenzie-Gregory B., Smale S. T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A. L., Meisterernst M., Pognonec P., Roeder R. G. Cooperative interaction of an initiator-binding transcription initiation factor and the helix–loop–helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 35.Roy A. L., Roeder R. G. Initiator element binding protein TFII-I: a tale of two sites. Indian J. Biochem. Biophys. 1994;31:14–19. [PubMed] [Google Scholar]
- 36.Du H., Roy A. L., Roeder R. G. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 1993;12:501–511. doi: 10.1002/j.1460-2075.1993.tb05682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy A. L., Du H., Gregor P. D., Novina C. D., Martinez E., Roeder R. G. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A. L., Malik S., Meisterernst M., Roeder R. G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 39.Lee J. S., Galvin K. M., Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 41.Bouwman P., Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 42.Solecki D., Schwarz S., Wimmer E., Lipp M., Bernhardt G. The promoters for human and monkey poliovirus receptors. Requirements for basic and cell type-specific activity. J. Biol. Chem. 1997;272:5579–5586. doi: 10.1074/jbc.272.9.5579. [DOI] [PubMed] [Google Scholar]
- 43.Kovarik A., Lu P. J., Peat N., Morris J., Taylor-Papadimitriou J. Two GC boxes (Sp1 sites) are involved in regulation of the activity of the epithelium-specific MUC1 promoter. J. Biol. Chem. 1996;271:18140–18147. doi: 10.1074/jbc.271.30.18140. [DOI] [PubMed] [Google Scholar]
- 44.Lucioni A., Womack C., Musch M. W., Rocha F. L., Bookstein C., Chang E. B. Metabolic acidosis in rats increases intestinal NHE2 and NHE3 expression and function. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G51–G56. doi: 10.1152/ajpgi.00529.2001. [DOI] [PubMed] [Google Scholar]
- 45.Torigoe T., Izumi H., Yoshida Y., Ishiguchi H., Okamoto T., Itoh H., Kohno K. Low pH enhances Sp1 DNA binding activity and interaction with TBP. Nucleic Acids Res. 2003;31:4523–4530. doi: 10.1093/nar/gkg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malakooti J., Dahdal R. Y., Schmidt L., Layden T. J., Dudeja P. K., Ramaswamy K. Molecular cloning, tissue distribution, and functional expression of the human Na+/H+ exchanger NHE2. Am. J. Physiol. 1999;277:G383–G390. doi: 10.1152/ajpgi.1999.277.2.G383. [DOI] [PubMed] [Google Scholar]
- 47.Hoogerwerf W. A., Tsao S. C., Devuyst O., Levine S. A., Yun C. H., Yip J. W., Cohen M. E., Wilson P. D., Lazenby A. J., Tse C. M., Donowitz M. NHE2 and NHE3 are human and rabbit intestinal brush-border proteins. Am. J. Physiol. 1996;270:G29–G41. doi: 10.1152/ajpgi.1996.270.1.G29. [DOI] [PubMed] [Google Scholar]