Abstract
Human α2M (α2-macroglobulin) and the complement components C3 and C4 are thiol ester-containing proteins that evolved from the same ancestral gene. The recent structure determination of human C3 has allowed a detailed prediction of the location of domains within human α2M to be made. We describe here the expression and characterization of three α2M domains predicted to be involved in the stabilization of the thiol ester in native α2M and in its activation upon bait region proteolysis. The three newly expressed domains are MG2 (macroglobulin domain 2), TED (thiol ester-containing domain) and CUB (complement protein subcomponents C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1) domain. Together with the previously characterized RBD (receptor-binding domain), they represent approx. 42% of the α2M polypeptide. Their expression as folded domains strongly supports the predicted domain organization of α2M. An X-ray crystal structure of MG2 shows it to have a fibronectin type-3 fold analogous to MG1–MG8 of C3. TED is, as predicted, an α-helical domain. CUB is a spliced domain composed of two stretches of polypeptide that flank TED in the primary structure. In intact C3 TED interacts with RBD, where it is in direct contact with the thiol ester, and with MG2 and CUB on opposite, flanking sides. In contrast, these α2M domains, as isolated species, show negligible interaction with one another, suggesting that the native conformation of α2M, and the consequent thiol ester-stabilizing domain–domain interactions, result from additional restraints imposed by the physical linkage of these domains or by additional domains in the protein.
Keywords: complement component C3, crystal structure, domain organization, α2-macroglobulin (α2M), receptor-binding domain, thiol ester
Abbreviations: α2M, α2-macroglobulin; CUB, complement protein subcomponents C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1; DSC, differential scanning calorimetry; HSQC, heteronuclear single-quantum coherence; IAF, 5-iodoacetamidofluorescein; IPTG, isopropyl β-D-thiogalactoside; ITC, isothermal titration calorimetry; LRP, low-density-lipoprotein-receptor-related protein; MG1, macroglobulin domain 1; Ni-NTA, Ni2+-nitrilotriacetate; PEG, poly(ethylene glycol); RBD, receptor-binding domain; rmsd, root mean square deviation; TED, thiol ester-containing domain
INTRODUCTION
Human α2M (α2-macroglobulin) is a homotetrameric plasma glycoprotein composed of polypeptide chains of 1451 residues [1]. It is found in all individuals, suggesting that it is essential for life, and it is highly abundant (1–2 g/l). In addition to being a pan-proteinase inhibitor capable of inhibiting proteinases of all mechanistic families by a remarkable physical sequestration mechanism [2] that has been likened to the closing of a Venus fly trap around its prey, α2M can tightly bind a number of growth factors in a non-covalent, reversible manner [3,4]. The massive conformational changes that result in proteinase entrapment are initiated by proteolytic cleavage of the bait region by the prey proteinase. The bait region is a stretch of approx. 30 residues located roughly in the middle of the polypeptide chain; it is a flexible region [5] containing many types of residues [6,7] that give it a wide spectrum of cleavage sites.
In addition to trapping the proteinase, the conformation changes result in activation of the thiol ester that is present between the side chains of residues Cys949 and Glx952 and in exposure of the four previously cryptic RBDs (receptor-binding domains), each of which is formed by the C-terminal 138 residues of the monomer (Figure 1) [8]. The activated thiol ester reacts rapidly with any available nucleophile [9]. If this is provided by a side chain from the attacking proteinase (usually a lysine ϵ-amino group), it results in covalent linkage between the proteinase and α2M [10]. Exposure of the RBD results in tight binding to the receptor LRP (low-density-lipoprotein-receptor-related protein) [11] and consequent internalization and, in some circumstances, signalling [12].
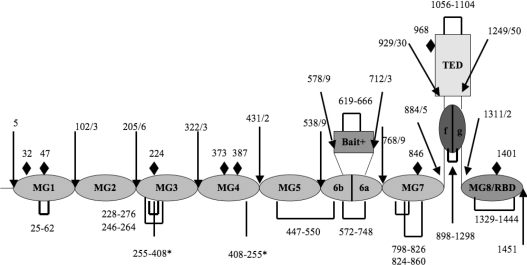
Figure 1. Schematic representation of the domain organization of the α2M monomer, based on alignment with the structure of C3 [17].
MG designates a macroglobulin domain; CUB, a domain found in a number of developmentally regulated proteins [32]; ‘bait’, bait region domain. Intramolecular disulfide bonds are shown as solid black rectangular loops. The two inter-chain disulfides (Cys255 and Cys408) [33,34] are shown as straight black lines and asterisked. The position of possible N-linked glycosylation sites are marked with black diamonds. Note that both MG6 and CUB are split domains whose primary structures are interrupted by other domains, the bait domain and TED respectively.
It was recognized many years ago that α2M is closely related to the complement proteins C3 and C4, with all probably arising from the same ancestral gene, though with features unique to each protein [13]. Thus C3 and C4 are monomers that contain a furin cleavage site that is absent from α2M, an excisable anaphylotoxin domain in C3, as well as a C-terminal domain designated C345 that lies beyond the RBD. Conversely, α2M contains a bait region domain that is located in a position equivalent to the C3a anaphylotoxin domain. The similarities in the remainder of the chain, however, are high enough to suggest that the remaining domains present in C3 are also present in α2M. Thus the recent determinations of the structures of C3, C3b and C3c by X-ray crystallography [14–17] provide a solid basis for predicting the location and structural type of the domains in α2M (Figure 1) and thus a means of examining domain–domain interactions in α2M that might be expected to exist in the native state.
We describe here the expression and characterization of three α2M domains that, together with RBD, are expected to interact intimately in native α2M and thereby play a role in the maintenance of the native conformation and of the integrity of the thiol ester. The ability to express each as a ‘well-behaved’ protein, especially for the spliced CUB (complement protein subcomponents C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1) domain, supports the predicted domain organization. However, the absence of strong interactions between any of them suggests that the conformation of one or more as isolated domains, most likely the thiol ester domain, differs from native α2M and that constraints imposed by being part of the intact α2M tetramer result in a ‘stressed’ native state that is critical for maintaining these interactions.
MATERIALS AND METHODS
Expression of MG2 (macroglobulin domain 2)
cDNA encoding MG2 (residues 103–205) of human α2M was cloned into pQE-30 (Qiagen) and transformed into Escherichia coli SG13009 cells. Cells were grown in 2YT medium [1.6% (w/v) tryptone/1% (w/v) yeast extract/0.5% (w/v) NaCl] to a D600 (attenuance) of 0.6–0.8 and induced for 5 h at 37 °C with 1 mM IPTG (isopropyl β-D-thiogalactoside). Cell lysate, obtained after sonication, was then loaded on to an Ni-NTA (Ni2+-nitrilotriacetate) Superflow (Qiagen) column, washed with 50 mM sodium phosphate buffer, 300 mM NaCl and 10 mM imidazole (pH 7.4) and eluted with 50 mM NaPO4, 300 mM NaCl and 250 mM imidazole (pH 7.4). Fractions collected from the Ni-NTA column were then dialysed against 20 mM Bis-Tris (pH 6.0) overnight, loaded on to an SP-Sepharose (sulfopropyl-Sepharose) HP column equilibrated with 20 mM Bis-Tris and eluted with a 500 mM to 1 M NaCl gradient. Selenomethionine-labelled MG2 was expressed by growing the cells in M9 medium followed, at a D600 of 0.3–0.5, by the addition of L-selenomethionine to a final concentration of 60 mg/l and L-isoleucine, L-leucine, L-lysine, L-phenylalanine, L-threonine and L-valine to final concentrations of 100 mg/l each. Induction and purification were the same as for non-labelled MG2. MG2 was concentrated to 10 mg/ml, dialysed against 20 mM Bis-Tris (pH 6.0) and kept frozen at −20 °C until crystallization.
Expression of TED (thiol ester-containing domain)
Expression of TED in E. coli and refolding of the material obtained from inclusion bodies yielded non-glycosylated TED with low solubility. As an alternative, wild-type and variant TEDs were expressed in yeast. cDNA encoding wild-type and variant TEDs (residues 935–1242) of human α2M was cloned into pPICZαA (Invitrogen) and transformed into Pichia pastoris. Cells were grown in BGC (10 g of yeast extract, 20 g of peptone, 20 ml of 50% glycerol, 3.4 g of yeast nitrogen base, 10 g of ammonium sulfate, 40 μg of biotin and 100 mM potassium phosphate, pH 6.0, in 1 litre) medium at 30 °C overnight. The cells were then transferred into BMC (as BGC, but with 5 ml of 100% methanol instead of glycerol) medium containing methanol for induction. Medium was supplemented with 0.5% methanol every 24 h. After 96 h, cells were spun down and discarded, while medium containing proteins of interest was loaded on to an Ni-NTA Superflow (Qiagen) column, washed with 50 mM sodium phosphate buffer, 300 mM NaCl and 10 mM imidazole (pH 7.4) and eluted with 50 mM sodium phosphate buffer, 300 mM NaCl and 250 mM imidazole (pH 7.4). Fractions collected from Ni-NTA column were then dialysed against 20 mM Tris and 500 mM NaCl (pH 8.0) buffer overnight and then concentrated to 6 mg/ml and stored at −20 °C.
Expression and purification of CUB
cDNA encoding CUB (residues 885–930 and 1250–1311) of human α2M was cloned as a single transcript into pQE-30 (Qiagen) and transformed into E. coli Origami cells (Novagen). Cells were grown in 2YT medium at 37 °C to a D600 of 0.6–0.8 and induced for 5 h at 20 °C with 0.5 mM IPTG. The cell lysate obtained after sonication was then loaded on to an Ni-NTA Superflow (Qiagen) column, washed with 50 mM sodium phosphate buffer, 300 mM NaCl and 10 mM imidazole (pH 7.4) and eluted with 50 mM sodium phosphate buffer, 300 mM NaCl and 250 mM imidazole (pH 7.4). Fractions collected from the Ni-NTA column were then dialysed against 20 mM Tris (pH 8.0) buffer overnight, loaded on to a Q-Sepharose HP column equilibrated with 20 mM Tris (pH 8.0) and eluted with a 0–500 mM NaCl gradient. Fractions from this Q-Sepharose HP column were then concentrated to 2 ml and loaded on to a Superdex-75 gel-filtration column to isolate monomers. Purified monomers were then concentrated to 10 mg/ml in 20 mM Tris (pH 8.0).
Expression, purification and refolding of RBD (MG8)
RBD was expressed, purified and refolded as previously described [18].
Structure determination of MG2
MG2 was crystallized in sitting drops from mother liquor containing 200 mM di-ammonium hydrogen citrate (pH 6.6), 11% (v/v) PEG [poly(ethylene glycol)] 3350. Crystals grew to 100 μm×80 μm×80 μm within 2 weeks. Crystals were flash-frozen in liquid nitrogen after cryoprotection in oil and data were collected at 100 K from a single crystal at the Argonne SER-CAT 22-BM beamline synchrotron. Three datasets at three different wavelengths were collected for selenomethionine-labelled MG2. Crystals had the space group of P43212 (a=68.81 Å, b=68.81 Å and c=120.07 Å; 1 Å=0.1 nm), contained two molecules per asymmetric unit and diffracted to 2.3 Å. Diffraction data were processed using HKL2000. Selenium MAD (multiwavelength anomalous dispersion) was used to obtain initial phases. Phase calculation and density modification were done by SOLVE and RESOLVE respectively. RESOLVE outputted the initial model and the remaining portions for the model were built manually using O. CNS was used for refinement and the refined structure of MG2 has a R factor of 24.44% and a Rfree of 28.11% at the resolution of 2.3 Å.
Crystallization of TED
Plate-like crystals of C949A mutant TED were obtained from hanging drops containing 20% PEG 3350, 200 mM ammonium formate and 50 mM MnCl2 after 5 days. There were many attempts to optimize the crystallization conditions to obtain three-dimensional crystals, but all failed to give crystals thicker than 20 μm. Crystals diffracted poorly even at the synchrotron (Argonne SER-CAT 22-BM beamline). SDS/PAGE of crystals confirmed the presence of TED protein. Only TED variants failed to yield crystals.
CD spectroscopy
Proteins were dialysed against 20 mM potassium phosphate buffer and concentrated to 1 μM for data measurement using a Jasco J-710 spectropolarimeter.
NMR spectroscopy
Variants of TED were dialysed against 50 mM sodium phosphate buffer and 100 mM NaCl, (pH 7.4) and concentrated to the specified concentrations. CUB and MG2 were dialysed against 20 mM sodium phosphate buffer (pH 7.4) and concentrated to specified concentrations. NMR data were recorded on a Bruker 900 MHz Avance NMR spectrometer equipped with a cryoprobe.
Fluorescence spectroscopy
Fluorescence experiments were performed on a PTI Quantamaster instrument equipped with double monochromators on both the excitation and emission sides. For spectra of the TED species, 1 μM protein in 20 mM sodium phosphate buffer (pH 7.4) was excited at 280 nm and emission was recorded from 300 to 380 nm in 1 nm steps. For examining binding between C949A and Q952C TED and RBD, the absence of tryptophan residues in RBD was exploited to permit selective monitoring of the tryptophan residues in TED by excitation at 295 nm. To detect possible interactions between MG2 and C949A TED, C949A TED was excited at 280 nm while titrating with MG2, with correction made for the fluorescence of the MG2, which contains one tryptophan residue. The ‘melting’ of CUB (5 μM) was followed by change in tyrosine fluorescence as a function of temperature, with excitation at 280 nm and emission at 307 nm.
ITC (isothermal titration calorimetry)
ITC measurements were performed with a VP-ITC calorimeter (Microcal). Portions of 10 μM C949A and Q952C TED were titrated with RBD or MG2 to a final concentration of 400 μM. Experiments were carried out in 20 mM sodium phosphate buffer (pH 7.4).
DSC (differential scanning calorimetry)
DSC measurements were performed using a VP-DSC calorimeter (Microcal). Melting temperatures were measured in 20 mM sodium phosphate buffer (pH 7.4) with proteins at 1 mg/ml for TED and 3 mg/ml for MG2. Data were analysed with Origin software (Microcal).
RESULTS AND DISCUSSION
Prior to the present study, the only discrete domain definitely known to exist in human α2M was the 138-residue RBD at the extreme C-terminal end of the polypeptide chain [8,19–21]. Using the revelation of the domain organization of C3 from its recent X-ray structure determination, together with the sequence alignment of α2M with C3 to give prediction of domain boundaries in α2M, we have now expressed three new domains from human α2M that represent an additional 513 residues out of the full-length chain of 1451 residues. Together with RBD, they represent approx. 42% of the whole polypeptide chain.
Thiol ester domain
One of the consequences of proteolytic activation of C3 and C4 is the formation in each case of a discrete ∼310-residue fragment, designated C3d and C4d respectively, that contains the thiol ester-forming residues CGEQ. X-ray structures of both C3d [22] and C4d [23] were determined even before those of the parent proteins C3 and C3b and they showed them to be all α-helical domains composed of 12 helices, with the thiol ester-forming residues being located at the surface of the domain in a turn between the first and second helices. We expressed in E. coli the equivalent region of α2M, representing residues 935–1242 (designated TED). This was expressed in two forms: one in which the thiol ester-forming Cys949 had been mutated to alanine, thus precluding formation of a thiol ester, and one in which the Gln952 had been mutated to cysteine, to permit formation of an approximately isosteric disulfide in place of the more labile thiol ester.
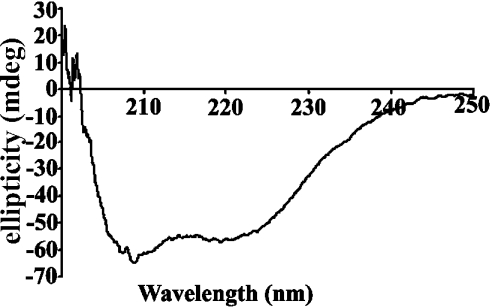
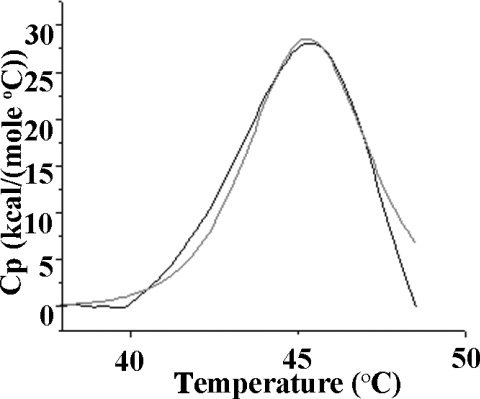
As expected from homology with C3d and C4d, as well as secondary-structure predictions of this region, the CD spectrum of this species was consistent with an all α-helical domain (Figure 2). Analysis of the spectrum (http://www.embl-heidelberg.de/~andrade/k2d/) using reference sets of proteins and homopolymers of known structure gave 100% α-helix [24,25]. DSC-monitored unfolding of TED gave a single peak (Figure 3), with a Tm (melting temperature) of 45 °C and a ΔHcalorimetric/ΔHvan't Hoff value of 1, confirming the presence of only a single domain [26]. The 1H-NMR spectrum showed upfield-shifted methyl and aromatic resonances (results not shown) consistent with the species being a discrete folded domain. Also consistent with the homogeneity of the species was the ability to obtain plate-like crystals that were shown by SDS/PAGE to contain TED and to diffract, albeit poorly (results not shown).
Figure 2. CD spectrum of TED, showing exclusive α-helical secondary structure.
The spectrum is of 1 μM TED in 20 mM potassium phosphate buffer (pH 7.4).
Figure 3. DSC-monitored unfolding of TED.
TED unfolds as a single domain with Tm of 45 °C (grey line). The fit to a single two-state unfolding is shown as a continuous black line. C949A TED (1 mg/ml) was in 20 mM sodium phosphate buffer (pH 7.4). 1 kcal=4.184 kJ.
The rationale for expressing one form of TED with C949A mutation and a second with the Q952C mutation was to be able to examine the effect on conformation of having no thiol ester (C949A variant) or a surrogate thiol ester (Q952C variant) that would be both resistant to facile cleavage by ambient nucleophiles and capable of being cleavable by reduction under controlled conditions. The presence of an intact disulfide bond between Cys949 and Cys952 in the unreduced Q952C variant (in addition to the buried natural disulfide) was confirmed by allowing the TED is react with the fluorescent cysteine-reactive compound IAF (5-iodoacetamidofluorescein). Overnight incubation of both native Q952C and C949A variants with IAF (200 μM) resulted in no label incorporation, consistent with no free accessible cysteine residues. Following reaction of both variants with 0.5 mM 2-mercaptoethanesulfonate, chosen to preferentially reduce surface-accessible disulfides, the C949A variant still gave no label incorporation, whereas the Q952C gave two labels per molecule, supporting the presence of an intact surface-accessible disulfide in the unreduced form of the Q952C variant. Finally, it was confirmed by reaction with DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] that the denatured and fully reduced Q952C variant contained four free cysteine residues [27].
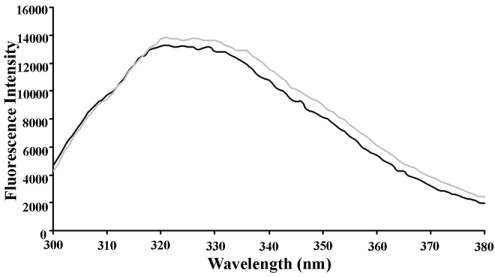
Since TED contains five tryptophan residues, two of which are positioned relatively close to the thiol ester-forming motif CGEQ in C3d and C4d and might therefore be very sensitive probes of conformational change in this region, we used tryptophan fluorescence to determine if there was a significant conformational difference between the two TED species. The fluorescence spectrum of C949A TED had a broad maximum centred at approx. 322 nm, consistent with the expected buried location of most of the tryptophan residues (from homology with C3d and C4d) (Figure 4). Spectra of C949A TED and oxidized Q952C TED were nearly superimposable (Figure 4), while reduction of Q952C TED caused no significant perturbation. This suggested that the presence of a covalent linkage between residues 949 and 952 did not influence the conformation of TED in the context of the isolated domain.
Figure 4. Fluorescence spectra of TED species.
Fluorescence emission spectra of C949A and Q952C (oxidized) TED in 20 mM sodium phosphate buffer (pH 7.4) showing very similar environments for the five tryptophan residues present.
Structure of MG2
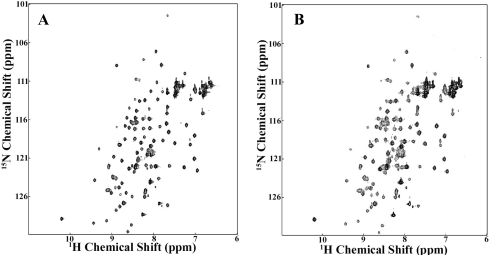
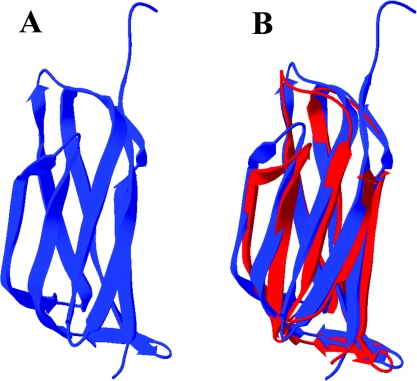
The various structures of C3 and C3 fragments all show the presence of eight domains with the fibronectin type-3 fold, each consisting of one three-strand and one four-strand antiparallel β-sheet stacked in an approximately parallel sandwich arrangement. These have been named MG (for macroglobulin) domains. The second macroglobulin domain (MG2) of human α2M is predicted to stretch from residue 103 to 205. We expressed a slightly longer version that had an N-terminal His tag. A 15N-labelled sample gave a well-dispersed two-dimensional HSQC (heteronuclear single-quantum coherence) spectrum (Figure 5A), consistent both with a homogeneous folded domain and with the expected β-sheet secondary structure. This construct with His tag still attached gave crystals that diffracted to 2.3 Å. The structure was determined using anomalous dispersion from incorporated selenomethionine for phasing, to ensure a completely unbiased structure, and was found to be the expected β-sandwich (Figure 6A and Table 1) present in the MG domains of C3. Superpositioning of the present MG2 domain from α2M with that from MG2 in C3 showed that the structures are almost identical, with an rmsd (root mean square deviation) of 1.35 Å (Figure 6B).
Figure 5. 1H-15N HSQC spectra of MG2.
(A) Uniformly 15N-labelled MG2 (100 μM) alone; (B) uniformly 15N-labelled MG2 (100 μM) after addition of 150 μM of unlabelled TED. Samples were in 20 mM sodium phosphate buffer (pH 7.4). Spectra were recorded at 900 MHz.
Figure 6. X-ray crystal structure of MG2 and comparison with MG2 from C3.
(A) Ribbon diagram of MG2 from α2M determined in the present study (Protein Data Bank accession code 2P9R). (B) Super-positioning of MG2 from α2M (blue) with MG2 from bovine C3 (red).
Table 1. Data collection and refinement statistics for MG2.
| Parameter | Value |
|---|---|
| Data collection | |
| Space group | P43212 |
| Unit cell dimensions (Å) | a=68.81, b=68.81, c=120.07 |
| Wavelength (Å) | 1.000 |
| Resolution* | 48.7–2.3 (2.38–2.3) |
| Measured reflections | 310398 (14526) |
| Unique reflections | 13500 (1297) |
| I/σI | 58.0 (4.7) |
| Redundancy | 23 (11.2) |
| Completeness (%) | 99.4 (98.6) |
| Rsym† | 6.1 (37) |
| Refinement | |
| Resolution | 20–2.3 |
| R‡/Rfree (%) | 24.4/28.1 |
| No. of atoms | |
| Protein | 1612 |
| Water | 65 |
| B factors (Å2) | 39.8 |
| rmsd | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.35 |
| Ramachandran plot | |
| Most favoured (%) | 86.8 |
| Additionally allowed (%) | 9.6 |
| Generously allowed (%) | 2.4 |
| Disallowed (%) | 1.2 |
* The numbers in parentheses are for the highest resolution shell.
† Rsym=∑∑i|Ii−〈I〉|/∑〈I〉, where 〈I〉 is the mean intensity of the N reflections with intensities Ii and common indices hkl.
‡ R=∑hkl||Fobs|−k|Fcalc||/∑hkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors respectively. Rfree was calculated using 10% of all reflections, which were excluded from refinement.
Expression and properties of the CUB domain
The primary structure of the CUB domain in C3 (two stacked interdigitated four-stranded β-sheets) is interrupted by the entire TED (Figure 1). The ability to express the CUB domain of α2M as an isolated folded species by constructing an artificial single stretch of polypeptide composed of the two separate moieties would therefore be a rigorous test of the secondary-structure prediction for this region. In the tertiary structure of C3, the points of exit from CUB into TED (residue 962) and re-entry into CUB from the C-terminus of TED (residue 1269) lie close together in space (9.9 Å). Sequence alignment predicted a similar spliced arrangement for the CUB domain in human α2M [17], but with the difference that CUB from human α2M has an intra-domain disulfide, whereas there are no such disulfides in C3. The disulfide in α2M is between Cys898 and Cys1298 and thus covalently links the N- and C-terminal moieties of the domain. These residues align with residues 925 and 1317 in C3, which are close enough together to permit disulfide bond formation in α2M CUB without conformational change of the domain relative to that in C3. To permit expression of CUB from α2M as a single species, we constructed cDNA in which the encoded C-terminal residue of the stretch of CUB preceding TED (residue 929) was linked to the N-terminal residue following TED (residue 1250). This was expressed in E. coli and refolded as described in the Materials and methods section.
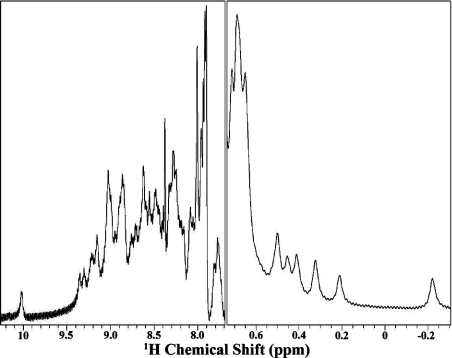
The resulting soluble species gave a well-dispersed 1H NMR spectrum (Figure 7), with amide protons extending down to 10.0 p.p.m., and a number of shifted aliphatic resonances upfield from 0.7 p.p.m., consistent with a folded, structured domain. Also consistent with this was a single unfolding transition at 69 °C, monitored by change in tyrosine fluorescence as a function of temperature, and a CD spectrum with a single minimum at 215 nm consistent with β secondary structure (results not shown).
Figure 7. 1H NMR spectrum of α2M CUB domain.
Residues 885–930 were joined to residues 1251–1311 to generate a spliced, single-chain CUB domain. Low- and high-field regions of the one-dimensional spectrum of CUB are shown. Evidence for this spliced domain being well folded are the good spread of amide protons in the region from 10 to 8 p.p.m. and a number of upfield-shifted resonances between 0.7 and −0.2 p.p.m.
Absence of domain–domain interactions
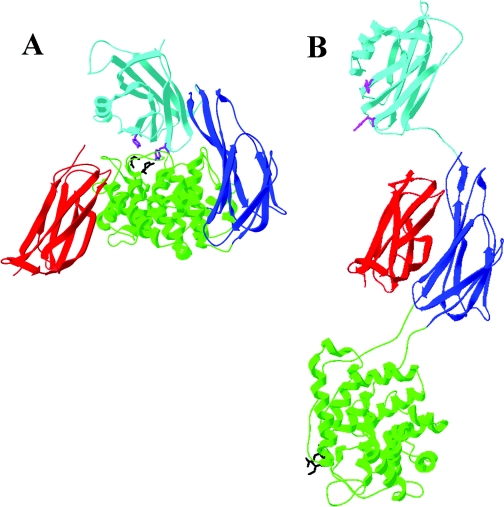
In the structure of C3, which can be considered to be equivalent to native α2M in terms of being the unactivated form, TED is in intimate contact with MG2, CUB and MG8 (Figure 8A). However, upon proteolytic activation of C3 to C3b, a series of major domain movements occurs that results, inter alia, in disruption of all of these contacts between TED and the domains MG2, CUB and MG8 (Figure 8B). In addition, the conformation of TED is significantly altered, such that it more closely resembles that of the fragment C3d. Whether the alteration in TED's conformation is a cause or consequence of the disruption of these interactions is not known. We were therefore interested in examining whether the equivalent domains from α2M could interact with one another as isolated species. For this purpose, we also expressed MG8, which is the RBD of α2M, which we have previously expressed and characterized structurally [18]. It should be noted that, although the TED used was glycosylated [approx. 3 kDa carbohydrate estimated from the MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) spectrum], the expected site of N-linked glycosylation is on the outer face of TED (bottom surface in Figure 8A), far from any predicted interfaces with MG2, CUB or MG8 and therefore very unlikely to interfere with domain–domain interaction.
Figure 8. Relationship of MG2, TED, CUB and MG8 to one another in C3 and C3b.
(A) Domains in intact C3 [17]. (B) Domains following activation of C3 to C3b [15]. MG2 is in red, TED in green, CUB in blue and MG8(RBD) in cyan. The thiol ester-forming residues are shown in black and the conserved tyrosine pair in magenta.
As a qualitative means of detecting possible pairwise or higher-order interactions between these domains, native PAGE was used on the combinations TED–RBD, MG2–TED, RBD–CUB–TED–MG2, CUB–TED and CUB–RBD. In no instance was there evidence of a gel shift that might indicate a strong interaction. Subsequently, fluorescence spectroscopy and ITC were used to examine possible interactions between TED, RBD and MG2. Again, no evidence of binding interactions were observed, even when using a 20:1 molar excess of the titrant. As a last attempt to observe even weaker interactions, we used NMR spectroscopy at much higher protein concentrations to examine the interaction of TED with MG2. 15N-labelled MG2 was titrated with unlabelled TED up to 1:1.5, without any significant perturbation of any of the backbone cross-peaks of MG2, or increases in line width suggestive of formation of a much-higher-molecular-mass species (Figure 5B). These negative findings suggest that the specific binding interactions that are present between these domains in intact α2M are greatly weakened as a result of the conformational changes that occur in TED subsequent to α2M activation.
Such a weakening, at least between TED and RBD, is not too surprising, since a principal function of the proteolytic activation of α2M and the subsequent conformational changes is to permit release of RBD from a tightly associated cryptic location within α2M so that it becomes accessible to its receptor, LRP. This contrasts with a demonstrated tightening of interactions elsewhere within α2M that makes it much harder to dissociate the non-covalently associated dimers following proteolysis within the bait region [28,29], as well as the finding of a shift in the dimer–tetramer equilibrium of the related human macroglobulin PZP (pregnancy zone protein) from mostly dimer to predominantly tetramer following bait region activation [30]. These observations, together with the transformation of tetrameric α2M into dimeric forms as a result of excision of residues within the ‘bait domain’ (Figure 1) that are immediately C-terminal to the exposed flexible bait region [31], suggest that the tightening involves strengthening of domain–domain interactions between the bait domains from monomers in opposing dimers. This may serve to help retain the proteinase within the α2M ‘trap’.
Conclusions
The ability to express three new domains from human α2M as folded, structured proteins, based on sequence alignment with complement C3, provides strong support for the predicted domain organization of α2M (Figure 1), both in terms of the existence of multiple discrete domains and in terms of their precise location and structure. However, the clear lack of a strong interaction between TED and MG8(RBD), whether examined by fluorescence, ITC or gel shift, suggested that this critical interaction in native C3 and α2M must depend on additional constraints. Since no strong interaction was found between TED and either CUB or MG2, alone or together with MG8, this suggests that these restraints do not arise from strong binding interactions between TED and these domains, but rather from additional restraints that occur only in the context of native α2M or C3. Such an interpretation is consistent with a number of observations on both C3 and α2M, and is also more in keeping with the need for an essentially irreversible activation as a result of proteolysis.
Acknowledgments
We thank Dr Alex Dementiev for help with X-ray diffraction data collection and analysis, Dr Klavs Dolmer for help with protein expression and for Figure 1, and for helpful discussions, and Dr Steven Olson for comments on this paper. This work was supported by grant GM54414 from the NIH (National Institutes of Health). The ITC and DSC instruments were purchased with shared instrumentation grant S10 RR15958 from the NIH; the 900 MHz NMR spectrometer was purchased with funds from NIH grant P41 GM68944.
References
- 1.Sottrup-Jensen L. α-Macroglobulins. Structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 1989;264:11539–11542. [PubMed] [Google Scholar]
- 2.Barrett A. J., Starkey P. M. The interaction of α2-macroglobulin with proteinases. Characteristics and specificity of the reaction and a hypothesis concerning its molecular mechanism. Biochem. J. 1973;133:709–724. doi: 10.1042/bj1330709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb D. J., Crookston K. P., Hall S. W., Gonias S. L. Binding of transforming growth factor-β1 to immobilized human α2-macroglobulin. Arch. Biochem. Biophys. 1992;292:487–492. doi: 10.1016/0003-9861(92)90020-w. [DOI] [PubMed] [Google Scholar]
- 4.Ronne H., Anundi H., Rask L., Peterson P. A. Nerve growth factor binds to serum α2-macroglobulin. Biochem. Biophys. Res. Commun. 1979;87:330–336. doi: 10.1016/0006-291x(79)91683-8. [DOI] [PubMed] [Google Scholar]
- 5.Gettins P., Cunningham L. W. Identification of 1H resonances from the bait region of human α2-macroglobulin and effects of proteases and methylamine. Biochemistry. 1986;25:5011–5017. doi: 10.1021/bi00366a007. [DOI] [PubMed] [Google Scholar]
- 6.Sottrup-Jensen L., Lønblad P. B., Stepanik T. M., Petersen T. E., Magnusson S., Jörnvall H. Primary structure of the ‘bait’ region for proteinases in α2-macroglobulin. FEBS Lett. 1981;127:167–173. doi: 10.1016/0014-5793(81)80197-4. [DOI] [PubMed] [Google Scholar]
- 7.Sottrup-Jensen L., Sand O., Kristensen L., Fey G. H. The α-macroglobulin bait region. Sequence diversity and localization of cleavage sites for proteinases in five mammalian α-macroglobulins. J. Biol. Chem. 1989;264:15781–15789. [PubMed] [Google Scholar]
- 8.Sottrup-Jensen L., Gliemann J., Van Leuven F. Domain structure of human α2-macroglobulin. Characterization of a receptor-binding domain obtained by digestion with papain. FEBS Lett. 1986;205:20–24. doi: 10.1016/0014-5793(86)80857-2. [DOI] [PubMed] [Google Scholar]
- 9.Sottrup-Jensen L., Petersen T. E., Magnusson S. Trypsin-induced activation of the thiol esters in α2-macroglobulin generates a short-lived intermediate (‘nascent’ α2M) that can react rapidly to incorporate not only methylamine or putrescine but also proteins lacking proteinase activity. FEBS Lett. 1981;128:123–126. doi: 10.1016/0014-5793(81)81096-4. [DOI] [PubMed] [Google Scholar]
- 10.Sottrup-Jensen L., Hansen H. F. Nascent α2-macroglobulin-trypsin complex: incorporation of amines and water at the thiol esterified Glx residues of α2-macroglobulin during activation with trypsin. Biochem. Biophys. Res. Commun. 1982;107:93–100. doi: 10.1016/0006-291x(82)91674-6. [DOI] [PubMed] [Google Scholar]
- 11.Kristensen T., Moestrup S. K., Gliemann J., Bendtsen L., Sand O., Sottrup-Jensen L. Evidence that the newly cloned low density lipoprotein receptor-related protein (LRP) is the α2-macroglobulin receptor. FEBS Lett. 1990;276:151–155. doi: 10.1016/0014-5793(90)80530-v. [DOI] [PubMed] [Google Scholar]
- 12.Bacskai B. J., Xia M. Q., Strickland D. K., Rebeck G. W., Hyman B. T. The endocytic receptor protein LRP also mediates neuronal calcium signaling via N-methyl-D-aspartate receptors. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11551–11556. doi: 10.1073/pnas.200238297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sottrup-Jensen L., Stepanik T. M., Kristensen T., Lønblad P. B., Jones C. M., Wierzbicki D. M., Magnusson S., Domdey P. B., Wetsel R. A., Lundwall Å., et al. Common evolutionary origin of α2-macroglobulin and complement components C3 and C4. Proc. Natl. Acad. Sci. U.S.A. 1985;82:9–13. doi: 10.1073/pnas.82.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesmann C., Katschke K. J., Jr, Yin J. P., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 15.Janssen B. J. C., Christodouliou A., McCarthy A., Lambris J. D., Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 16.Ajees A., Gunasekaran K., Narayana S. V. L., Kotwal G. J., Murthy H. M. K. The structure of complement C3b provides insights into complement activation and regulation. Nature. 2006;444:221–225. doi: 10.1038/nature05258. [DOI] [PubMed] [Google Scholar]
- 17.Janssen B. J. C., Huizinga E. G., Raaijmakers H. C. A., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 18.Huang W., Dolmer K., Liao X. B., Gettins P. G. W. Localization of basic residues required for receptor binding to the single α-helix of the receptor binding domain of human α2-macroglobulin. Protein Sci. 1998;7:2602–2612. doi: 10.1002/pro.5560071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Leuven F., Marynen P., Sottrup-Jensen L., Cassiman J.-J., Van den Berghe H. The receptor-binding domain of human α2-macroglobulin. Isolation after limited proteolysis with a bacterial proteinase. J. Biol. Chem. 1986;261:11369–11373. [PubMed] [Google Scholar]
- 20.Jenner L., Husted L., Thirup S., Sottrup-Jensen L., Nyborg J. Crystal structure of the receptor-binding domain of α2-macroglobulin. Structure. 1998;6:595–604. doi: 10.1016/s0969-2126(98)00061-6. [DOI] [PubMed] [Google Scholar]
- 21.Huang W., Dolmer K., Liao X., Gettins P. G. W. NMR solution structure of the receptor binding domain of human α2macroglobulin. J. Biol. Chem. 2000;275:1089–1094. doi: 10.1074/jbc.275.2.1089. [DOI] [PubMed] [Google Scholar]
- 22.Nagar B., Jones R. G., Diefenbach R. J., Isenman D. E., Rini J. M. X-ray crystal structure of C3d: A C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 23.van den Elsen J. M. H., Martin A., Wong V., Clemenza L., Rose D. R., Isenman D. E. X-ray crystal structure of the C4d fragment of human complement component C4. J. Mol. Biol. 2002;322:1103–1115. doi: 10.1016/s0022-2836(02)00854-9. [DOI] [PubMed] [Google Scholar]
- 24.Yang J. T., Wu C.-S. C., Martinez H. M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 25.Chang C. T., Wu C.-S. C., Yang J. T. Circular dichroic analysis of protein conformation: inclusion of the β-turns. Anal. Biochem. 1978;91:13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- 26.Privalov P. L. Stability of proteins. Proteins which do not present a single cooperative system. Adv. Protein Chem. 1982;35:1–104. [PubMed] [Google Scholar]
- 27.Ellman G. L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 28.Pochon F., Barray M., Delain E. Functional α2-macroglobulin half-molecules induced by cadmium. Biochem. Biophys. Res. Commun. 1987;149:488–492. doi: 10.1016/0006-291x(87)90394-9. [DOI] [PubMed] [Google Scholar]
- 29.Pochon F., Barray M., Delain E. Dissociation of α2-macroglobulin into functional half-molecules by mild acid treatment. Biochim. Biophys. Acta. 1989;996:132–138. doi: 10.1016/0167-4838(89)90105-2. [DOI] [PubMed] [Google Scholar]
- 30.Jensen P. E. H., Stigbrand T. Differences in the proteinase inhibition mechanism of human a2-macroglobulin and pregnancy zone protein. Eur. J. Biochem. 1992;210:1071–1077. doi: 10.1111/j.1432-1033.1992.tb17513.x. [DOI] [PubMed] [Google Scholar]
- 31.Gettins P. G. W., Hahn K.-H., Crews B. C. α2Macroglobulin variants. A role for the bait region in tetramer formation. J. Biol. Chem. 1995;270:14160–14167. doi: 10.1074/jbc.270.23.14160. [DOI] [PubMed] [Google Scholar]
- 32.Bork P., Beckmann G. The CUB domain: a widespread module in developmentally regulated proteins. J. Mol. Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 33.Jensen P. E. H., Sottrup-Jensen L. Primary structure of human α2-macroglobulin. Complete disulfide bridge assignment and localization of two intrachain bridges in the dimeric proteinase binding unit. J. Biol. Chem. 1986;261:15863–15869. [PubMed] [Google Scholar]
- 34.Oxvig C., Gleich G. J., Sottrup-Jensen L. Localization of disulfide bridges and free sulfhydryl groups in human eosinophil granule major basic protein. FEBS Lett. 1994;341:213–217. doi: 10.1016/0014-5793(94)80459-1. [DOI] [PubMed] [Google Scholar]