Abstract
We have developed a technique to replica plate mammalian cells grown on plastic dishes using low melt agarose. This method is simpler than previously described methods that use polyester membranes to grow and transfer cells. We have tested the effectiveness of this technique on normal and immortal cell lines and have found that we can transfer cells with an efficiency of 80–90%. We have used this technique to rapidly screen clones for insertion of a lentivirally encoded gene without a selectable marker.
Keywords: Cell culture, Replica plating, Ring cloning, Screening, Transfection
Introduction
Replica plating has been widely used in bacterial and yeast genetics to screen for desired phenotypes since its invention by Lederberg (Lederberg and Lederberg 1952). The original technique used velveteen pads to transfer cells onto Petri dishes containing different types of media to aid screening large numbers of colonies for clones occurring at low frequency. Mammalian cells do not lend themselves to being picked up by a velveteen surface. One previous report suggested the use of polyester membranes on which mammalian cells can be grown. Transferring part of the colony involved placing another membrane of smaller pore size on top, and allowing some of the cells to grow onto the second membrane, which may take 1–7 days for complete transfer (Hornsby et al. 1992). The only other methods currently available to screen mammalian colonies for a desirable phenotype are ring cloning, cloning by limiting dilution or picking individual colonies with a toothpick, which are cumbersome for processing large numbers of colonies. We have developed a simpler and quicker method to replica plate mammalian cells grown on plastic dishes using low-melt agarose to facilitate the transfer. Ring cloning can then be used to pick positive clones. The transfer is immediate and the original dish can be used to screen for the desired phenotype. Since we do not depend on cell growth to facilitate transfer, one eliminates the waiting period of 1–7 days for screening in the technique developed by Hornsby et al. (1992). The ability to assay large numbers of clones prior to picking an individual clone greatly reduces the amount of tissue culture time and effort to isolate rare phenotypes.
Materials and methods
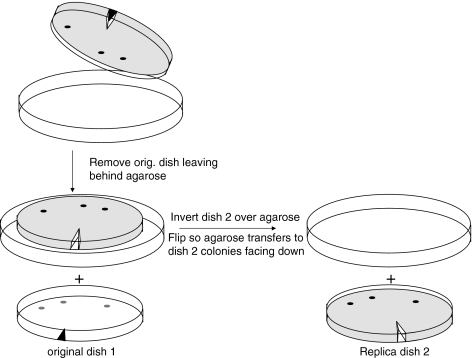
The method is outlined in Fig. 1. Cells are plated at clonal density on 10 cm diameter tissue culture dishes and allowed to grow until visible colonies of ≥1,000 cells form. We prefer to plate at a density that yields 20–50 colonies per dish to minimize the risk of cross contamination between clones. Most immortal cells plate at clonal density with an efficiency of ∼5–15%. Each dish requires 20 mL of 3% low-melt agarose (Invitrogen, Carlsbad, CA) cooled to 37 °C then mixed with 2.5 mL 10 × concentrated growth medium and 2.5 mL serum (final concentration 1 × medium and 10% serum). After removing the culture media and rinsing once with PBS to remove traces of serum, 1 mL of Trypsin-EDTA (Invitrogen, Carlsbad, CA) is added at room temperature to loosen some of the cells from each colony. The dish is closely observed under the microscope. As soon as some cells in an average-sized colony begin to round up, the trypsin is removed and replaced with the freshly mixed agarose solution (containing growth medium plus serum) for 10–15 min. The serum stops the action of trypsin and prevents those cells that are still well attached in the original colonies from detaching from the dish. The agarose is allowed to solidify. The trypsinization procedure should be monitored for every dish to avoid over-trypsinizing and completely detaching cells that could cause colony cross-contamination. A small V-shape is cut with a sterile spatula in the agarose as a reference mark. The position of this reference cut is marked on the bottom of the original dish, then the dish is inverted over a 15 cm dish so that the agarose falls out onto a sterile surface. This positions the rounded easily-detached cells from each of the colonies facing up (Fig. 1). A sterile spatula can be used to initiate removal of the agarose. Growth medium is meanwhile added back to the first dish to allow the residual cells of each colony (consisting of most of the cells) left behind to continue growing. A second 10 cm dish is then inverted on the agarose and the plates are flipped so that the agarose drops into the 10 cm dish, colony side down. The reference cut is then marked on the second dish. Any trapped air bubbles are released by poking the agarose with a Pasteur pipette. This plate is incubated at 37 °C for 16–24 h in order to permit the transferred cells to adhere. No additional medium is added during this period, since the agarose itself contains complete medium. The absence of free liquid permits good contact between the agarose and the bottom of the dish, which facilitates adhesion of the transferred cells. The agarose is then removed as described above and growth media added to allow the transferred cells to form colonies.
Fig. 1.
Replica plating using low-melt agarose. A mixture of 3% low melt agarose containing complete growth medium and serum is poured on a dish with partially trypsinized colonies and allowed to solidify. As shown in the figure, the agarose is removed from the original dish after marking the dish and agarose for orientation and flipped over onto a second dish to transfer part of the colony from the original dish. Only three rather than 50 colonies are shown for illustrative purposes. The dishes are shown prior to marking the replica dish for orientation. After 1 day, the agarose is removed from the replica dish 2
Results
We have tested this method using SW39 (T-antigen immortalized IMR90 lung fibroblasts), MDAOH41 (breast cancer fibroblasts from a Li Fraumeni patient), HeLa cells and BJ (normal diploid foreskin fibroblasts). We have been able to transfer these cell lines with an efficiency of 80–90%. Small colonies of 100 cells transfer with an efficiency of 30–50%.
We have used this method to rapidly screen clones of SW26 (T-antigen immortalized IMR90 lung fibroblasts) and MDAOH87 (breast cancer fibroblasts from a Li Fraumeni patient) for the presence of a tetratcycline-regulated transactivator that was introduced into a population of cells by lentiviral infection. The original plate was transduced with a lentiviral vector expressing a tetracycline-regulated dsRED gene. Only those clones that have the transactivator expressed dsRED. The colonies that were dsRED-positive on the original plate were then identified on the replica plate to recover the desired clone. This method is semi-quantitative and can be used to get a rough estimate of the percentage of desired clones.
Discussion
The replica plate can also be stained with antibodies to identify clones with differential levels of expression of a target gene, selected with antibiotics, placed at restrictive temperatures to identify sensitive clones, or subjected to other manipulations depending on the application and the specific phenotype involved. It is important to plate cells at the appropriate density (20–50 clones per dish) to minimize the risk of cross contamination, although much larger numbers per plate can be screened as needed if one reclones the selected cells. It is also important to allow the clones to grow until there are about 1,000 cells per clone in order to get high-efficiency replica plating of the colonies. While this procedure has worked well with the cell-types that we tested, it is possible that it may not work with all cell types and preliminary experiments will have to be done when working with different cell lines.
References
- Lederberg J, Lederberg EM (1952) Replica plating and indirect selection of bacterial mutants. J Bacteriol 63(3):399–406 [DOI] [PMC free article] [PubMed]
- Hornsby PJ, Yang L, Lala DS, Cheng CY, Salmons B (1992) A modified procedure for replica plating of mammalian cells allowing selection of clones based on gene expression. BioTechniques 12(2):244–251 [PubMed]