Abstract
A long-standing question in Quaternary paleontology is whether climate-induced, population-level phenotypic change is a result of large-scale migration or evolution in isolation. To directly measure genetic variation through time, ancient DNA and morphologic variation was measured over 2,400 years in a Holocene sequence of pocket gophers (Thomomys talpoides) from Lamar Cave, Yellowstone National Park, Wyoming. Ancient specimens and modern samples collected near Lamar Cave share mitochondrial cytochrome b sequences that are absent from adjacent localities, suggesting that the population was isolated for the entire period. In contrast, diastemal length, a morphologic character correlated with body size and nutritional level, changed predictably in response to climatic change. Our results demonstrate that small mammal populations can experience the long-term isolation assumed by many theoretical models of microevolutionary change.
Keywords: population genetics, phenotypic plasticity, body size
The Greater Yellowstone ecosystem in the northern Rockies has experienced marked climatic fluctuations during the past few thousand years, with extremes during the Medieval Warm Period (450 to 1350 yr B.P.) and the Little Ice Age (150 to 450 yr B.P.) (1, 2). An exceptional record of small mammal populations was preserved during this period in Lamar Cave, where more than 10,000 mammal specimens have been recovered, defining a sequence of populations spanning the last 3,000 years (3). Morphologic studies of the pocket gopher, an abundant small mammal throughout this deposit, have documented change in size-related craniodental characters corresponding with climatic change (4, 5).
To discriminate between migration and within-locality causes of morphologic change, we used single-stranded DNA conformation polymorphism analysis (SSCP) and DNA sequencing to measure variation in a 164-bp segment of the mitochondrial cytochrome b gene in 73 ancient specimens§ from 10 stratigraphic levels spanning the past 2,400 years (Fig. 1). We also analyzed DNA from seven extant pocket gophers near Lamar Cave and six individuals from adjacent localities (Fig. 2). Because pocket gopher populations are strongly differentiated with regard to cytochrome b sequences (6, 7), migration from nearby populations would be revealed as the appearance of new sequences over time. We compared the cytochrome b results to the pattern of morphologic change over time in diastemal length, a developmentally plastic character correlated with body size and plane of nutrition (4, 8) (Fig. 1). As a morphologic control, we compared the genetic results to changes in toothrow length, which in pocket gophers is a conservative indicator of differences between subspecies (9).
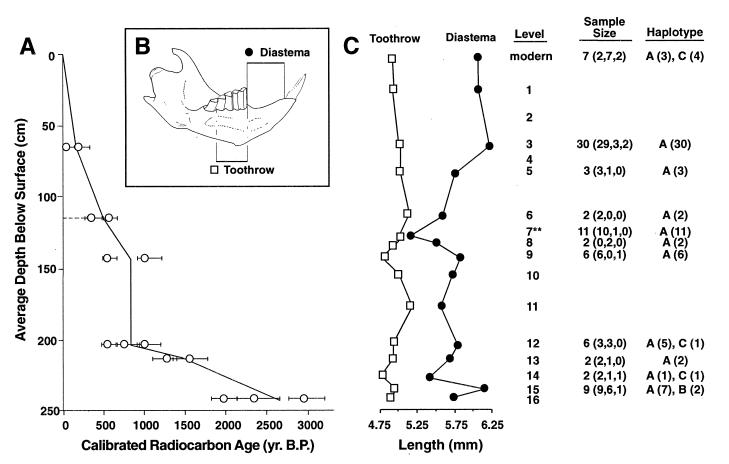
Figure 1.
Average depth of stratigraphic levels in Lamar Cave, number of modern (n = 7 specimens of T. t. tenellus and T. t. fuscus) and ancient specimens (n = 73) from Lamar Cave that were subjected to DNA analysis, respective age of the specimens, and summary of SSCP and sequence data. (A) The age-versus-depth curve for the deposits based on 18 calibrated radiocarbon dates with 95% confidence intervals shown (3). (B) Illustration of a lower left jaw with the measurements taken to analyze morphologic characters. (C) Changes in diastemal length (•) and toothrow length (□) for comparison with haplotypes A–C identified by SSCP analysis. We examined 88 ancient specimens spanning all 16 stratigraphic levels. In 73 cases (83%) we obtained a 164-bp PCR product of the mitochondrial cytochrome b gene (cyp b) 5′ end. Level 16 (2,400 to 3,000 years) did not yield any PCR product. The sample size column lists the number of specimens that were examined, and in parentheses, the number screened for 164 bp by using SSCP (first number), individuals directly sequenced for 63 bp (second number), and individuals cloned and sequenced for 134 bp (third number). Note: cloned individuals also have been sequenced directly with the exception of level 6. At level 7 morphologic change is most pronounced, coincident with changes in temperature and/or moisture regimes in the region.
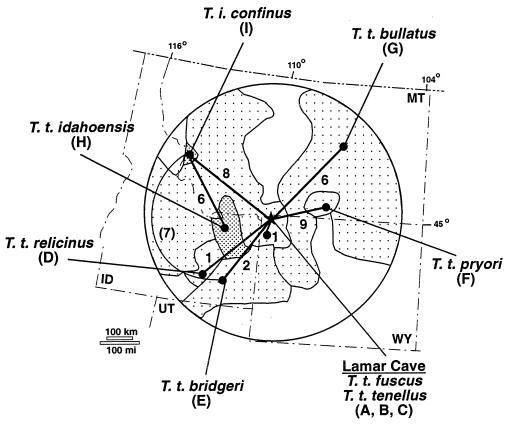
Figure 2.
Sampling localities and genetic relationships of the modern neighboring populations of pocket gophers to the Lamar Cave population. Shaded areas illustrate the present range of six nominal subspecies of T. talpoides (light stipple), and two subspecies of T. idahoensis (dense stipple) in and around Yellowstone National Park. Numbers above lines denote the number of observed substitutions between haplotypes connected by a minimum spanning network (6) based on 63 bp of cytochrome b sequence. An alternative link is shown in parentheses. Haplotype numbers are indicated at localities where they are found (Table 1).
DNA from ancient teeth was extracted by a silica-based purification method (10). Single teeth were wrapped in aluminum foil, dipped into liquid nitrogen, and ground to a fine powder before extraction. DNA from skins was isolated by cell lysis followed by organic solvent purification (11). Contamination was monitored by extraction controls (mock extractions) subjected to PCR and procedures to avoid and monitor contamination were followed carefully (12–14). Notably, no DNA from recent pocket gophers was amplified before the work on ancient samples was completed.
We designed cytochrome b primers Tta 1 (L 5′-ACCCCACCTAACATCTCAGG-3′, 5′ at position 66 of GenBank sequence L38467) and Tta 2 (CAGCCATAGTTTACATCTCGG-3′, 5′ at position 230) based on an alignment of Thomomys talpoides and T. bottae cytochrome b sequences (GenBank accession nos. L38467 and L38476), and unpublished sequences of T. t. monoensis (MVZ 176455); T. monticola (PAG 17); and T. mazama (MVZ 176431) kindly provided by M. F. Smith from the University of California, Berkeley. Amplification from 5 μl of the extract was carried out for 40 cycles in a 30-μl volume using wax-mediated hot starts (10). Amplification cycles in a programmable thermal cycler (Perkin–Elmer Cetus 480) were as follows; denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min. For SSCP analysis, primers were labeled with 32P following a standard kinase reaction (15). PCR controls (PCR without extract) were included in each amplification to monitor for contamination. Samples that yielded PCR product were screened for variation with SSCP, a method that accurately detects single nucleotide substitutions (15–17). Additionally, to test the accuracy of SSCP typing, partial DNA sequences were determined from 24 individuals from the ancient and recent Lamar Cave population and one individual from each of six neighboring localities (Fig. 2). Sequence comparisons revealed that haplotypes A, B, and C found in modern and ancient specimens collected at Lamar differ by at least one substitution. The other six localities each exhibited a unique haplotype. Haplotypes that differ by only one substitution were resolved by SSCP analysis (Fig. 3). PCR products either were sequenced directly by using the Applied Biosystems PRISM Dye Terminator Cycle sequencer (Perkin–Elmer Cetus) or were sequenced by the Sanger method (11) after cloning (TA cloning kit, Invitrogen).
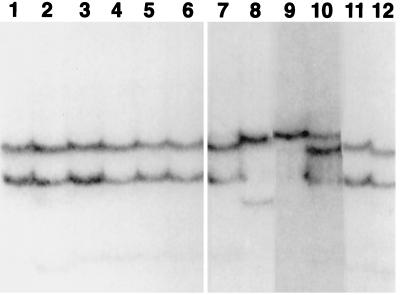
Figure 3.
SSCP typing of T. talpoides from Lamar Cave and surrounding localities. Lanes 1 and 2, level 3, haplotype A; lanes 3 and 4, level 6, haplotype A; lanes 5 and 6, level 12, haplotype A; lanes 7 and 11, recent Lamar Cave, haplotype A; lane 10, recent Lamar Cave, haplotype C; lanes 8, 9, and 12, recent from localities outside Lamar Cave, all having unique haplotypes differing by 1–8 substitutions.
The authenticity of ancient DNA sequences often is questioned because contaminating DNA and PCR products readily outcompete lower copy number and potentially damaged ancient templates (12–14, 18–20). A critical concern in our population-level study is the possibility of accidental carryover of a common sequence among experiments giving the appearance of genetic stasis. We approached this problem by using four types of controls (12–14). First, we monitored for contamination through inclusion of extraction blanks and PCR blanks in all experiments. None of these controls were found to be contaminated. Second, DNA extraction and PCR setup were undertaken in a spatially separate facility dedicated to low-copy DNA research. Third, our results were independently replicated in the laboratory of Charles Marshall in a separate building (Department of Geology, University of California, Los Angeles). For this replication, eight ancient samples were provided to a technician (J.A.L.) otherwise not associated with this project. Using equipment and reagents unique to the Marshall laboratory and a different extraction protocol (21), she succeeded in extracting, amplifying, and sequencing DNA from six of eight ancient specimens provided. One sequence was amplified from level 3, two from level 8, and three from level 12. All were identical to the common A haplotype except one from level 12, which was identical to haplotype C otherwise found in level 15 and in recent Lamar Cave pocket gophers. Finally, for ancient and recent samples that provided 164-bp products we tried to amplify a region of 346 bp in cytochrome b by using a second H primer we designed (5′-TAACCTACAAAGGCTGTACG-3′; 5′ at position 408 in GenBank sequence L38467). Sequences longer than a few hundred base pairs should be rare in ancient extracts (12–14). As expected, only in one ancient sample from level 3 and recent samples could we obtain the 346-bp product. Conceivably, nuclear-transposed copies of mtDNA could have been amplified by our procedure although this is unlikely considering that nuclear sequences are much less common than mitochondrial sequences in ancient extracts (12–14, 20). To determine whether our PCR products were a mixture of sequences, we cloned products from five and two, ancient and modern Lamar Cave specimens, respectively, and sequenced three clones from each (Fig. 1, Table 1). All clones had the common haplotype A except for two, which differed by a T-C transition and A-G transition, respectively. Such limited differences among clones are consistent with DNA polymerase incorporation errors (18, 19). Moreover, mtDNA sequences that have been transposed into the nucleus have a different pattern of substitution, reflecting relaxed constraints on codon evolution (20). Translation of the 134-bp fragments that we obtained from clones revealed a single ORF without any stops. Variable sites were restricted to third codon positions as is expected in intraspecific comparisons. Finally, phylogenetic analysis using paup (22) identified haplotypes A, B, and C as a monophyletic, derived clade. As expected, this clade was sister to a T. talpoides sequence (GenBank no. L38467).
Table 1.
DNA sequence variation in the mitochondrial cytochrome b gene
| Haplotype | 1 | 3 | 4 | 5 | 7 | 10 | 16 | 19 | 22 | 25 | 28 | 31 | 34 | 43 | 46 | 52 | 55 | 58 | 61 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | T | T | C | A | C | T | C | A | A | T | C | T | T | A | T | T | T | T |
| B | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C |
| C | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . |
| D | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . |
| E | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | A | . | . | . |
| F | A | . | C | . | . | T | C | T | . | . | . | . | . | . | C | . | . | C | C |
| G | T | C | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | C | C |
| H | T | . | C | . | . | T | C | T | G | . | C | T | C | C | . | A | . | . | . |
| I | A | . | . | . | . | T | C | . | . | . | C | T | . | C | . | C | C | . | . |
| T. talpoides* | A | . | C | . | . | T | . | . | . | . | C | . | . | . | . | A | C | C | . |
| T. bottae* | A | . | . | T | G | . | C | T | . | T | . | . | A | C | . | A | A | C | C |
Position 1 corresponds to position 147 in T. bottae (6). Sequences denoted with an asterisk were retrieved from GenBank.
Analysis of cytochrome b haplotypes in the ancient specimens suggests that the Lamar Cave population was not replaced or augmented by migration from nearby populations over a 2,400-year time period (Fig. 1). One haplotype, A, was found throughout the deposit. Only in the oldest levels (levels 12, 14, and 15) did we find one of six, one of two, and two of nine individuals, respectively, with different haplotypes B and C (Fig. 1). Haplotype B was not found in any modern specimens and is unique to level 15 of Lamar Cave. Haplotype C was not detected in the 53 specimens from the upper levels of the cave (levels 3 and 5–9). However, haplotype C was present in four of seven specimens from the modern population of pocket gophers presently living near Lamar Cave. The common haplotype A in the ancient population was found in remaining specimens from the modern Lamar population and was detected in one specimen from approximately 150 km to the south. All other haplotypes found in six neighboring localities were distinct (Fig. 2).
As we have determined through SSCP analysis and DNA sequencing of ancient and recent pocket gophers (Fig. 1), haplotypes found in neighboring localities differed from those found in living and ancient Lamar Cave samples by at least 1–12 nucleotide substitutions (1.6%–19.0%). In contrast, haplotypes A, B, and C found in Lamar Cave differ by a single third-position nucleotide substitution (1.6%). Consequently, our results suggest that the ancient and recent samples from Lamar Cave and proximity belong to a single, temporally persistent population that did not exchange female migrants with nearby populations for 2,400 years or longer. The spatial pattern of genetic isolation in pocket gophers is supported by cytochrome b analyses of a related pocket gopher species, T. bottae, in which all extant populations had unique haplotypes (6, 7).
To confirm the genetic isolation of the Lamar Cave population, we measured variation in toothrow length, a taxonomically conservative character. Previous research has found that toothrow length is relatively uniform within pocket gopher populations, but differs among subspecies in a pattern not related to changes in temperature or other environmental variables (9). Consequently, in the absence of significant migration, no systematic change in the mean value of toothrow length would be expected to arise in response to changing climatic conditions through time. Our results show that toothrow length changed little, less than 8% throughout the 2,400-year period despite changes in climate (Fig. 1). In contrast, pocket gophers differ by as much as 45% in toothrow length between subspecies in this species group, whereas the within-population variation is less than 25% in T. talpoides tenellus (4). Therefore, the toothrow data confirm the genetic stasis implied by the cytochrome b data.
Diastemal length, a developmentally plastic character that increases with body size and nutritional plane (4, 8, 9), changed more dramatically than toothrow length, as much as 17%, throughout the same period (Fig. 1). During the Medieval Warm Period, pocket gophers had a significantly shorter diastema (89% of mean value) and presumably smaller body size than in colder times. This result accords with Bergmann’s rule, which states that animals from warmer parts of a geographic range tend to be smaller. However, diastemal length change may reflect both gene frequency fluctuations and developmental plasticity. Previous research indicates that body size in pocket gophers can be increased as much as 90% by a change in nutritional quality of food (8). Moreover, the degree of diastemal length change that has occurred in Lamar Cave through time is less than that between modern adjoining pocket gopher populations in low- and high-nutrient-level habitats (8). Consequently, the observed response could be accomplished by developmental changes alone.
Phenotypic responses to environmental change can be caused either by large-scale migration of individuals who have adaptations more appropriate for the altered climate or by genetic and nongenetic changes within populations (23–26). Our approach, involving the measurement of population diagnostic markers and features that respond to changing conditions, has identified within-population processes as critical in the phenotypic response of populations to changes in climate. Ancient DNA technology now enables the application of this approach to populations a few hundred to a few thousand years old (19, 27), thus providing a temporal dimension on the study of genetic variation in natural populations. As in our analysis, genetic markers may be more informative as population diagnostic markers than morphologic measurements because we could distinguish sample localities based on cytochrome b sequences that were similar in toothrow length (e.g., T. t. tenellus vs. T. idahoensis confinus). Nonetheless, judicious choice of morphologic measurements enables our approach to be applied to much older populations (23, 24).
Our results show that isolated populations can persist over several thousand years of climatic change without extinction, suggesting that recolonization in a metapopulation framework (28, 29) at the spatial and temporal scale represented by our study is not a prerequisite for population persistence. However, populations threatened with extinction that are as isolated as the Lamar Cave pocket gophers may not be recolonized through immigration. Given that many species of endangered small vertebrates are strongly subdivided genetically (30), a better understanding of within-population developmental and genetic responses to changing environmental conditions is needed to predict how well the species will survive climatic changes resulting from global warming. In some species, the primary response to environmental change may be developmental rather than genetic (8, 26, 31), a limitation not well appreciated by the focus on genetic variation in species conservation (32).
Acknowledgments
We are indebted to M. F. Smith and J. L. Patton for access to unpublished sequence information and samples. We thank David Jacobs for providing some laboratory equipment for the ancient DNA facility at the University of California, Los Angeles. We are grateful to Charles Marshall for the use of his lab to independently verify our results. We acknowledge support from National Science Foundation Grant EPS 96-40667 (to E.A.H.).
ABBREVIATION
- SSCP
single-stranded DNA conformation polymorphism analysis
Footnotes
Specimens used in the study (note: some sample identifications represent more than a single specimen). Fossils from Lamar Cave: level 3, EH1348, EH1259, EH1281, EH88–243, EH90–61, EH90–67, and EH90–70; level 5, EH1173; level 6, EH88–271A; level 7, EH1011 and EH1116; level 8, EH989; level 9, EH976; level 12, EH92–54 and EH92–87; level 13, EH92–166, EH92–179, and EH92–183; level 14, EH93–101; level 15, EH93–123; and level 16, EH93–164. Total of 88 fossil specimens. One sample of Neotoma cinerea (EH92–77) from level 12 was used in the study. Modern specimens: T. t. bullatus (MVZ 25660), T. t. bridgerii (MVZ 51886), T. i. confinus (MVZ 93439), T. t. fuscus (MVZ 72044), T. i. idahoensis (MVZ 67542), T. t. pryori (MVZ 107000), T. t. relicinus (MVZ 64664), and T. t. tenellus (MVZ 135375 and MVZ 182723–182727). Total of 13 modern pocket gopher specimens. For the number of specimens examined for each level refer to Fig. 1. Excavation details and analysis of stratigraphic completeness are described elsewhere (3, 5).
References
- 1.Lamb H H. Climate: Present, Past, and Future. London: Methuen; 1977. [Google Scholar]
- 2.Meyer G A, Wells S G, Balling R C, Jr, Jull A J T. Nature (London) 1992;357:147–150. [Google Scholar]
- 3.Hadly E A. Q Res. 1996;46:298–310. [Google Scholar]
- 4.Hadly E A. Biol J Linn Soc. 1997;60:277–296. [Google Scholar]
- 5.Hadly, E. A. (1998) Palaeogeog. Palaeoclim. Palaeoecol., in press.
- 6.Patton J L, Smith M F. Syst Biol. 1994;43:11–26. [Google Scholar]
- 7.Ruedi M, Smith M F, Patton J L. Mol Ecol. 1997;6:453–462. doi: 10.1046/j.1365-294x.1997.00210.x. [DOI] [PubMed] [Google Scholar]
- 8.Patton J L, Brylski P V. Am Nat. 1987;130:493–506. [Google Scholar]
- 9.Smith M F, Patton J L. Syst Zool. 1988;37:163–178. [Google Scholar]
- 10.Höss M, Pääbo S. Nucleic Acids Res. 1993;21:3913–3914. doi: 10.1093/nar/21.16.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook A J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Handt O, Krings M, Ward R H, Pääbo S. Am J Hum Genet. 1996;59:368–376. [PMC free article] [PubMed] [Google Scholar]
- 13.Austin J J, Smith A B, Thomas R H. Trends Ecol Evol. 1997;12:303–306. doi: 10.1016/S0169-5347(97)01102-6. [DOI] [PubMed] [Google Scholar]
- 14.Ward R, Stringer C. Nature (London) 1997;388:225–226. doi: 10.1038/40746. [DOI] [PubMed] [Google Scholar]
- 15.Lessa E P, Applebaum G. Mol Ecol. 1993;2:119–129. doi: 10.1111/j.1365-294x.1993.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 16.Ravnik-Glavac M, Ravnik-Glavac D, Dean M. Hum Mol Genet. 1994;3:801–807. doi: 10.1093/hmg/3.5.801. [DOI] [PubMed] [Google Scholar]
- 17.Friesen V L, Congdon B C, Walsh H E, Birt T P. Mol Ecol. 1997;6:1047–1058. doi: 10.1046/j.1365-294x.1997.00277.x. [DOI] [PubMed] [Google Scholar]
- 18.Thomas W K, Pääbo S, Villablanca F X, Wilson A C. J Mol Evol. 1990;31:101–112. doi: 10.1007/BF02109479. [DOI] [PubMed] [Google Scholar]
- 19.Höss M, Vereshchagin N K, Pääbo S. Nature (London) 1994;370:333. doi: 10.1038/370333a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D X, Hewitt G M. Trends Ecol Evol. 1996;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- 21.Cooper A, Mourer-Chauviré C, Chambers G K, von Haeseler A, Wilson A C, Pääbo S. Proc Natl Acad Sci USA. 1992;89:8741–8744. doi: 10.1073/pnas.89.18.8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swofford D L. Phylogenetic Analysis Using Parsimony (paup, version 3.0) Urbana, IL: Illinois Natural History Survey; 1990. [Google Scholar]
- 23.Smith F A, Betancourt J, Brown J H. Science. 1995;270:2012–2014. [Google Scholar]
- 24.Bell M A, Baumgartener J V, Olson E C. Palaeobiology. 1985;11:258–271. [Google Scholar]
- 25.Graham R W, Grimm E C. Trends Ecol Evol. 1990;5:289–292. doi: 10.1016/0169-5347(90)90083-P. [DOI] [PubMed] [Google Scholar]
- 26.Scheiner S M. Annu Rev Ecol Syst. 1993;24:35–68. [Google Scholar]
- 27.Pääbo S. Sci Am. 1993;269:86–92. doi: 10.1038/scientificamerican1193-86. [DOI] [PubMed] [Google Scholar]
- 28.Hanski I, Gilpin M. Biol J Linnean Soc. 1991;41:3–16. [Google Scholar]
- 29.McCaugley D E. Trends Ecol Evol. 1991;6:5–8. doi: 10.1016/0169-5347(91)90139-O. [DOI] [PubMed] [Google Scholar]
- 30.Avise J C, Arnold J, Ball R M, Bermingham E, Lamb T, Neigel J E, Reeb C A, Saunders N C. Annu Rev Ecol Syst. 1987;18:449–522. [Google Scholar]
- 31.Via S, Lande R. Evolution. 1985;39:505–523. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 32.Lacy R C. J Mamm. 1996;78:321–335. [Google Scholar]