Abstract
Significant strides have been made in mammalian cell based biopharmaceutical process and cell line development over the past years. With several established mammalian host cell lines and expression systems, optimization of selection systems to reduce development times and improvement of glycosylation patterns are only some of the advances being made to improve cell culture processes. In this article, the advances pertaining to cell line development and cell engineering strategies are discussed. An overview of the cell engineering strategies to enhance cellular characteristics by genetic manipulation are illustrated, focusing on the use of genomics and proteomics tools and their application in such endeavors. Included in this review are some of the early studies using the ‘omic’ technique to understand cellular mechanisms of product synthesis and secretion, apoptosis, cell proliferation and the influence of the physicochemical environment. The article highlights the significance of integrating genomics and proteomics data with the vast amounts of bioprocess data for improved analysis of the biological pathways involved. Further improvements of the techniques and methodologies used are needed but ultimately, the new cell engineering strategies should provide great insight into the regulatory networks within the cell in a bioprocess environment and how to manipulate them to increase overall productivity.
Keywords: Cell engineering, Proteomics, Genomics, Bioinformatics, Biopharmaceuticals
Introduction
It has been over two decades since the first FDA-approved biopharmaceutical process was established and since then significant strides have been made in the development of process strategies and technological platform for optimal production of biopharmaceuticals. One such technological platform is based on the use of mammalian cell lines for the production of complex biomolecular products. In 2006 it was reported that over 165 new biopharmaceutical entities received FDA approval (Walsh 2006). Mammalian cell lines command an effective monopoly towards production of complex therapeutic proteins that require post-translational modifications (Walsh 2006; Walsh and Jefferis 2006). This unique advantage outweighs the costs associated with mammalian cell culture, which are far greater in terms of development time and manufacturing when compared to bacterial culture. Moreover, in comparison to bacteria the scale up and optimization of mammalian cell cultures to meet production demands has been problematic due to the low productivity and instability of the cell lines used.
The development of cell lines has undergone several advances over the years, essentially to meet the requirement to cut the time and costs associated with using such a complex hosts as production platforms. This paper will review the aspects involved in the development of cell lines and the cell engineering approach that can be employed to enhance productivity, improve cellular metabolism, control proliferation and apoptosis, and reduce instability.
Direct cell engineering has been applied for years to enhance cell lines through the manipulation of single genes that play important roles in key metabolic and regulatory pathways but recently consideration has been given to the use of indirect cell engineering approach, utilizing genomic and proteomic techniques and tools, to aid the discovery process of novel targets for metabolic manipulation within the host cell line. It is hoped that the use of these new techniques and tools will greatly advance our understanding of cellular mechanics in conjunction with improvement of overall productivity in the manufacturing environment and lead to efficient and safe processing of protein products.
Cell line development
Host cell lines
The most common host cell line used for the production of therapeutics is Chinese Hamster Ovary (CHO) (Cockett et al. 1990, Milbrandt et al. 1983) but apart from this cell line, others that are commonly used for the production of recombinant proteins are: mouse myeloma derived (NS0) (Barnes et al. 2001, Bebbington et al. 1992), human embryonic kidney (HEK-293) (Baldi et al. 2005), baby hamster kidney (BHK) (Wurm 2004) and more recently, the human retina derived (PerC6) (Jones et al. 2003) cell lines. For the purpose of process homogeneity, reproducibility and safety, chemically defined protein free medium has become a necessity. However, some media formulations still contain one or two recombinant proteins (such as recombinant human insulin) depending on the cell requirement. The NS0 cell line is the only exception to anchorage dependency and will only need to undergo adaptation to serum free or chemically defined protein free media while the other cell lines maintain their anchorage dependent characteristics unless adapted to suspension culture.
The use of these common cell lines, has led to several sub-cell lines utilizing specific expression technology that have become an industry standard such as CHO DHFR (dihydrofolate reductase system) (Lucas et al. 1996), where a mutant CHO DHFR- cell line is transfected with a vector containing the target gene along with the DHFR marker gene. Selection occurs in the absence of the metabolites hypoxanthine and thymidine and gene amplification is accomplished by exposing the cell line to MTX concentrations, which inhibit DHFR enzyme activity. CHO GS (Glutamine Synthetase) (Cockett et al. 1990) and NS0 GS (Bebbington et al. 1992) are another standardized selection system, where cells are transfected with a vector containing the gene of interest in addition to the glutamine synthetase (GS) gene. GS is an enzyme that allows the cell to synthesize its own intracellular glutamine from glutamate and the ammonia group provided from asparagines (Barnes et al. 2000). NS0 cells are preferred for this system because they have no endogenous GS activity unlike its CHO counterpart. GS cells are selected in glutamine free media and gene amplification can occur by the addition of the GS inhibitor methionine sulphoximine (MSX). The GS cells require lower gene copy numbers, when compared to the DHFR system, for a similar expression level and thus give the added advantage of a shorter selection period. Another advantage of the GS selection system is that the cultures produce less of the toxic metabolite, ammonia, which can negatively affect the glycosylation of the produced recombinant protein (Yang and Butler 2000a; b).
Expression engineering
Most of the expression systems used are inserted into the cellular chromosome by means of random transgene integration events (Wurm 2004). In this process a positional effect can occur where transgene expression is dependent on the position of integration. With standard vector systems, if integration occurs in the heterochromatin, no or minimal transgene expression occurs while integration into the euchromatin will not guarantee maximal transgene expression (Kwaks and Otte 2006). This is supported by evidence that CHO cell line clones with amplified genes located on specific chromosomal regions were found to be more productive and stable than other type of clones (Yano et al. 2006). Thus the copy number of an integrated gene may not indicate a stable expression since deterioration over time may occur due to repeat induced silencing. Targeting transgene integration by the use of the Cre/loxP recombination system and Flp/FRT (Flp-recognition target) in combination with recombinase mediated cassette exchange (RCME) (Baer and Bode 2001) means it is possible to create cell lines which can be selected for high expression of a specific tag, such as Green Fluorescent Protein (GFP). The selected clones can be assessed for stability and are transfected with the gene of interest so that RCME takes place and a high producing cell line with the gene of interest is obtained (Baer and Bode 2001, Feng et al. 1999).
Over the years the use of epigenetics to augment gene expression has been gaining momentum. Several new technologies have been introduced to overcome positional dependent inactivation. For example, using chemicals such as sodium butyrate and sodium propionate, which inhibit histone deacteylases since histone actylation is generally related to enhanced expression, but these compounds tend to have cytotoxic effects as well (Chun et al. 2003, Davie 2003). An alternative method to overcome positional dependent inactivation is the use of vectors with a matrix attachment region (MAR) allowing for the repression of some of the silencing. This allows an open chromatin structure to be maintained, although it doesn’t directly reflect the copy number, and its molecular basis is still not fully understood (Barnes and Dickson 2006, Kwaks and Otte 2006). The discovery of gene regulatory elements which give dominant chromatin opening and resist silencing have started to be used in an industrial setting (Trish et al. 2002). These DNA fragments composed of methylation free-CpG islands are known as ubiquitous chromatin opening elements (UCOE) which have been isolated from ubiquitously expressed housekeeping genes (Antoniou et al. 2003), which can be resistant to heterochromatin mediated silencing. When linked to the human cytomegalovirus (hCMV) promoter, UCOE’s can give stable high level expression with resistance to silencing of the transgene associated with these elements (Williams et al. 2005).
The use of licensed compounds for the induction/repression of gene transcription, such as antibiotic resistance operons including macrolide inducible resistance operons and pharmacological agents that can be used to regulate gene expression, have been exploited for biopharmaceutical production (Weber et al. 2002). An example of such an approved compound is tetracycline in the common tetracycline regulatory system (Gossen and Bujard 1992). The use of vectors that use these compounds, for multiregulated multigene metabolic engineering (Fussenegger et al. 1998a; Fux et al. 2004) will be useful with our increased greater understanding of the complex metabolic and gene regulation networks within the host cell, giving opportunities for application of these technologies to increase product yields.
Optimization of selection procedures
Traditionally, selection of a high producing cell line is a costly process due to the time and labor-intensive procedures involved. It has been referred to as one of the main bottlenecks of getting a producer cell line out of development and into the manufacturing environment (Borth et al. 2000; Carroll and Al-Rubeai 2004). A heterogeneous cell population that undergoes selection for high producers have the disadvantage that the high producing clones mainly have a reduced proliferation rate due to the energy being diverted towards production (Borth et al. 2000; Kim and Lee 1999; Kromenaker and Srienc 1994). However, recently we found from genomics and metabolomics data that the increased specific recombinant protein productivity in arrested or slow growing cells is the result from overall increase in cell growth rather than from a diversion of cellular energy (unpublished data). As a consequence, cell selection using the limited dilution method is usually employed which results in multiple screens of hundreds of wells to select out the appropriate high producing subclone.
Several high throughput techniques for selection of high producing subclones have been conceived in the past few years, some have utilized fluorescence activated cell sorting (FACS) technique (Böhm et al. 2004; Borth et al. 2000; Carroll and Al-Rubeai 2004, 2005; Holmes and Al-Rubeai 1999). A strategy for selection of high producers described by Holmes et al. (1999) using an affinity surface display matrix consisting of avidin and neutravidin, allowed the capture antibody and cell to become irreversibly linked. This complex forms the primary means of capturing the secreted target antibody product which can then be fluorescently labeled for sorting by flow cytometry. This method significantly speeds up the selection process (Carroll and Al-Rubeai 2004), where the critical path activity of commercial cell lines can be minimized to 31 weeks compared to a conventional 40 (Racher 2006).
Optimisation of glycosylation patterns
Mammalian cells have the unique ability to produce therapeutic proteins with oligosaccharides attached to their serine/threonine (O-linked glycosylation) or asparagines (N-linked glycosylation) residues (Butler 2005; Seth et al. 2006a). The functional biological activity of the glycoprotein is directly affected by glycosylation, including its trafficking and folding within the host cell (Scallon et al. 2006; Walsh and Jefferis 2006; Willey 1999). Once secreted, solubility, aggregation, stability and immunogenicity of the protein may be affected by its glycosylation pattern (Kobata 1992; Sinclair and Elliott 2005; Willey 1999; Wyss and Wagner 1996).
N-glycosylation is initiated in the ER, and O-glycosylation can be initiated in either the ER or Golgi apparatus, but due to processing inconstancies glycans can have frequent structural heterogeneities (Patel et al. 1992; Seth et al. 2006a; Varki 1998). The most common therapeutic monoclonal antibody approved biopharmaceutical is of the immunoglobulin G (IgG) class (Walsh and Jefferis 2006). This glycoprotein has several isoforms which are cell line and clone dependent, plus the structure is influenced by the culture environment (Chee Furng et al. 2005; Patel et al. 1992; Raju et al. 2000). This structure or the pattern of glycosylation is dependent on the glycosyltransferases present in the Golgi apparatus. Mouse cells have different glycosylation patterns to humans because they generate Galα1,3-Galβ1,4-GlcNAc residues due to the presence of α1,3 galactosyltransferase. These residues are highly immunogenic in humans and can have an impact on the decision of which cell line to use when this type of glycosylation is present in the end product (Butler 2005; Jenkins et al. 1996). In addition, most host mammalian cell lines (other than the human derived) may not possess the same set of glycosylation enzymes present in humans which may pose an immunogenic threat. It is known that human glycans predominately contain terminal N-acetylneuraminic (NANA) sialylation of the glycoproteins, found to vary in other species (Raju et al. 2000). In fact, most species predominantly have N-glycolyl-neuraminic acid (NGNA) rather than NANA. CHO cells have the advantage of the presence of a high percentage of NANA sialylation, but still include some NGNA sialylation (Baker et al. 2001).
Stability
The consistency of growth, productivity, or product characteristics with increasing generation number defines cell line stability and contributed to overall process consistency. With this in mind, the stability of a genetic, hence physiological, heterogeneic biopharmaceutical cell line is reflected by no deviation of the recombinant product quality and quantity across its manufacturing window (Racher 2004). Therefore, the recombinant product undergoes extensive quality control by assessment of product physiochemical, biochemical, and immunochemical properties under stringent analytical tests since it is the key feature for regulatory approval of a bioprocess (Crommelin et al. 2003; Katterle et al. 2006). In terms of cell line productivity, studies have reported that copy number may not be a viable indicator for productivity and cell line stability rather that mRNA level can be more of a predictive indicator (Barnes et al. 2003, 2004). The location of the amplified genes also determines the stability since studies on CHO cells during prolonged culture showed that in the absence of selective agent such as Methotrexate (MTX), genes have higher stability when located in the telomeric regions of chromosomes (Kim and Lee 1999; Yoshikawa et al. 2000). Recently research has shown that over-expression of human telomerase reverse transcriptase (hTERT), which adds hexameric repeats to the chromosome ends preventing telomeric loss, enhances chromosomal stability in CHO cell lines and possibly production stability (Crea and Al-Rubeai 2006).
Cell engineering
Direct approach to cell engineering
Cell engineering has benefited greatly from recombinant DNA technology and the ability to design complex expression systems. Apart from epigenetic strategies; inducible vectors can be used to regulate gene transcription for over-expression or silencing of key regulatory genes. The direct approach to cell engineering is to introduce to or omit from the cell a gene or genes by genetic engineering methods to endow a particular phenotype in order to improve the cellular processes. Table 1, illustrates some of the cell engineering strategies attempted to control apoptosis, proliferation and productivity within cell lines.
Table 1.
Summary of some of the successful cell engineering strategies carried out over the past years
| Reference | Year | Modification | Cell type |
|---|---|---|---|
| Anti-Apoptotic Engineering | |||
| Itoh et al. | 1995 | bcl -2 | hybridoma |
| Singh et al. | 1996 | bcl -2 | myeloma |
| Tey et al. | 2000a | bcl -2 | NSO |
| Tey et al. | 2000b | bcl -2 | CHO |
| Simpson et al. | 1997 | bcl -2 | hybridoma |
| Perani et al. | 1998 | bcl -2 | hybridoma |
| Ifandi and Al-Rubeai | 2005 | bcl -2 | CHO |
| Meents et al. | 2002 | bcl -2, bcl -xL | CHO |
| Mastrangelo et al. | 2000b | bcl -2, bcl -xL | CHO, BHK |
| Figueroa et al. | 2001 | bcl -2, bcl -2 deletion mutant | CHO |
| Figueroa et al. | 2004 | aven, bcl -xL | CHO |
| Figueroa et al. | 2006 | aven, e1B-19K | CHO |
| Chung et al. | 1998 | bcl -2 | hybridoma |
| Jung et al. | 2002 | bcl -xL | hybridoma |
| Gauthier et al. | 1996 | bcl -xL | hybridoma |
| Charbonneau et al. | 2003 | bcl -xL | hybridoma |
| Sauerwald et al. | 2002 | XIAP | CHO, HEK292 |
| Sauerwald et al. | 2003 | XIAP, CrmA | CHO, HEK293 |
| Lee and Lee | 2003 | bcl -2 | CHO |
| Mercille and Massie | 1999 | elB-19K | NSO,BHK |
| Lasunskaia et al. | 2003 | hsp70 | NSO |
| Terada et al. | 1997 | bag-1, bcl -2 | hybridoma |
| Crea et al. | 2006 | htert | CHO |
| Veraitch and Al-Rubeai | 2005 | neo, bcl -2 | NSO |
| Lim et al. | 2006 | RNAi to Bak, RNAi to Bak | CHO |
| Wong et al. | (2006b) | siRNA to Alg2, siRNA to Requim | CHO |
| Cell-Cycle Engineering | |||
| Koester et al. | 1995 | IRF-1 | BHK |
| Kirchhoff et al. | 1996 | IRF-1 | BHK |
| Geserick et al. | 2000 | IRF-1 | BHK |
| Mazur et al. | 1998 | p21CIPI, p27KIPI,P53275p | CHO |
| Fussenegger et al. | 1998b | p21CIPI, p27KIPI, bclXL | CHO |
| Watanabe et al. | 2002 | p21CIPI | NSO |
| Bi et al. | 2004 | p21CIPI | CHO |
| Ifandi and Al-Rubeai | 2003 | c-myc | CHO |
| Glycosylation Engineering | |||
| Mori et al. | 2004 | siRNA to α2,3-siayltransferase | CHO |
| Weikert et al. | 1999 | siRNA to α1,6-fucosyltransferase | CHO |
| Umaña et al. | 1999 | β1-4-N-acteylglucosaminyltransferase III | CHO |
| Davies et al. | 2001 | β1-4-N-acteylglucosaminyltransferase III | CHO |
Apoptosis engineering
Some of the first direct approach strategies for cellular engineering looked at prolonging the viability of cells in culture by delaying the onset of apoptosis (Al-Rubeai 1998; Al-Rubeai and Singh 1998; Cotter and Al-Rubeai 1995). One anti-apoptotic protein known to suppress the mitochondrial apoptotic pathway is bcl-2 (Gupta 2003). The over-expression of the bcl-2 gene in plasmocytoma (myeloma) (Rabinder 1996), NS0 (Tey et al. 2000b), CHO (Tey et al. 2000a), and hybridoma (Simpson et al. 1998) cells has imparted improved robustness to nutrient deprivation and toxin exposure, including longer survival in intensified culture systems (Tey and Al-Rubeai 2004). Some of the first studies showed that when hybridoma cells over-expressed the Bcl-2 protein it significantly increases cell viability and productivity (Simpson et al. 1997). When myeloma cells, in perfusion culture, were engineered to express E1B-19K (the adenovirus Bcl-2 homologue) the resulted 2-fold increase in viable cell densities allowed for a 40% increase in monoclonal antibody yield (Mercille and Massie 1999). Another approach utilized small interfering RNA (siRNA) (Brummelkamp et al. 2002; Elbashir et al. 2001) constructs directed at mRNA sequences of two pro-apoptotic proteins, Bax and Bak (Lim et al. 2006). Although, silencing was not optimal it was enough to give extended survival in culture and increase IFNγ-product yield in CHO cells by 35% when compared to the control.
Cell cycle engineering
This direct approach has also been used to manipulate the cell cycle genes to increase or decrease proliferation rate and thereby to enhance productivity. When c-myc gene transfected into CHO cells a significant increase in proliferative rate of CHO cell line is demonstrated (Ifandi and Al-Rubeai 2005). Over-expression of both c-myc and bcl-2 resulted in a cell line with increased proliferation and maximum cell densities while a decrease in apoptosis was achieved (Ifandi and Al-Rubeai 2005). Another cell cycle associated gene, p21, has had its potential for inducing cytostasis exploited. Research has shown that by using an inducible p21CIP1 vector a 4 fold increase in antibody productivity resulted in a suspension NS0 culture (Watanabe et al. 2002). Further work showed that this engineered cell line had an increased cell volume, ribosomal protein S6, mitochondrial activity and mitochondrial mass when induced suggesting that p21CIP1induced cell cycle arrest uncouples cell growth from cell cycle progression (Bi et al. 2004). This in-turn was directly related with productivity. Early work with inducing cell arrest was done utilising the interferon regulatory factor 1 (IRF-1) to regulate cell growth (Kirchhoff et al. 1993; Koester et al. 1995) and increase production in BHK-21 cells via estradiol induction (Kirchhoff et al. 1996). However, this work demonstrated that when using the IRF-1 cytostatic gene the main task is keeping the viability high as this has a tendency to decrease with time. By controlling the addition and removal of estradiol, hence modulate IRF-1 activity, it is possible to overcome the loss in viability (Carvalhal et al. 2001). Other studies have also investigated the use of cytostatic genes on CHO cells secreting alkaline phosphatase (SEAP) with similar effects of higher productivity when cell are growth arrested (Carvalhal et al. 2003; Mazur et al. 1998). An example is the use of the p27KIP1 gene induced by acetylaldehyde gas which allowed increased SEAP production in HEK.EBNA cells (Werner et al. 2006). Apart from the control of cell cycle by the p27KIP1 gene, the use of acetylaldehyde as an inducing agent has several advantages in terms of low cost, precise fine tuning of transgene expression (Weber et al. 2004), the use of concentrations below observable toxicities (Hartenbach and Fussenegger 2005; Weber et al. 2005b), and ease of removal prior to downstream processing (Weber et al. 2005a).
Glycosylation engineering
Cell engineering of glycosylation patterns (Davies et al. 2001; Natsume et al. 2006; Shields et al. 2002; Shinkawa et al. 2003; Umaña et al. 1999; Warner 1999; Weikert et al. 1999) and in vitro glycosylation (Butler 2005; Raju et al. 2001) has provided new possibilities for biopharmaceutical design and optimization thus lowering immunogenicity effects, increasing stability and enhancing functional activity. Weikert et al. (1999) engineered CHO cells that secreted either tissue necrosis factor receptor–IgG1 fusion protein (TNFR-IgG) or tissue plasminogen activator (TNK-tPA) to over-express α2,3-sialyltransferase and the resultant glycoproteins had a considerable longer pharmacokinetic mean residence time in rabbit models. Bisected oligosaccharides have been implicated in enhanced antibody dependant cellular cytotoxicity (ADCC) activity. When CHO cells were engineered to over express β1,4 N-acetyl glucosaminoyltransferase (GnTIII), the bisected oligosaccharide glycoforms, ADCC activity was increased 20–100 fold (Davies et al. 2001; Umaña et al. 1999). A silencing approach has also been used in CHO cells to increase the antibody effector function of ADCC by introducing siRNA targeting α1,6 fucosyltransferase (FUT8) mRNA (Mori et al. 2004). Recently, one of the first biopharmaceuticals was approved for the market that included engineered modifications of glycosylation which extent its half life in human patients (Walsh and Jefferis 2006).
Engineering of metabolic pathways
Apart from manipulation of the apoptosis, cell cycle and glycosylation pathways the focus has been on glycolysis to enhance energy use and reduce cytotoxic by-product formation. For instance, by enhancing pyruvate carboxylase expression, which converts pyruvate to oxaloacetate (OAA). The increase in oxalacetate conversion to malate via the malate-aspartate shuttle for entry into the tricarboxylic acid cycle decreased pyruvate concentration which in turn reduced lactate formation (Irani et al. 2002, Seth et al. 2006a); however, no data have been presented on the effect of oxygen uptake rate which should give a more conclusive picture. In another study, antisense technology has been employed to knockdown the enzyme LDH in CHO cells. LDH is involved in the reversible reaction of lactate acid from pyruvate and interestingly, this strategy reduced the overall glycolysis rate, likely due to reduced NADH to NAD+ formation, which is coupled to the pyruvate-lactate reaction (Seth et al. 2006a). This work illustrates the complexity of metabolic pathway engineering and how multiple gene engineering in a single host might be a possible future strategy to overcome some of these problems. The drawback of the antisense and RNA interference (RNAi) technologies is that they lack the precision in controlling expression levels with the consequence that using this technology for metabolic flux control may prove to be difficult.
Other engineering routes
Cell engineering research has also started to focus on other regulatory elements of the cell such as mRNA processing pathways and microRNA (miRNA). Deciphering of the mRNA processing pathways is still in its infancy. It is known that some RNA processing events can affect cell productivity. Recently, it has been discovered that if X-Box binding protein 1 spliced (S) (XBP-1(S)) was obtained from the transcript XBP-1 by mRNA processing, it had the possibility to lead to increased secretion, cell size, membrane synthesis, and energy production (Brewer and Hendershot 2005, Yoshida et al. 2006). Tigges et al. (2006) have transfected a CHO cell line with XBP-1(S) and demonstrated a 6-fold increase in SEAP production compared to the control cell line (Tigges and Fussenegger 2006). On the contrary, this may not pertain to all cases as observed when CHO cells expressing antithrombin III (AT-III) had decreased productivity by 0.7 fold when over-expressing XBP-1(S) (Hayashi et al. 2006). When it comes to the use of miRNA, for now, it is limited since many miRNA and the pathways they affect are still being identified (Ford 2006). These small, 17–24 nucleotide (debatable), non-coding, regulatory RNAs, may comprise a network that regulates 30% of genes within the cell, thus controlling a substantial amount of protein production (Bentwich et al. 2005; Cheng et al. 2005; Ford 2006; Hwang and Mendell 2006). Genes may have several target sites for either one miRNA or a few different miRNAs which makes it possible for combinatorial regulation to fine tune activities (Plasterk 2006). Recent work by Cheng et al. (2005) found that two miRNA; miR-21 and miR-24, profoundly increased cellular proliferation while miR-214 resulted in decreasing apoptosis in HeLa cells. However, the effect of these miRNA seems to be cell specific. Since the complexity of miRNA targeting also gives it the power to regulate several genes at the same time, it might prove to be very useful in engineering cell lines which can bypass apoptosis, have optimal productivity and proliferation by using only 1 or 2 miRNA constructs. Non-transcription events, such as miRNA and mRNA processing pathways, will still require further study but they will nevertheless represent another tool in the cell engineers’ arsenal.
Indirect approach to cell engineering
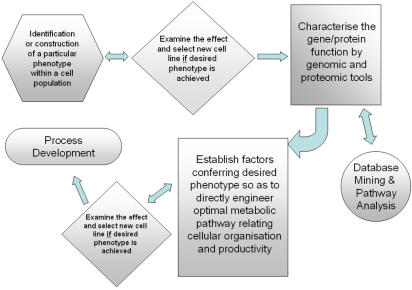
The indirect approach takes a more global view of the cellular pathways and its regulation. By identifying or constructing of the desired phenotype into the host, the factors conferring the phenotype can be discovered and used to engineer improved cell lines (Bailey et al. 2002). A cell line with a particular phenotype either produced by selection, silencing or over-expression can be employed for characterisation according to global gene and protein expression profiles. Thanks to the growth of information from bioinformatics, data mining and pathway analysis of resultant profiles, important genetic and protein expression information pertaining to the phenotype can now be obtained with relative ease. Once the factors that establish the conferred phenotype are elucidated, a direct cell engineering approach may be applied to the pathway in question, an approach that may eventually contribute to overall process optimisation (see Fig. 1). Using this method a greater understanding of the cellular biochemical pathway interactions is gained and it is hoped that the discovery of novel targets can contribute to an overall increase in cell productivity. Even if the target cell line does not respond well to the first attempt of this approach the valuable knowledge gained should aid future engineering strategies.
Fig. 1.
Example of workflow for an indirect engineering strategy employing the use of omic tools for development of cell lines
Genomics and proteomics-tools for indirect cell engineering
In order to apply the indirect approach, new tools have emerged to aid this endeavor. In fact, great efforts have been made in the last ten years to identify genes and pathways relevant to cells enhancing productivity, to identify the metabolic bottlenecks, to understand the mechanism of protein synthesis, to develop better nutrients and media formulation, to reduce apoptosis and increase growth rate, and to design processes with the aim to increase overall productivity. Two new technologies have recently become available which allow for a thorough high throughput assessment of changes at gene (genomics) and protein (proteomics) levels involved in determining productivity in different environmental conditions and to establish functional relationships between cellular organisation and productivity. By using these two new methodologies, genomics and proteomic analysis, together in combination with cell engineering strategies we are one step closer towards a better understanding of cellular behavior during bioprocessing.
Two dimensional polyacrylamide gel electrophoresis (2D-PAGE or 2DE) is the most common proteomic technique and with the recent advent of 2D-differential gel electrophoresis (DIGE) (Lilley and Friedman 2004) combined with advances in quantitative analysis, in image warping and spot detection algorithms (Miller et al. 2006), followed by MALDI-TOF (matrix-assisted laser-desorption/ionization time-of-flight) mass spectrometry (MS) or LC-MS/MS (liquid chromatography) for protein identification (Aebersold and Mann 2003; Gorg et al. 2004), a powerful proteomic tool has been generated. In genomics the use of high-density oligonucleotide GeneChip microarrays for transcriptomics analysis has become a common method to analyse gene expression. These technologies allow for deeper insights into cell metabolism and physiology under process and manufacturing conditions. In addition, these two approaches seem to be a more precise analytical tools for monitoring hundreds to thousands expression changes at gene and protein levels and could significantly contribute to cell culture and cell line development efforts which may, in the near future, achieve specific productivities of over 90 pg cell-1 day−1 (Wurm 2004).
Before discussing some of the applications of this technology and reviewing the progress made, some of the alternative underlying technologies available for such development should be mentioned. When looking at the platforms that can be used for expression analysis in the genomics field or gene transcriptome analysis, the area has undergone significant development from early cDNA microarrays (Schena et al. 1995). The available platforms today are varied (reviewed in Kuo et al. 2006) and the general conclusion is that, overall, most platforms today show good agreement on expression profile data; however, commercial platforms consistently out-performed in-house platforms for internal and external consistency with the best platforms being one-dye, such as Applied Biosystems (ABI), Affymetrix, and Amersham (now GE Healthcare) systems (Kuo et al. 2006).
In proteomics, apart from 2D-gel techniques, the use of MS (as the primary quantitative step) has become a powerful and alternative tool over the last years, with the ability to analyse large numbers of proteins in complex biological samples through development of methods as MudPIT (multi-dimensional protein identification technology), SILAC (stable isotope labeling by amino acids), ICAT (isotope-coded affinity tags), iTRAQ, or 16O-to-18O for functional and quantitative proteomics (Ong and Mann 2005; Roe and Griffin 2006). For example, more than 2000 proteins could be analyzed by SILAC labeling (de Godoy et al. 2006). However, the problem is that in applying these new methods adequate high performance MS machines and properly skilled scientists to operate these systems are necessary. Therefore, 2D-electrophoresis will probably be the method of choice, for the moment, in most of the academic research laboratories in order to carry out quantitative proteome profiling.
Recent studies have shown the potential of applying genomic and proteomic techniques to gain insights into intracellular changes when environmental conditions are altered (e.g. changes in pH, osmolarity and temperature). Further on, expression changes of genes and proteins directly or indirectly involved in the metabolic processes have been identified in different cell clones and cell lines. However, the field of studying the genome and proteome of mammalian cell culture are still in its infancy (Table 2) in comparison with microbial culture; nevertheless, it has a clear objective in identifying new targets for cell line engineering strategies to improve productivity, proliferation, and viability.
Table 2.
Published papers dealing with genomic and proteomic analysis in mammalian cell culture
| Cell line | Comparison | Culture Condition | Analysis | Source | |
|---|---|---|---|---|---|
| Genomic | Proteomic | ||||
| Hybridoma Cell line | Temperature, sparging, agitation | Bioreactor/suspension | − | + | Passini et al. (1989) |
| CHO-K1 | 10% FCS, insulin, basic fibroblast growth factors | adherent/T-flask/batch | − | + | Lee et al. (1996) |
| NS0/NS0_r (cholesterol independent) | Cholesterol-dependent/-independent | Serum-free/suspension/spinner flask/batch | + | + | Seth et al. (2005) |
| CHO | Exponential growth phase—reference map | Serum-free/suspension/2L bioreactor/fed-batch | − | + | Champion et al. (1999) |
| CHO-K1 | 70–80% confluence—reference map | Adherent/T-flask/batch | − | + | Hayduk et al. (2004) |
| CHO-K1 | 0.5 mM n-butyrate acid, 80 μM zinc sulphate | Adherent/T-flask/batch | − | + | van Dyk et al. 2003) |
| Murine hybridoma cells | Hyperosmotic—390 mOsm/kg | Suspension/T-flask/batch | + | − | Shen et al. (2005) |
| CHO | Hyperosmotic—300, 350, 400, 450 mOsm/kg | Adherent/6-well plates/batch | − | + | Lee et al. (2003) |
| MAK mouse-mouse hybridoma cell line | Nutrient feeding—metabolic shift | Serum-free/suspension/fed-batch to continuous culture | − | + | Seow et al. (2001) |
| CHO | 0–2.5% DMSO | Adherent/T-flask/batch | − | + | Li et al. (2006) |
| CHO-K1 | Temperature shift—37°C to 31°C | Adherent/T-flask/batch | − | + | Kaufmann et al. (1999) |
| CHO | Temperature shift—37°C to 33°C | Adherent/T-flask/batch | + | + | Baik et al. (2006) |
| GS-NS0 | Temperature—37°C, 34°C and 22°C | Suspension/T-flask/batch | + | − | Swiderek et al. (2006) |
| CHO-SEAP | High vs low producer | Adherent/T-flask/batch | − | + | Hayduk et al. (2005) |
| GS-NS0/wild type NS0 | Producer vs non-producer | Suspension/spinner flask/batch | + | − | Khoo et al. (2007) |
| GS-NS0 | High vs low producer | Suspension/T-flask/batch | − | + | Alete et al. (2005) |
| GS-NS0 | High vs low producer | Suspension/T-flask/batch | − | + | Smales et al. (2004) |
| GS-NS0 | Mid-exponential phase—reference map | Serum-free/shaker flask/batch | − | + | Smales et al. (2003) |
| GS-NS0 | High vs low producer | Suspension/T-flask/batch | − | + | Dinnis et al. (2006) |
| Murine hybridoma cells | Zinc as insulin replacement | Serum-free/shaker flask/batch | + | − | Wong et al. (2006c) |
| CHO | Batch and fed-batch culture | Serum-free/suspension/5L bioreactor/batch&fed-batch | + | − | Wong et al. (2006a) |
The first application of a large-scale protein analysis approach for mammalian cell culture was reported by (Passini and Goochee 1989). The cellular response of hybridoma cell lines grown in a bioreactor system experienced different environmental stresses (heat shock, agitation and sparging) were analysed by 2DE. Later, work done by Lee and co-workers (Lee et al. 1996a) took the initial steps towards the use of large-scale quantitative proteome analysis as part of an indirect cell engineering strategy for the improvement of specific recombinant protein production and proliferation rates in mammalian cell lines. Detailed 2DE profile maps were established for CHO cells cultivated in media supplemented with foetal calf serum, insulin, or basic fibroblast growth factors. Twenty-four gene products involved in growth factor signaling were identified as differentially regulated. Additionally, the regulation and interaction of transcription factor E2f-1 with cyclin A and E, as well as involvement of cyclin D1 in cell cycle regulation has been found to have an impact on controlling cell growth stimulation. Subsequently studies indicated that over-expression of transcription factor E2f-1 resulted in an increased proliferation in serum free and protein free media (Lee et al. 1996b).
Elucidating the biological pathways regulated by different environmental conditions may contribute to a further understanding of the mechanisms that control cell survival and productivity. Studying the role of these pathways may identify putative targets to interfere with these events, allowing the development of more robust and productive cell lines. Temperature is a significant parameter which can be manipulated to alter the production of recombinant proteins. Kaufmann et al. (1999) have characterized the synthesis of specific cold-induced proteins and molecular changes at posttranslational level resulted from cultivation of recombinant CHO cells at 30°C. Under this hypothermic condition accumulation of cells in G1 phase occurred, with an increased specific productivity and delayed onset of apoptosis. A preliminary analysis of the CHO transcriptome and proteome induced by low temperature have recently been published (Baik et al. 2005), where commercially available rat and mouse cDNA microarrays were used. Overall, transcriptome analysis revealed that low culture temperature could lead to changes in gene expression in various cellular processes such as metabolism, transport and signaling pathways. Complementary to the earlier findings (Kaufmann et al. 1999) 26 different kinds of proteins induced by low-culture temperature were identified by the work of Baik et al. (2006).
Recently, the transcription profile of a NS0 6A1 cell line cultivated at 37°C, 34°C and 22°C has been compared (Swiderek and Al-Rubeai 2006). Microarray analysis revealed that the altered transcription at different temperatures was associated with a broad spectrum of functions within a wide range of regulated genes that were implicated in metabolic pathways such as the urea cycle and metabolism of amino groups. Also affected were the signaling pathways: apoptosis, death receptor, endoplasmatic reticulum stress pathway, and cell cycle checkpoints. Major changes occurred at the transcription level when NS0 6A1 cells were grown at 22°C leading to repression of genes involved in metabolic pathways in contrast from cells grown at 37°C and 34°C where the latter condition had the highest product yield. These observations indicate that the beneficial effect of reduced temperature can only be obtained if the cells are cultured at a condition that does not reduce the catabolic and anabolic needs of cells.
Transcriptional profiling of apoptotic pathways in batch and fed-batch CHO cultures (Wong et al. 2006a) revealed that during periods of high viability, most pro-apoptotic signaling genes were down-regulated but upon loss in viability, several early pro-apoptotic signaling genes were up-regulated. At later stages of viability loss, late pro-apoptotic effector genes such as caspases and DNases were up-regulated. These findings enabled the development of cell lines which are apoptosis resistant (Wong et al. 2006b).
Van Dyk et al. (2003) have presented the protein expression profiles of CHO-K1 cells under normal growth conditions and when exposed to media additives such as zinc sulphate and n-butyrate acid, which are known to have a positive effect on recombinant protein production. Comparison of the expression patterns has revealed a number of cellular proteins induced by thioredoxin, GRP75 and enolase. Surprisingly, there was no increased intracellular response of stress proteins (chaperones) to the addition of zinc sulphate and n-butyrate. Beyond, 2DE analysis has been contributed to gain insight into the cellular changes of recombinant antibody-expressing CHO cells to hyperosmotic stress (Lee et al. 2003). Change of osmolarity from 300 to 450 mOsm/kg in culture media through addition of salts and sugars resulted in a decrease in cell proliferation but increase in specific productivity. Only a few significant proteins such as pyruvate kinase and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) were differentially regulated, as a consequence of increased glucose consumption and subsequently energy metabolism. Down-regulation of tubulin reflected the reduced cell growth. Further, a genome-wide analysis of a murine hybridoma systems (Shen and Sharfstein 2005) subjected to hyperosmotic stress revealed changes on gene transcription levels of several functional groups, such as transcription regulation, cell cycle regulation and apoptosis. In this study, the immunoglobulin heavy chain gene showed no significant change in its mRNA level; however, the immunoglobulin kappa chain gene (light chain) expression was significantly up-regulated, which may contribute to the enhanced antibody production. Many of differentially deregulated genes under osmotic shock might be suggested as candidates for cellular engineering to improve culture viability and longevity.
By maintaining mammalian cultures at low lactate to glucose ratios high cell concentration with high viability were achieved (Europa et al. 2000). It is suggested that the identification of specific proteins or genes involved in the manipulated metabolism could contribute to cell engineering of improved cell lines. Therefore, 2DE has been utilized to analyze the proteome of mouse hybridomas during high consumption of glucose and amino-acids which resulted in a large formation of toxic metabolites (lactate and ammonia) in comparison to a shift in metabolism by sustaining low nutrient feeding in a continuous mode. The cellular response to a metabolic shift showed at least eight proteins to be differentially expressed including metabolic enzymes (GAG, NDUS8) and proteins involved in protein degradation (L1) (Seow et al. 2001). Another investigation focused on the effect of 0–2.5% DMSO presence in culture medium by protein expression profiling to view intracellular changes in cell physiology. DMSO as a medium additive was shown to prolong culture duration, enhance protein production and simultaneously decrease nutrient consumption of CHO cells. Four enzymes related to glycolysis were down-regulated suggesting the enhancement of redistribution of substrate metabolism flux (Li et al. 2006). Limiting steps for further improvements of recombinant protein expression were attributed towards protein post-translational modification and secretion machineries. This was assumed due to the lack of over-expressed chaperon proteins in the presence of DMSO which support protein folding and secretion.
The genetic basis for differentiating a high producer clone from low producer is widely still unclear whether expression is limited by energy metabolism, metabolic regulation or protein folding and secretion capacity (Khoo et al. 2007). Unexpectedly, differences in gene expression profiling of producer and non-producer NS0 cell lines suggested that the selection process actually resulted in a producer clone that did not have higher number of expressed genes in relation to protein synthesis. In mammalian cells, specific productivity seems to be clone dependent as a consequence of random integration sites of foreign genes in the host chromosome (Yoon et al. 2004). Thus it may not be plausible to expect to find a set of genes that have their expression levels always associated with high productivity across the different clones and cell lines. The complexity of this relationship was demonstrated in several studies on the proteome maps of NS0 clones varying in their specific recombinant antibody production rates where the identification of a total of 79 proteins and their categorization into groups suggested that high producer cells are more functionally equipped in some cellular areas, together facilitating improved recombinant protein production instead of only single genes (Alete et al. 2005; Dinnis et al. 2006; Smales et al. 2004). In a study that looked at the proteomic comparison of different productivities of recombinant CHO clones selected under high MTX concentrations, it has been suggested that actin capping protein, one of 21 differentially expressed proteins, might play an important role in heterologous protein secretion as well (Hayduk and Lee 2005). High producer clones expressing SEAP showed as much as a 4-fold increase in actin capping protein expression. Subsequently, studies of CHO clones treated with the small drug molecule cytochalasin D, mimicking the actin cytoskeleton protein, showed an increase of 2- to 3-fold of SEAP productivity within these cells postulated due to modifications in the secretion pathways.
A transcriptomic and proteomic analysis of NS0 cells grown at different densities in perfusion culture have identified a total of 47 genes and 53 proteins that were deregulated at high cell density (Krampe and Al-Rubeai 2006). Specifically, it was found, that up-regulation of gene and protein expression involved in energy metabolism, anti-apoptosis, and cell cycle checkpoints ensured cell survival in high cell density populations. Although no change in expression of protein folding and assembly was observed, expression of chaperone proteins was significantly increased. Another proteomic study looked at differences between an over-expressing c-myc CHO cell line and its control (Kuystermans and Al-Rubeai 2006). This proteomic approach was used to discover factor(s) involved in c-myc-CHO cells that promote the increased proliferative capacity and to gain insights into the mechanism by which c-myc modulates cell growth and apoptosis. Unique changes in protein expression level for 23 proteins were identified including proteins associated with proliferation such as Hsp70 and proteins implicated in exocytosis and cell-to-cell adhesion such as Annexin A2. A large number of proteins identified were involved in the cytoskeletal network, such as actin like protein, β-actin, and F-actin (also involved in the secretory pathway (Hayduk and Lee 2005)) and others in energy metabolism such as ATP synthetase, phosphoglycerate mutase-1 and triophosphate isomerase. It was postulated that a balance exists between proliferation and protein production most likely integrated into the predicted extensive c-myc sub network. The increase in F-actin (which is known to hold up the secretory pathway) and the decreased expression in ER proteins have resulted in reduced secretion of heterologous protein as was subsequently demonstrated by the transfection of the cell line with the hSEAP-hFc construct.
The first combined transcriptomic and proteomic analysis was accomplished on a NS0 cell line by Seth et al. (2005). In this study, the gene and protein expression profile of NS0 cholesterol-dependent phenotype, cultivated under standard cholesterol-dependent growth conditions (NS0) was compared to cells adapted to cholesterol-independent conditions (NS0 revertant) (Seth et al. 2005). Overall, a large number of genes were expressed differentially and mostly down-regulated in the cholesterol biosynthesis, lipid metabolism and central energy metabolism. It was deduced that the deficient expression of a single gene, Hsd17b7, allowed for cholesterol auxotrophy. Subsequent work confirmed, through the use of a cell-engineering route, the role of Hsd17b7 in cholesterol dependency of NS0 cells (Seth et al. 2006b).
Integration of proteomic and genomics analysis
We are continually finding ways to optimise the use of these tools effectively. Further improvements of the techniques and methodologies which address the variation in biological samples and lack of standard procedures (number of replicates and statistical methods) are being addressed including the merging of the “omic” disciplines to give a more complete global understanding of a biological system. With a well established transcriptomic technology and a well advancing proteomic technology, the next few years will most likely see the further integration of these two technological platforms to describe complex biological systems. Since intracellular expression information does not flow in a unidirectional path from transcriptome to proteome it is more likely that a better view would be obtained by examining an integrated view of transcriptomic and proteomic profiles of the same system. At the mRNA level, observations in gene expression give only information on some of the effectors of biological functions while proteins are usually seen as the effectors of biological functions (Chan 2006), although specific time point snapshots of both these systems (transcriptome and proteome) might not be enough to fully understand the regulatory complexity of the biological systems.
Proteins and mRNA transcripts have varied half-life, where mRNA is known to have a half-life from minutes to hours, the half-life of proteins can be from minutes to days. This can give discrepancies in the data, which must be accounted for when analyzing profiles and integrating the two data sets. Temporal integrative profiling experiments might shed some light on these discrepancies and give us a better understanding of the regulatory pathways that interact within the cell (Ideker et al. 2001). At the same time bioinformatics analyses that allow for the interpretation of both data sets on one platform will be advantageous (Tian et al. 2004).
Conclusions
Direct cell engineering approaches have resulted in several current and potential improvements in cell lines of biopharmaceutical importance. With the advent of genomic and proteomic tools, indirect cell engineering for the improvement of biopharmaceutical cell lines is becoming a useful strategic approach. In combination with advances in expression engineering, selection and media development it is hoped to decrease development time and dramatically increase cell line productivity reducing overall costs for the next generation of approved products.
The analyses of gene and protein expression profiles, alone, in cells undergoing the physiological changes provide great insight into the regulatory systems. The examples discussed, showed clearly that by interaction of cells with their extracellular environment some specific pathways are activated. The comparisons of genes and metabolic pathways whose changes in expression profiles influence the phenotype lead to new targets for bioprocess engineering. Therefore, the tools of functional genomics and proteomics could be applied directly in cell and metabolic engineering to identify new targets for improved phenotypes. An additional benefit from this is the knowledge gained to have a fundamental understanding of genetic mechanisms involved in recombinant protein production.
Acknowledgement
This work is funded by Science Foundation Ireland (SFI).
References
- Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207 [DOI] [PubMed]
- Al-Rubeai M (1998) Apoptosis and cell culture technology. Adv Biochem Eng Biotechnol 59:225–249 [DOI] [PubMed]
- Al-Rubeai M, Singh RP (1998) Apoptosis in cell culture. Curr Opin Biotechnol 9:152–156 [DOI] [PubMed]
- Alete DE, Racher AJ, Birch JR, Stansfield SH, James DC, Smales CM (2005) Proteomic analysis of enriched microsomal fractions from GS-NS0 murine myeloma cells with varying secreted recombinant monoclonal antibody productivities. Proteomics 5:4689–4704 [DOI] [PubMed]
- Antoniou M, Harland L, Mustoe T, Williams S, Holdstock J, Yague E, Mulcahy T, Griffiths M, Edwards S, Ioannou PA, Mountain A, Crombie R (2003) Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics 82:269–279 [DOI] [PubMed]
- Baer A, Bode J (2001) Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr Opin Biotechnol 12:473–480 [DOI] [PubMed]
- Baik JY, Lee MS, An SR, Yoon SK, Joo EJ, Kim YH, Park HW, Lee GM (2006) Initial transcriptome and proteome analyses of low culture temperature-induced expression in CHO cells producing erythropoietin. Biotechnol Bioeng 93:361–371 [DOI] [PubMed]
- Bailey JE, Sburlati A, Hatzimanikatis V, Lee K, Renner WA, Tsai PS (2002) Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol Bioeng 79:568–579 [DOI] [PubMed]
- Baker KN, Rendall MH, Hills AE, Hoare M, Freedman RB, James DC (2001) Metabolic control of recombinant protein N-glycan processing in NS0 and CHO cells. Biotechnol Bioeng 73:188–202 [DOI] [PubMed]
- Baldi L, Muller N, Picasso S, Jacquet R, Girard P, Thanh HP, Derow E, Wurm FM (2005) Transient gene expression in suspension HEK-293 cells: application to large-scale protein production. Biotechnol Prog 21:148–153 [DOI] [PubMed]
- Barnes LM, Bentley CM, Dickson AJ (2001) Characterization of the stability of recombinant protein production in the GS-NS0 expression system. Biotechnol Bioeng 73:261–270 [DOI] [PubMed]
- Barnes LM, Bentley CM, Dickson AJ (2003) Stability of protein production from recombinant mammalian cells. Biotechnol Bioeng 81:631–639 [DOI] [PubMed]
- Barnes LM, Bentley CM, Dickson AJ (2004) Molecular definition of predictive indicators of stable protein expression in recombinant NS0 myeloma cells. Biotechnol Bioeng 85:115–121 [DOI] [PubMed]
- Barnes LM, Dickson AJ (2006) Mammalian cell factories for efficient and stable protein expression. Curr Opin Biotechnol 17:381–386 [DOI] [PubMed]
- Barnes LM, Bentley CM, Dickson AJ (2000) Advances in animal cell recombinant protein production: GS-NS0 expression system. Cytotechnology 32:109–123 [DOI] [PMC free article] [PubMed]
- Bebbington CR, Renner G, Thomson S, King D, Abrams D, Yarranton GT (1992) High-level expression of a recombinant antibody from myeloma cells using a glutamine synthetase gene as an amplifiable selectable marker. Biotechnology (NY) 10:169–175 [DOI] [PubMed]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37:766–770 [DOI] [PubMed]
- Bi JX, Shuttleworth J, Al-Rubeai M (2004) Uncoupling of cell growth and proliferation results in enhancement of productivity in p21CIP1-arrested CHO cells. Biotechnol Bioeng 85:741–749 [DOI] [PubMed]
- Böhm E, Voglauer R, Steinfellner W, Kunert R, Borth N, Katinger H (2004) Screening for improved cell performance: selection of subclones with altered production kinetics or improved stability by cell sorting. Biotechnol Bioeng 88:699–706 [DOI] [PubMed]
- Borth N, Zeyda M, Kunert R, Katinger H (2000) Efficient selection of high-producing subclones during gene amplification of recombinant Chinese hamster ovary cells by flow cytometry and cell sorting. Biotechnol Bioeng 71:266–273 [DOI] [PubMed]
- Brewer JW, Hendershot LM (2005) Building an antibody factory: a job for the unfolded protein response. Nat Immunol 6:23–29 [DOI] [PubMed]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553 [DOI] [PubMed]
- Butler M (2005) Animal cell cultures: recent achievements and perspectives in the production of biopharmaceuticals. Appl Microbiol Biotechnol 68:283–291 [DOI] [PubMed]
- Carroll S, Al-Rubeai M (2004) The selection of high-producing cell lines using flow cytometry and cell sorting. Expert Opin Biol Ther 4:1821–1829 [DOI] [PubMed]
- Carroll S, Al-Rubeai M (2005) ACSD labelling and magnetic cell separation: a rapid method of separating antibody secreting cells from non-secreting cells. Journal of Immunological Methods 296:171–178 [DOI] [PubMed]
- Carvalhal AV, Marcelino I, Carrondo MJ (2003) Metabolic changes during cell growth inhibition by p 27 overexpression. Appl Microbiol Biotechnol 63:164–173 [DOI] [PubMed]
- Carvalhal AV, Moreira JL, Carrondo MJ (2001) Strategies to modulate BHK cell proliferation by the regulation of IRF-1 expression. J Biotechnol 92:47–59 [DOI] [PubMed]
- Champion KM, Arnott D, Henzel WJ, Hermes S, Weikert S, Stults J, Vanderlaan M, Krummen L (1999) A two-dimensional protein map of Chinese hamster ovary cells. Electrophoresis 20:994–1000 [DOI] [PubMed]
- Chan E (2006) Integrating Transcriptomics and Proteomics. Genomics and Proteomics [April 2006]. 1-4-2006. Advantage Media. Ref Type: Magazine Article
- Charbonneau JR, Furtak T, Lefebvre J, Gauthier ER (2003) Bcl-xL expression interferes with the effects of L-glutamine supplementation on hybridoma cultures. Biotechnol Bioeng 81:279–290 [DOI] [PubMed]
- Chee Furng WD, Tin Kam WK, Tang GL, Kiat HC, Gek Sim YM (2005) Impact of dynamic online fed-batch strategies on metabolism, productivity and N-glycosylation quality in CHO cell cultures. Biotechnol Bioeng 89:164–177 [DOI] [PubMed]
- Cheng AM, Byrom MW, Shelton J, Ford LP (2005) Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33:1290–1297 [DOI] [PMC free article] [PubMed]
- Chun BH, Park SY, Chung N, Bang WG (2003) Enhanced production of recombinant B-domain deleted factor VIII from Chinese hamster ovary cells by propionic and butyric acids. Biotechnol Lett 25:315–319 [DOI] [PubMed]
- Chung JD, Sinskey AJ, Stephanopoulos G (1998) Growth factor and bcl-2 mediated survival during abortive proliferation of hybridoma cell line. Biotechnol Bioeng 57:164–171 [PubMed]
- Cockett MI, Bebbington CR, Yarranton GT (1990) High level expression of tissue inhibitor of metalloproteinases in Chinese hamster ovary cells using glutamine synthetase gene amplification. Biotechnology (NY) 8:662–667 [DOI] [PubMed]
- Cotter TG, Al-Rubeai M (1995) Cell death (apoptosis) in cell culture systems. Trends Biotechnol 13:150–155 [DOI] [PubMed]
- Crea F, Al-Rubeai M (2006) Does Telomerase Over-Expression Enhance Chromosomal Stability and Possibly Production Stability. Unpublished results
- Crea F, Sarti D, Faleiani F, Al-Rubeai M (2006) Over-expression of hTERT in CHO KI results in decreased apoptosis and reduced serum dependency. J Biotechnol 121:109–123 [DOI] [PubMed]
- Crommelin DJ, Storm G, Verrijk R, de LL, Jiskoot W, Hennink WE (2003) Shifting paradigms: biopharmaceuticals versus low molecular weight drugs. Int J Pharm 266:3–16 [DOI] [PubMed]
- Davie JR (2003) Inhibition of histone deacetylase activity by butyrate. J Nutr 133:2485S-2493S [DOI] [PubMed]
- Davies J, Jiang L, Pan LZ, LaBarre MJ, Anderson D, Reff M (2001) Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnol Bioeng 74:288–294 [PubMed]
- de Godoy LM, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol 7:R50 [DOI] [PMC free article] [PubMed]
- Dinnis DM, Stansfield SH, Schlatter S, Smales CM, Alete D, Birch JR, Racher AJ, Marshall CT, Nielsen LK, James DC (2006) Functional proteomic analysis of GS-NS0 murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng 94:830–841 [DOI] [PubMed]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15:188–200 [DOI] [PMC free article] [PubMed]
- Europa AF, Gambhir A, Fu PC, Hu WS (2000) Multiple steady states with distinct cellular metabolism in continuous culture of mammalian cells. Biotechnol Bioeng 67:25–34 [DOI] [PubMed]
- Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE (1999) Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol 292:779–785 [DOI] [PubMed]
- Figueroa B, Jr, Ailor E, Osborne D, Hardwick JM, Reff M, Betenbaugh MJ (2006) Enhanced cell culture performance using inducible anti-apoptotic genes E1B-19K and Aven in the production of a monoclonal antibody with Chinese hamster ovary cells. Biotechnol Bioeng 2006 Nov 10, [Epub ahead of print] [DOI] [PubMed]
- Figueroa B Jr., Chen S, Oyler GA, Hardwick JM, Betenbaugh MJ (2004) Aven and Bcl-xL enhance protection against apoptosis for mammalian cells exposed to various culture conditions. Biotechnol Bioeng 85:589–600 [DOI] [PubMed]
- Figueroa B Jr., Sauerwald TM, Mastrangelo AJ, Hardwick JM, Betenbaugh MJ (2001) Comparison of Bcl-2 to a Bcl-2 deletion mutant for mammalian cells exposed to culture insults. Biotechnol Bioeng 73:211–222 [DOI] [PubMed]
- Ford LP (2006) Using synthetic miRNA mimics for diverting cell fate: a possibility of miRNA-based therapeutics? Leuk Res 30:511–513 [DOI] [PubMed]
- Fussenegger M, Mazur X, Bailey JE (1998a) pTRIDENT, a novel vector family for tricistronic gene expression in mammalian cells. Biotechnol Bioeng 57:1–10 [DOI] [PubMed]
- Fussenegger M, Schlatter S, Datwyler D, Mazur X, Bailey JE (1998b) Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat Biotechnol 16:468–472 [DOI] [PubMed]
- Fux C, Langer D, Kelm JM, Weber W, Fussenegger M (2004) New-generation multicistronic expression platform: pTRIDENT vectors containing size-optimized IRES elements enable homing endonuclease-based cistron swapping into lentiviral expression vectors. Biotechnol Bioeng 86:174–187 [DOI] [PubMed]
- Gauthier ER, Piche L, Lemieux G, Lemieux R (1996) Role of bcl-X(L) in the control of apoptosis in murine myeloma cells. Cancer Res 56:1451–1456 [PubMed]
- Geserick C, Bonarius HP, Kongerslev L, Hauser H, Mueller PP (2000) Enhanced productivity during controlled proliferation of BHK cells in continuously perfused bioreactors. Biotechnol Bioeng 69:266–274 [DOI] [PubMed]
- Gorg A, Weiss W, Dunn MJ (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665–3685 [DOI] [PubMed]
- Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89:5547–5551 [DOI] [PMC free article] [PubMed]
- Gupta S (2003) Molecular signaling in death receptor and mitochondrial pathways of apoptosis (Review). Int J Oncol 22:15–20 [PubMed]
- Hartenbach S, Fussenegger M (2005) Autoregulated, bidirectional and multicistronic gas-inducible mammalian as well as lentiviral expression vectors. J Biotechnol 120:83–98 [DOI] [PubMed]
- Hayashi T, Ohya T, Kiyama E, Miki H, Kobayashi K, Honda Kn Omasa, Ohtake H (2006) Enhancement of AT-III production in Chinese Hamster ovary cells by modulating transcription factors activated in unfolded protein response. Presented at the 19th JAACT Meeting, Kyoto, 25–28 Sept 2006, Proceedings in press
- Hayduk EJ, Choe LH, Lee KH (2004) A two-dimensional electrophoresis map of Chinese hamster ovary cell proteins based on fluorescence staining. Electrophoresis 25:2545–2556 [DOI] [PubMed]
- Hayduk EJ, Lee KH (2005) Cytochalasin D can improve heterologous protein productivity in adherent Chinese hamster ovary cells. Biotechnol Bioeng 90:354–364 [DOI] [PubMed]
- Holmes P, Al-Rubeai M (1999) Improved cell line development by a high throughput affinity capture surface display technique to select for high secretors. J Immunol Methods 230:141–147 [DOI] [PubMed]
- Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776–780 [DOI] [PMC free article] [PubMed]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934 [DOI] [PubMed]
- Ifandi V, Al-Rubeai M (2003) Stable transfection of CHO cells with the c-myc gene results in increased proliferation rates, reduces serum dependency, and induces anchorage independence. Cytotechnology 41:1–10 [DOI] [PMC free article] [PubMed]
- Ifandi V, Al-Rubeai M (2005) Regulation of cell proliferation and apoptosis in CHO-K1 cells by the coexpression of c-Myc and Bcl-2. Biotechnol Prog 21:671–677 [DOI] [PubMed]
- Irani N, Beccaria AJ, Wagner R (2002) Expression of recombinant cytoplasmic yeast pyruvate carboxylase for the improvement of the production of human erythropoietin by recombinant BHK-21 cells. J Biotechnol 93:269–282 [DOI] [PubMed]
- Jenkins N, Parekh RB, James DC (1996) Getting the glycosylation right: implications for the biotechnology industry. Nat Biotechnol 14:975–981 [DOI] [PubMed]
- Jones D, Kroos N, Anema R, van MB, Vooys A, van der KS, van der HE, Smits S, Schouten J, Brouwer K, Lagerwerf F, van BP, Opstelten DJ, Logtenberg T, Bout A (2003) High-level expression of recombinant IgG in the human cell line per.c6. Biotechnol Prog 19:163–168 [DOI] [PubMed]
- Jung D, Cote S, Drouin M, Simard C, Lemieux R (2002) Inducible expression of Bel-XL restricts apoptosis resistance to the antibody secretion phase in hybridoma cultures. Biotechnol Bioeng 79:180–187 [DOI] [PubMed]
- Katterle B, Muller K, Dippel M (2006) A question of endurance. Eur Biopharmaceutical Rev Bioformulation Manufacturing 108–112
- Kaufmann H, Mazur X, Fussenegger M, Bailey JE (1999) Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol Bioeng 63:573–582 [DOI] [PubMed]
- Khoo G, Falciani F, Al-Rubeai M (2007) A genome-wide transcriptional analysis of producer and non-producer NS0 myeloma cell lines. Biotechnol Appl Biochem 2007, Jan 16 [Epub ahead of print] [DOI] [PubMed]
- Kim SJ, Lee GM (1999) Cytogenetic analysis of chimeric antibody-producing CHO cells in the course of dihydrofolate reductase-mediated gene amplification and their stability in the absence of selective pressure. Biotechnol Bioeng 64:741–749 [PubMed]
- Kirchhoff S, Kroger A, Cruz H, Tummler M, Schaper F, Koster M, Hauser H (1996) Regulation of cell growth by IRF-1 in BHK-21 cells. Cytotechnology 22:147–156 [DOI] [PubMed]
- Kirchhoff S, Schaper F, Hauser H (1993) Interferon regulatory factor 1 (IRF-1) mediates cell growth inhibition by transactivation of downstream target genes. Nucleic Acids Res 21:2881–2889 [DOI] [PMC free article] [PubMed]
- Kobata A (1992) Structures and functions of the sugar chains of glycoproteins. Eur J Biochem 209:483–501 [DOI] [PubMed]
- Koester M, Kirchhoff S, Schaper F, Hauser H (1995) Proliferation control of mammalian cells by the tumor suppressor IRF-1. Cytotechnology 18:67–75 [DOI] [PubMed]
- Krampe B, Al-Rubeai M (2006) Transcriptomic and proteomic analysis of antibody producing NS0 cells cultivated in a high cell density perfusion culture. Global mRNA and protein expression analysis. Meeting at the National Institute for Cellular Biotechnology, Dublin, Ireland
- Kromenaker SJ, Srienc F (1994) Stability of producer hybridoma cell lines after cell sorting: a case study. Biotechnol Prog 10:299–307 [DOI] [PubMed]
- Kuo WP, Liu F, Trimarchi J, Punzo C, Lombardi M, Sarang J, Whipple ME, Maysuria M, Serikawa K, Lee SY, McCrann D, Kang J, Shearstone JR, Burke J, Park DJ, Wang X, Rector TL, Ricciardi-Castagnoli P, Perrin S, Choi S, Bumgarner R, Kim JH, Short GF III, Freeman MW, Seed B, Jensen R, Church GM, Hovig E, Cepko CL, Park P, Ohno-Machado L, Jenssen TK (2006) A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat Biotechnol 24:832–840 [DOI] [PubMed]
- Kuystermans D, Al-Rubeai M (2006) Proteomic analysis of differential protein expression induced by c-myc over-expression in CHO cells. Global mRNA and protein expression analysis. Meeting at the National Institute for Cellular Biotechnology, Dublin, Ireland
- Kwaks TH, Otte AP (2006) Employing epigenetics to augment the expression of therapeutic proteins in mammalian cells. Trends Biotechnol 24:137–142 [DOI] [PubMed]
- Lasunskaia EB, Fridlianskaia II, Darieva ZA, da Silva MS, Kanashiro MM, Margulis BA (2003) Transfection of NS0 myeloma fusion partner cells with HSP70 gene results in higher hybridoma yield by improving cellular resistance to apoptosis. Biotechnol Bioeng 81:496–504 [DOI] [PubMed]
- Lee KH, Harrington MG, Bailey JE (1996a) Two-dimensional electrophoresis of proteins as a tool in the metabolic engineering of cell cycle regulation. Biotechnol Bioeng 50:336–340 [DOI] [PubMed]
- Lee KH, Sburlati A, Renner WA, Bailey JE (1996b) Deregulated expression of cloned transcription factor E2F-1 in Chinese hamster ovary cells shifts protein patterns and activates growth in protein-free medium. Biotechnology and Bioengineering 50:273–279 [DOI] [PubMed]
- Lee MS, Kim KW, Kim YH, Lee GM (2003) Proteome analysis of antibody-expressing CHO cells in response to hyperosmotic pressure. Biotechnol Prog 19:1734–1741 [DOI] [PubMed]
- Lee SK, Lee GM (2003) Development of apoptosis-resistant dihydrofolate reductase-deficient Chinese hamster ovary cell line. Biotechnol Bioeng 82:872–876 [DOI] [PubMed]
- Li JH, Huang Z, Sun XM, Yang PY, Zhang YX (2006) Understanding the enhanced effect of dimethyl sulfoxide on hepatitis B surface antigen expression in the culture of Chinese hamster ovary cells on the basis of proteome analysis. Enzyme Microbial Technol 38:372–380
- Lilley KS, Friedman DB (2004) All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics 1:401–409 [DOI] [PubMed]
- Lim SF, Chuan KH, Liu S, Loh SO, Chung BY, Ong CC, Song Z (2006) RNAi suppression of Bax and Bak enhances viability in fed-batch cultures of CHO cells. Metab Eng 8:509–522 [DOI] [PubMed]
- Lucas BK, Giere LM, DeMarco RA, Shen A, Chisholm V, Crowley CW (1996) High-level production of recombinant proteins in CHO cells using a dicistronic DHFR intron expression vector. Nucleic Acids Res 24:1774–1779 [DOI] [PMC free article] [PubMed]
- Mastrangelo AJ, Hardwick JM, Bex F, Betenbaugh MJ (2000) Part I. Bcl-2 and Bcl-x(L) limit apoptosis upon infection with alphavirus vectors Biotechnol Bioeng 67:544–554 [PubMed]
- Mazur X, Fussenegger M, Renner WA, Bailey JE (1998) Higher productivity of growth-arrested Chinese hamster ovary cells expressing the cyclin-dependent kinase inhibitor p27. Biotechnol Prog 14:705–713 [DOI] [PubMed]
- Meents H, Enenkel B, Eppenberger HM, Werner RG, Fussenegger M (2002) Impact of coexpression and coamplification of sICAM and antiapoptosis determinants bcl-2/bcl-x(L) on productivity, cell survival, and mitochondria number in CHO-DG44 grown in suspension and serum-free media. Biotechnol Bioeng 80:706–716 [DOI] [PubMed]
- Mercille S, Massie B (1999) Apoptosis-resistant E1B-19K-expressing NS/0 myeloma cells exhibit increased viability and chimeric antibody productivity under perfusion culture conditions. Biotechnol Bioeng 63:529–543 [DOI] [PubMed]
- Milbrandt JD, Azizkhan JC, Hamlin JL (1983) Amplification of a cloned Chinese hamster dihydrofolate reductase gene after transfer into a dihydrofolate reductase-deficient cell line. Mol Cell Biol 3:1274–1282 [DOI] [PMC free article] [PubMed]
- Miller I, Crawford J, Gianazza E (2006) Protein stains for proteomic applications: which, when, why? Proteomics 6:5385–5408 [DOI] [PubMed]
- Mori K, Kuni-Kamochi R, Yamane-Ohnuki N, Wakitani M, Yamano K, Imai H, Kanda Y, Niwa R, Iida S, Uchida K, Shitara K, Satoh M (2004) Engineering Chinese hamster ovary cells to maximize effector function of produced antibodies using FUT8 siRNA. Biotechnol Bioeng 88:901–908 [DOI] [PubMed]
- Natsume A, Wakitani M, Yamane-Ohnuki N, Shoji-Hosaka E, Niwa R, Uchida K, Satoh M, Shitara K (2006) Fucose removal from complex-type oligosaccharide enhances the antibody-dependent cellular cytotoxicity of single-gene-encoded bispecific antibody comprising of two single-chain antibodies linked to the antibody constant region. J Biochem (Tokyo) 140:359–368 [DOI] [PubMed]
- Ong SE, Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1:252–262 [DOI] [PubMed]
- Passini CA, Goochee CF (1989) Response of a mouse hybridoma cell-line to heat-shock, agitation, and sparging. Biotechnol Prog 5:175–188
- Patel TP, Parekh RB, Moellering BJ, Prior CP (1992) Different culture methods lead to differences in glycosylation of a murine IgG monoclonal antibody. Biochem J 285(Pt 3):839–845 [DOI] [PMC free article] [PubMed]
- Perani A, Singh RP, Chauhan R, Al-Rubeai M (1998) Variable functions of bcl-2 in mediating bioreactor stress-induced apoptosis in hybridoma cells. Cytotechnology 28:177–188 [DOI] [PMC free article] [PubMed]
- Plasterk RH (2006) Micro RNAs in animal development. Cell 124:877–881 [DOI] [PubMed]
- Rabinder PS (1996) Enhancement of survivability of mammalian cells by overexpression of the apoptosis-suppressor gene bcl-2. Biotechnol Bioeng 52:166–175 [DOI] [PubMed]
- Racher AJ (2004) GS Gene Expression System: GS, biochemical comparability, and cell line stability. Waterside Workshop Presentation Slide, 1–9–2004. Lonza Biologics. (http://www.lonza.com/group/en/company/news/tradefairs). Ref Type: Slide
- Racher AJ (2006) FACS-based method for shortening cell line selection timelines: direct comparison with a conventional approach. Cell Culture Engineering X. Whistler, BC, Canada, 23–4–2006. Engineering Conferences International, 99. Ref Type: Conference Proceeding
- Raju TS, Briggs JB, Borge SM, Jones AJ (2000) Species-specific variation in glycosylation of IgG: evidence for the species-specific sialylation and branch-specific galactosylation and importance for engineering recombinant glycoprotein therapeutics. Glycobiology 10:477–486 [DOI] [PubMed]
- Raju TS, Briggs JB, Chamow SM, Winkler ME, Jones AJ (2001) Glycoengineering of therapeutic glycoproteins: in vitro galactosylation and sialylation of glycoproteins with terminal N-acetylglucosamine and galactose residues. Biochemistry 40:8868–8876 [DOI] [PubMed]
- Roe MR, Griffin TJ (2006) Gel-free mass spectrometry-based high throughput proteomics: Tools for studying biological response of proteins and proteomes. Proteomics 6:4678–4687 [DOI] [PubMed]
- Sauerwald TM, Betenbaugh MJ, Oyler GA (2002) Inhibiting apoptosis in mammalian cell culture using the caspase inhibitor XIAP and deletion mutants. Biotechnol Bioeng 77:704–716 [DOI] [PubMed]
- Sauerwald TM, Oyler GA, Betenbaugh MJ (2003) Study of caspase inhibitors for limiting death in mammalian cell culture. Biotechnol Bioeng 81:329–340 [DOI] [PubMed]
- Scallon BJ, Tam SH, McCarthy SG, Cai AN, Raju TS (2006) Higher levels of sialylated Fc glycans in immunoglobulin G molecules can adversely impact functionality. Mol Immunol 44:1524–1534 [DOI] [PubMed]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470 [DOI] [PubMed]
- Seow TK, Korke R, Liang RC, Ong SE, Ou K, Wong K, Hu WS, Chung MC (2001) Proteomic investigation of metabolic shift in mammalian cell culture. Biotechnol Prog 17:1137–1144 [DOI] [PubMed]
- Seth G, Hossler P, Yee JC, Hu WS (2006a) Engineering cells for cell culture bioprocessing–physiological fundamentals. Adv Biochem Eng Biotechnol 101:119–164 [DOI] [PubMed]
- Seth G, Ozturk M, Hu WS (2006b) Reverting cholesterol auxotrophy of NS0 cells by altering epigenetic gene silencing. Biotechnol Bioeng 93:820–827 [DOI] [PubMed]
- Seth G, Philp RJ, Denoya CD, McGrath K, Stutzman-Engwall KJ, Yap M, Hu WS (2005) Large-scale gene expression analysis of cholesterol dependence in NS0 cells. Biotechnol Bioeng 90:552–567 [DOI] [PubMed]
- Shen D, Sharfstein ST (2005) Genome-wide analysis of the transcriptional response of murine hybridomas to osmotic shock. Biotechnol Bioeng 93:132–145 [DOI] [PubMed]
- Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, Presta LG (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 277:26733–26740 [DOI] [PubMed]
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem 278:3466–3473 [DOI] [PubMed]
- Simpson HS, Milner AE, Al-Rubeai M (1997) Prevention of hybridoma cell death by bcl-2 during suboptimal culture conditions. Biotechnol Bioeng 54:1–16 [DOI] [PubMed]
- Simpson NH, Singh RP, Perani A, Goldenzon C, Al-Rubeai M (1998) In hybridoma cultures, deprivation of any single amino acid leads to apoptotic death, which is suppressed by the expression of the bcl-2 gene. Biotechnol Bioeng 59:90–98 [DOI] [PubMed]
- Sinclair AM, Elliott S (2005) Glycoengineering: the effect of glycosylation on the properties of therapeutic proteins. J Pharm Sci 94:1626–1635 [DOI] [PubMed]
- Singh RP, Emery AN, Al-Rubeai M (1996) Enhancement of survivability of mammalian cells by overexpression of the apoptosis-suppressor gene bel-2. Biotechnol Bioeng 52:166–175 [DOI] [PubMed]
- Smales CM, Birch JR, Racher AJ, Marshall CT, James DC (2003) Evaluation of individual protein errors in silver-stained two-dimensional gels. Biochem Biophys Res Commun 306:1050–1055 [DOI] [PubMed]
- Smales CM, Dinnis DM, Stansfield SH, Alete D, Sage EA, Birch JR, Racher AJ, Marshall CT, James DC (2004) Comparative proteomic analysis of GS-NS0 murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng 88:474–488 [DOI] [PubMed]
- Swiderek H, Al-Rubeai M (2006) Functional Genome-Wide Analysis of Antibody Producing NS0 Cell Line Cultivated at Different Temperatures. Biotechnol Bioeng (in press) [DOI] [PubMed]
- Terada S, Fukuoka K, Fujita T, Komatsu T, Takayama S, Reed JC, Suzuki E (1997) Anti-apoptotic genes, bag-1 and bel-2, enabled hybridoma cells to survive under treatment for arresting cell cycle. Cytotechnology 25:17–23 [DOI] [PMC free article] [PubMed]
- Tey BT, Al-Rubeai M (2004) Suppression of apoptosis in perfusion culture of Myeloma NS0 cells enhances cell growth but reduces antibody productivity. Apoptosis 9:843–852 [DOI] [PubMed]
- Tey BT, Singh RP, Piredda L, Piacentini M, Al-Rubeai M (2000a) Influence of bcl-2 on cell death during the cultivation of a Chinese hamster ovary cell line expressing a chimeric antibody. Biotechnol Bioeng 68:31–43 [DOI] [PubMed]
- Tey BT, Singh RP, Piredda L, Piacentini M, Al-Rubeai M (2000b) Bcl-2 mediated suppression of apoptosis in myeloma NS0 cultures. J Biotechnol 79:147–159 [DOI] [PubMed]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE (2004) Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics 3:960–969 [DOI] [PubMed]
- Tigges M, Fussenegger M (2006) Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab Eng 2006, Apr 3 [Epub ahead of print] [DOI] [PubMed]
- Trish B, Tim C, Michele M, Brian F, David K, Robert C, Thomas CT, Christopher B (2002) The use of UCOE vectors in combination with a preadapted serum free, suspension cell line allows for rapid production of large quantities of protein. Cytotechnology V38:43–46 [DOI] [PMC free article] [PubMed]
- Umaña P, Jean-Mairet J, Moudry R, Amstutz H, Bailey JE (1999) Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol 17:176–180 [DOI] [PubMed]
- Van Dyk DD, Misztal DR, Wilkins MR, Mackintosh JA, Poljak A, Varnai JC, Teber E, Walsh BJ, Gray PP (2003) Identification of cellular changes associated with increased production of human growth hormone in a recombinant Chinese hamster ovary cell line. Proteomics 3:147–156 [DOI] [PubMed]
- Varki A (1998) Factors controlling the glycosylation potential of the Golgi apparatus. Trends Cell Biol 8:34–40 [DOI] [PubMed]
- Veraitch FS, Al-Rubeai M (2005) Enhanced growth in NS0 cells expressing aminoglycoside phosphotransferase is associated with changes in metabolism, productivity, and apoptosis. Biotechnol Bioeng 92:589–599 [DOI] [PubMed]
- Walsh G (2006) Biopharmaceutical benchmarks 2006. Nat Biotechnol 24:769–776 [DOI] [PubMed]
- Walsh G, Jefferis R (2006) Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol 24:1241–1252 [DOI] [PubMed]
- Warner TG (1999) Enhancing therapeutic glycoprotein production in Chinese hamster ovary cells by metabolic engineering endogenous gene control with antisense DNA and gene targeting. Glycobiology 9:841–850 [DOI] [PubMed]
- Watanabe S, Shuttleworth J, Al-Rubeai M (2002) Regulation of cell cycle and productivity in NS0 cells by the over-expression of p21CIP1. Biotechnol Bioeng 77:1–7 [DOI] [PubMed]
- Weber W, Marty RR, Keller B, Rimann M, Kramer BP, Fussenegger M (2002) Versatile macrolide-responsive mammalian expression vectors for multiregulated multigene metabolic engineering. Biotechnol Bioeng 80:691–705 [DOI] [PubMed]
- Weber W, Rimann M, de Glutz FN, Weber E, Memmert K, Fussenegger M (2005a) Gas-inducible product gene expression in bioreactors. Metab Eng 7:174–181 [DOI] [PubMed]
- Weber W, Rimann M, Spielmann M, Keller B, oud-El BM, Aubel D, Weber CC, Fussenegger M (2004) Gas-inducible transgene expression in mammalian cells and mice. Nat Biotechnol 22:1440–1444 [DOI] [PubMed]
- Weber W, Spielmann M, oud El-Baba M, Keller B, Aubel D, Fussenegger M (2005b) Tobacco smoke as inducer for gas phase-controlled transgene expression in mammalian cells and mice. Biotechnol Bioeng 90:893–897 [DOI] [PubMed]
- Weikert S, Papac D, Briggs J, Cowfer D, Tom S, Gawlitzek M, Lofgren J, Mehta S, Chisholm V, Modi N, Eppler S, Carroll K, Chamow S, Peers D, Berman P, Krummen L (1999) Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat Biotechnol 17:1116–1121 [DOI] [PubMed]
- Werner NS, Weber W, Fussenegger M, and Geisse S (2006) A gas-inducible expression system in HEK.EBNA cells applied to controlled proliferation studies by expression of p27(Kip1). Biotechnol Bioeng 2006, Oct 20 [Epub ahead of print] [DOI] [PubMed]
- Willey KP (1999) An elusive role for glycosylation in the structure and function of reproductive hormones. Hum Reprod Update 5:330–355 [DOI] [PubMed]
- Williams S, Mustoe T, Mulcahy T, Griffiths M, Simpson D, Antoniou M, Irvine A, Mountain A, Crombie R (2005) CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnol 5:17 [DOI] [PMC free article] [PubMed]
- Wong DC, Wong KT, Lee YY, Morin PN, Heng CK, Yap MG (2006a) Transcriptional profiling of apoptotic pathways in batch and fed-batch CHO cell cultures. Biotechnol Bioeng 94:373–382 [DOI] [PubMed]
- Wong DC, Wong KT, Nissom PM, Heng CK, Yap MG (2006b) Targeting early apoptotic genes in batch and fed-batch CHO cell cultures. Biotechnol Bioeng 95:350–361 [DOI] [PubMed]
- Wong VV, Nissom PM, Sim SL, Yeo JH, Chuah SH, Yap MG (2006c) Zinc as an insulin replacement in hybridoma cultures. Biotechnol Bioeng 93:553–563 [DOI] [PubMed]
- Wurm FM (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol 22:1393–1398 [DOI] [PubMed]
- Wyss DF, Wagner G (1996) The structural role of sugars in glycoproteins. Curr Opin Biotechnol 7:409–416 [DOI] [PubMed]
- Yang M, Butler M (2000a) Effect of ammonia on the glycosylation of human recombinant erythropoietin in culture. Biotechnol Prog 16:751–759 [DOI] [PubMed]
- Yang M, Butler M (2000b) Effects of ammonia on CHO cell growth, erythropoietin production, and glycosylation. Biotechnol Bioeng 68:370–380 [DOI] [PubMed]
- Yano H, Takagi Y, Cao Y, Omasa T, Asakawa S, Shimizu N, Ohtake H (2006) Analysis of specific chromosomal region in Chinese Hamster ovary-dihydrofolate reductase gene amplification. Presented at the 19th JAACT Meeting, Kyoto 25–28 Sept 2006, Proceedings in press
- Yohito Itoh, Hiroshi Ueda, Eiji Suzuki (1995) Overexpression of bcl-2, apoptosis suppressing gene: Prolonged viable culture period of hybridoma and enhanced antibody production. Biotechnol Bioeng 48:118–122 [DOI] [PubMed]
- Yoon SK, Hwang SO, Lee GM (2004) Enhancing effect of low culture temperature on specific antibody productivity of recombinant Chinese hamster ovary cells: clonal variation. Biotechnol Prog 20:1683–1688 [DOI] [PubMed]
- Yoshida H, Oku M, Suzuki M, Mori K (2006) pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 172:565–575 [DOI] [PMC free article] [PubMed]
- Yoshikawa T, Nakanishi F, Ogura Y, Oi D, Omasa T, Katakura Y, Kishimoto M, Suga K (2000) Amplified gene location in chromosomal DNA affected recombinant protein production and stability of amplified genes. Biotechnol Prog 16:710–715 [DOI] [PubMed]