Abstract
Considerable increases in productivity have been achieved in biopharmaceutical production processes over the last two decades. Much of this has been a result of improvements in media formulation and process development. Though advances have been made in cell line development, there remains considerable opportunity for improvement in this area. The wealth of transcriptional and proteomic data being generated currently hold the promise of specific molecular interventions to improve the performance of production cell lines in the bioreactor. Achieving this—particularly for multi-gene modification—will require specific, targeted and controlled genetic manipulation of these cells. This review considers some of the current and potential future techniques that might be employed to realise this goal.
Keywords: Targeted, Homologous recombination, Recombinases, Site-specific, Inducible, Transgene, Recombinant protein, CHO
Introduction
Accurate and reproducible modification of genomic DNA is desirable in many cell and molecular biology applications. This is not a facile undertaking. There are significant challenges in achieving precise (as little as a single base pair) modification at a particular locus on a 3 billion base pair template. More so when this must be achieved in as many, if not all, cells in a population—and ultimately in a whole organ or organism as in the case of gene therapy. Researchers have been inserting exogenous sequences in cells for many years but most often in an unsupervised, random manner. Depending on the application this is often sufficient but there is always the risk of unwanted ‘side-effects’ such as oncogene activation or disruption of important endogenous loci.
In light of this a range of tools and techniques have been developed in an attempt to gain more precision in genetic engineering applications for biomedicine, biotechnology and basic research. Techniques such as gene deletion (knockout) by homologous recombination were developed over 20 years ago to study gene function in the mouse (Smithies et al. 1985). In the research laboratory this method remains the definitive means of silencing genes (notwithstanding the widespread use of RNAi for knockdown) though many additional features can now be included in the common-or-garden knockout construct—these will be discussed in more detail later. Improvements have come about in the methods of delivering the modifying sequence to the cell(s) (Hirata et al. 2002), more efficient promoter/enhancer combinations have led to more efficient transgene expression with less tendency toward silencing, better selection strategies have increased targeting efficiency; the incoming sequences may contain recognition sites for recombinases to remove or insert other features subsequently (Kilby et al. 1993) or even rare-endonuclease sites to facilitate further rounds of modification (Cohen-Tannoudji et al. 1998).
Several recent reviews have addressed these advances in the context of gene therapy in particular as well as for basic research (Coates et al. 2005; Vasquez et al. 2001; Sorrell 2005). In this review, we will focus on the application of these tools in cell lines relevant to the biopharmaceutical industry.
Therapuetic protein production
The first commercial use of mammalian cells as bioreactors came in 1986 with the introduction of Activase™—recombinant human tissue plasminogen activator. By 2004 the recombinant therapeutic protein market had expanded to over 70 products and a combined value of $35.8bn—with many more in the research pipeline or trials (Pavlou 2004). Many of these are complex molecules with post-translational modifications that necessitate their production in mammalian cells. The most widely used cell line in the Biopharma industry at the moment is Chinese Hamster Ovary (CHO). These are typically grown in non-adherant, serum-free medium, frequently in large batch processes up to of 10,000 l.
In the 20 years since the first product came on the market, the greatest leaps in productivity have been made first and foremost by refining the culture medium and secondly, by improving the scale-up process. Other gains have been made via improved promoter/enhancer constructs for transgene expression as well as clone selection procedures (Wurm 2004). These improvements have led to increases in yield from 10–50 mg/l in the 1980s to 500–5,000 mg/l today. Though there remains room for further improvements via medium and process optimisation, many believe that future increases in yield will come through metabolic engineering to create cell lines with improved bioreactor performance. Already, strategies such as engineered overexpression of anti-apopototic genes have demonstrated the value of this approach (Meents et al. 2002).
The cells that manufacture the protein product are engineered to do so using methods that have not changed significantly in the last 2 decades—though they have been modified in various ways—mostly due to ‘in-house’ company preferences—that have evolved over the years. The following is a brief outline of the steps involved in generating these cell lines:
The coding sequence for the protein (product) is cloned into an expression vector usually under the control of some viral promoter/enhancer combination. This vector also contains the cDNA for a resistance marker (e.g. the dihydrofolate reductase gene (DHFR)) for selection and amplification purposes.
CHO cells are transfected with the expression vector and incubated in low-nucleotide media containing the DHFR inhibitor, methotrexate (Mtx), or appropriate selective agent.
Only cells that have incorporated the exogenous DHFR gene will survive in the absence of essential nucleotide supplementation (1,000s of clones generated at this point).
In order to increase the expression level of the protein product, the product sequence is co-amplified with the DHFR gene by increasing the selective pressure with high concentrations of Mtx (100s clones generated).
The Mtx levels are further elevated to select for very high yield clones (10s). Frozen cell banks of each clone are created.
These candidates are adapted for growth in serum-free, suspension culture and assessed for stable product yield and growth characteristics.
The ‘production’ clone is chosen and a master cell bank generated before expansion to large-scale production vessels.
Though protocols based on this type of approach continue to be favoured by the industry there are significant disadvantages associated with it:
Step 2. The incorporation of the exogenous plasmid into the genome occurs in a random manner and can potentially disrupt important endogenous sequences or locate in transcriptionally silent regions leading to unpredictable phenotypes and low product yield, respectively (Sutter et al. 2003).
Step 4. Amplification of integrated sequence in this manner involves large areas of chromosomal DNA with unforeseeable consequences for cellular physiology. This kind of amplification has been shown to generate long, tandem repeats which are genetically unstable and may even induce chromosomal rearrangement (Wurm and Petropoulos 1994).
Step 5. As in 4, further amplification increases the likelihood of genetic instability and clone failure. In addition, amplified sequences (in particular viral promoters) are frequently silenced by methylation or other epigenetic modification (Derouazi et al. 2006).
Step 6. This part of the process can take many months with no guarantee that clones that performed well when attached to plastic in serum-supplemented media during initial screening will continue to do so or that they will maintain stable growth rates and product yields.
Step 7. Any change in clone behavior, such as specific yield (pg/cell/hr), growth rate/density, apoptosis, at this stage can have huge cost implications and may even affect regulatory approval for release into the market.
In future, the problems associated with achieving high product yields via gene amplification may be further complicated by the challenges of doing so in a cellular background where the host genome may already be carrying a ‘genetic load’ due to metabolic engineering. Clearly, an upper maximum will be reached in terms of how many transgenes (including selection markers) that a cell can sustain. Indeed many groups are actively involved in providing solutions to this problem. Most agree that this will involve a combination of inducible control of transcription (May et al. 2006; Weber et al. 2004; Hartenbach and Fussenegger 2005) with targeted insertion in the genome (Koduri et al. 2001; Coates et al. 2005).
First, let’s consider the options currently available to the molecular biologist when considering engineering cells in a specific manner.
Site-directed insertion.
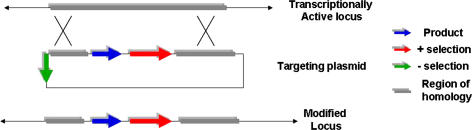
As mentioned, gene deletion by homologous recombination (HR) has been used in reverse genetics experiments for many years (Smithies et al. 1985). In its most basic form it comprises a drug selection marker and a DNA sequence with homology to the locus of interest in the target cell (Fig. 1). Typically the target cell is an embryonic stem (ES) cell that is used to generate chimeric mice by microinjection into harvested blastocysts. Upon introduction into the nucleus of the ES cell, the cellular DNA repair machinery catalyses its introduction at the chromosomal site of homology. Cells that have successfully acquired the incoming sequence can be selected for by their resistance to the selective agent. However, far more frequently (orders of magnitude) the process results in clones that have acquired resistance by randomly inserting the exogenous sequence in a non-homologous manner. This necessitates a screening procedure to identify correctly targeted clones. In somatic cells where HR occurs at much lower frequencies than in ES cells the number of HR events can be vanishingly low against a background of random insertion (Yamane-Ohnuki et al. 2004).
Fig. 1.
Targeting a transcriptionally active locus by homologous recombination
Several strategies have been developed in order to overcome this problem. Increasing the length of the homologous sequence (6–10 kb) in the targeting vector can improve efficiencies by as much as 190-fold (Hasty et al. 1991). Furthermore, using isogenic DNA also achieves greater efficiency in HR (Te Riele et al. 1992; Ward et al. 1993) than non-isogenic sequence of similar length. Enrichment of clones can be improved by using promoter-less selection cassettes that only confer resistance when placed downstream of an endogenous promoter (that of the gene being deleted/modified) (Bhat et al. 1988). Random insertion in this case would most often lead to a non-transcribed drug resistance gene, although this ‘promoter-trap’ design is only suitable firstly, when the target is a gene (which may not always be the case) and secondly, if the gene is actively transcribed in the host cell.
The inclusion of a negative selection marker provides another means of enriching for correctly targeted cells. In this approach (usually combined with a positive marker) a gene such as Herpes Simplex Virus thymidine kinase is included outside the region of homology. In the event of authentic HR, this sequence is removed and the cell will survive in medium supplemented with an agent (such as ganciclovir) that is only converted to a cytotoxic form in the presence of the negative selection marker. Those cells that incorporate the foreign DNA in a random manner will usually, though not always, have gained the negative marker also and will be killed.
Rouet et al. (1994) reported on another method for increasing incidence of HR upon introduction of exogenous DNA—the introduction of double stranded breaks (DSB) at the locus of interest. This was found to stimulate cellular DNA repair and resulted in HR efficiencies many orders of magnitude higher than in the absence of a DSB. A recognition site for an extremely rare-cutting endonuclease (I-SceI in this case) is introduced into the genome that could be snipped upon transient expression of the enzyme. The break is then repaired using exogenous DNA with a short region of homology. An attractive aspect of this system is that it can be used to repeatedly target the same locus once the recognition site is in place (Cohen-Tannoudji et al. 1998). However, one limitation is that the initial insertion of the site relies on standard HR techniques with lower efficiencies. This is a recurring theme for many of the tools that have been developed for site-specific genome engineering.
Recombinases
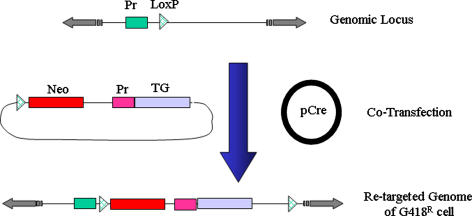
One of the most exciting and novel developments in mammalian genome modification has been the use of recombinase enzymes. These proteins catalyse recombination between two, double-stranded sequences of DNA in a site-dependent manner. The recognition site is usually quite long (>30 bp) and consists of a central spacer region surrounded by inverted repeat sequences. The best known of these, Cre, was discovered in the bacteriophage P1 (Abremski and Hoess 1984). Others include Flp from S.cerevisiae and the ϕC31 phage intergase (Kilby et al. 1993; Groth et al. 2000). The recognition sequence for Cre is 34 bp long comprising an eight bp spacer flanked by two 13 bp inverted repeats (Hoess and Abremski 1984). This sequence is referred to as a LoxP site (locus of cross-over in P1). By placing a LoxP site in the genome of a cell it is possible to target exogenous sequences to that locus by inclusion of another LoxP site in the incoming vector. In the presence of the Cre enzyme these sites are brought into close proximity and recombination results in insertion of the vector sequence flanked by two LoxP sites (Fig. 2). However, early studies revealed this process to be quite inefficient due to the thermodynamic bias towards the excision reaction. The proximity of the resulting LoxP sites favours their recombination over that of the exogenous vector and the genomic LoxP. In order to overcome this, two main approaches can be used. Firstly, Cre has been shown to catalyse recombination between a wild-type LoxP site and a mutant site with one or more base changes (Abremski et al. 1986; Lee and Saito 1998). However the resulting pair of mutant LoxP sites cannot recombine, preventing immediate removal of the inserted sequence. A further refinement to this approach that is much more versatile in terms of re-targeting the locus is referred to as Recombinase-Mediated Cassette Exchange (RMCE) (Seibler and Bode 1997; Seibler 2007). In this system, two mutant, non-compatible LoxP site are located adjacent to each other. Cre can not (or only very inefficiently) catalyse recombination between these sites. However, exogenous sequence flanked by complementary LoxP sites will be exchanged at the locus for the existing intervening sequence.
Fig. 2.
Cre-mediated genomic targeting. Insertion of a transgene at a LoxP site in the cell genome is accomplished by co-transfection of a cre-expression vector (pCre) and a second construct containing a LoxP site, a section marker (neo) and a transgene (TG) downstream of a promoter (Pr)
These recombinase-based approaches have been used in several ingenious ways to manipulate the genomes of mammalian cells. Strategic placement of loxP sites has been used to both activate and inactivate transcription. By placing the expression of Cre under the control of an inducible promoter, gene deletion or activation has been achieved in a temporal manner (Sauer 1998). Others have combined recombinase systems to place LoxP and FRT (substrate for Flp) sites adjacent to each other. This has a similar outcome to using mutant LoxP sites, i.e. recombination between compatible (and not the adjacent) sequences may only occur. Alternatively the ϕC31 recombinase will only insert incoming sequence in a unidirectional manner due to the heterologous nature of its recognition sequences (known as att sites) (Thorpe and Smith 1998). A further potential for this particular integrase is the existence of att-like sites in the genomes of both human and mouse, facilitating the insertion of transgenic sequences without first having to introduce a recombinase-recognition sequence (Sorrell 2005). There are also likely to be similar sites in the hamster, though this has not yet been investigated to our knowledge.
Novel DNA-modifying enzymes:
The discovery that HR could be greatly enhanced by introducing a DSB at the target locus prompted an interest in creating chimeric enzymes that could do so in a sequence specific manner (Durai et al. 2005). Indeed the ability to target other enzymatic or functional activities to predetermined sites has led to the emergence of a specialist field in designer zinc finger (ZF) proteins (Mandell and Barbas 2006). These proteins are made up of distinct zinc finger motifs—each of which binds to a specific DNA triplet. By combining several of these motifs a molecule can be designed that will bind a sequence of 15–18 bp in a highly selective and specific manner. The expertise required to design these proteins is still exclusive to a small number of labs though recently some web-based tools have been made available to help those outside the field take advantage of this technology (Mandell and Barbas 2006). By fusing one of these ZF proteins to a generic endonuclease a DSB can be introduced at a predetermined, unique site in the genome (Bibikova et al. 2001). Modifications at the locus can then be implemented by HR at high efficiency with an incoming sequence. There is exciting potential for these novel, designer proteins in terms of their application in biomedical research as well as gene therapy though further refinements in the design algorithms will be required to avoid ‘off-target’ effects and ensure safety.
Other tools:
Triplex-forming Oligos (TFOs)
These are short oligodeoxynucleotide sequences that bind tightly to purine-rich sites in duplex DNA (Helene 1991). They have been successfully used to disrupt genes by targeting damaging agents to specific loci (Giovannangeli et al. 1992). It is estimated that unique sites for TFOs occur approximately every 1 kb in the genome (Perkins 1998) which would suggest that most genes should be amenable to targeting in this manner. It has also been shown that these agents can stimulate HR with exogenous sequence at the APRT locus in CHO cells (Vasquez et al. 2001). Culver et al. (1999) demonstrated correction of point mutations in the ADA and p53 genes in human cells at high frequency using TFOs linked to a short donor oligo.
Transposons
Transposons are small mobile genetic elements found in prokaryotic and some eukaryotic genomes. There are no active transposons known in mammalian species but elements such as Sleeping Beauty from fish have been successfully used to integrate transgenes in mammalian cells (Kaufman et al. 2004). There is also a group of non-long terminal repeat retrotransposon elements found in mammals known as Long Interspersed Elements (LINE-1) that constitute approximately 20% of the genome (Soares et al. 1985). These elements are not generally active in somatic cells but transcripts have been detected in germ cells and some cancer cell lines and have been implicated in some single-gene diseases in humans where retrotransposition has resulted in disruption of genes such as dystrophin (Ferlini and Muntoni 1998). In terms of their use as a genome modification tool, a major drawback of transposons is the random nature of the insertion event.
Adeno-associated virus
So far, we have not mentioned viruses as agents for manipulating the genome. The reasons for this omission are twofold. Firstly, their use has been reviewed extensively in recent years (Ghosh 2006; Liu (2006) and secondly, the introduction of viral material into cells producing therapeutic proteins destined for biopharmaceutical use has yet to meet regulatory approval. However, this situation may change in the future and these types of vectors can have some distinct advantages over other delivery methods. One in particular is a combination of viral delivery of HR constructs by the adeno-associated virus (Miller et al. 2006; Hirata et al. 2002). This virus is naturally occurring and asymptomatic in humans and the wild-type strain normally integrates at a specific locus on chromosome 19 (Kotin et al. 1990). However, by including relatively short (1–4 kb) homologous sequence in the viral genome, targeting of specific loci can be achieved in a large percentage of transduced cells—often at both alleles and in the absence of any selective pressure (Chamberlain et al. 2004). This delivery method has obvious advantages in gene therapy applications but it holds great potential for modification of commercial cell lines also.
Current limitations:
One of the main limitations with many of the tools described so far is that the first step must include a HR targeting event, whether to create the desired phenotype or to insert elements that will facilitate further targeted manipulation. Though some of the techniques have improved the efficiency of HR in mammalian cells it remains the bottleneck in the targeted engineering of the genome. More efficient vectors such as retroviral or adenoviral vectors result in random or episomal delivery of the transgenic payload. Recent developments such as AAVs combined with HR targeting or designer zinc finger nucleases aspire to marry the best attributes of different approaches.
Implication for recombinant protein production processes:
As stated earlier, future improvements in the bioreactor performance of commercial cell lines, such as CHO, will likely be contributed to by metabolic engineering strategies. There are two points to consider here. Firstly, there is the question of the recombinant product itself. It would be desirable (and indeed some companies may already have pursued this route) to generate producer cell lines with minimal if not zero amplification of the transgenic sequence. Expression vectors have been developed that are efficient enough to generate excess mRNA transcripts from a single integration event. It is not apparent whether this integration must be at a specific, known transcriptional ‘hotspot’ or whether random insertion is sufficient (Koduri et al. 2001). Aside from the advantage of reducing the risk of genetically unstable clones, there is also the potential to reduce the time required to generate new producer cell lines by developing ‘master’ cell lines that have been pre-engineered to contain say, recombinase-recognition sites at a known locus.
The second consideration is improving the ‘master’ cell line to improve performance irrespective of the product being made. This is analogous to pre-adaptation to serum-free or anchorage-independent growth. There are concerted efforts underway by several groups to identify genes that would best influence the performance of cells in culture using transcriptional and proteomic profiling (Wlaschin and Seth 2006; Smales et al. 2004). To date, most metabolic engineering strategies reported entail transient and/or randomly integrated overexpression or knockdown (short hairpin RNA) constructs to confer resistance to apoptosis (Figueroa et al. 2004), control growth characteristics (Mazur et al. 1998; b) or modify the protein product (Wong and Yap 2006). In addition, most studies involve modifying the expression of a single gene. Significant improvements in cellular productivity will likely entail controlling multiple gene expression levels, some upwards and some down, raising the prospect of ‘over-engineered’ cell lines. A limited number of studies, notably by Fussenegger et al., (Gonzalez-Nicolini and Fussenegger 2005; Kramer and Fussenegger 2005; Malphettes and Fussenegger 2006) have demonstrated the possibility of controlled, artificial gene networks where expression of several transgenes is dependent upon exogenous signals (inducers/repressors) as well as the relative expression of other genes in the network.
Inducible systems in particular confer temporal as well as quantitative control over gene expression, both of which may be desirable in a process where different phenotypes (high growth rate/high productivity) are required at different times. In instances where expression of a particular gene is known to be actively detrimental to the process, then genomic knockout might be more attractive than knockdown of transcript levels by exogenous expression of shRNA (plus whatever selection marker is necessary to engineer in the shRNA expression cassette) in addition questions remain regarding long term stability of shRNA expression. For this purpose, techniques such as AAV-mediated HR make gene disruption, at more than one allele, a genuine prospect.
In summary, it is clear that the tools and techniques available now, both old and new and combinations thereof, give researchers much more options in planning cell engineering strategies. Implementing these will hopefully lead to the development of ‘master’ producer cell lines where metabolic flux can be controlled during the term of the process and the recombinant product can be manufactured in an efficient and stable manner.
Acknowledgements
Work in this laboratory is supported by funding from Science Foundation Ireland and Enterprise Ireland.
References
- Abremski K, Hoess R (1984) Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem 259:1509–1514 [PubMed]
- Abremski K, Wierzbicki A, Frommer B, Hoess RH (1986) Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J Biol Chem 261:391–396 [PubMed]
- Bhat K, McBurney MW, Hamada H (1988) Functional cloning of mouse chromosomal loci specifically active in embryonal carcinoma stem cells. Mol Cell Biol 8:3251–3259 [DOI] [PMC free article] [PubMed]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S (2001) Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21:289–297 [DOI] [PMC free article] [PubMed]
- Chamberlain JR, Schwarze U, Wang PR, Hirata RK, Hankenson KD, Pace JM, Underwood RA, Song KM, Sussman M, Byers PH, Russell DW (2004) Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science 303:1198–1201 [DOI] [PubMed]
- Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD, Kolb AF (2005) Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol 23:407–409 Review [DOI] [PubMed]
- Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F (1998) I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol Cell Biol 18:1444–1448 [DOI] [PMC free article] [PubMed]
- Culver KW, Hsieh WT, Huyen Y, Chen V, Liu J, Khripine Y, Khorlin A (1999) Correction of chromosomal point mutations in human cells with bifunctional oligonucleotides. Nat Biotechnol 17:989–993 [DOI] [PubMed]
- Derouazi M, Martinet D, Besuchet Schmutz N, Flaction R, Wicht M, Bertschinger M, Hacker DL, Beckmann JS, Wurm FM (2006) Genetic characterization of CHO production host DG44 and derivative recombinant cell lines, Biochem Biophys Res Commun 340:1069–1077 [DOI] [PubMed]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc-finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33:5978–5990 [DOI] [PMC free article] [PubMed]
- Ferlini A, Muntoni F (1998) The 5’ region of intron 11 of the dystrophin gene contains target sequences for mobile elements and three overlapping ORFs. Biochem Biophys Res Commun 242:401–406 [DOI] [PubMed]
- Figueroa B, Chen SL, Oyler GA et al (2004) Aven and Bcl-X-L enhance protection against apoptosis for mammalian cells exposed to various culture conditions. Biotechnol Bioeng 85:589–600 [DOI] [PubMed]
- Ghosh SS, Gopinath P, Ramesh A (2006) Adenoviral vectors: a promising tool for genetherapy. Appl Biochem Biotechnol 133:9–29 [DOI] [PubMed]
- Giovannangeli C, Thuong NT, Helene C (1992) Oligodeoxynucleotide-directed photo-induced cross-linking of HIV proviral DNA via triple-helix formation. Nucleic Acids Res 20:4275–4281 [DOI] [PMC free article] [PubMed]
- Gonzalez-Nicolini V, Fussenegger M (2005) A novel binary adenovirus-based dual-regulated expression system for independent transcription control of two different transgenes. J Gene Med 7:1573–1585 [DOI] [PubMed]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP (2000) A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA 97:5995–6000 [DOI] [PMC free article] [PubMed]
- Hartenbach S, Fussenegger M (2005) Autoregulated, bidirectional and multicistronic gas-inducible mammalian as well as lentiviral expression vectors. J Biotech 120:83–98 [DOI] [PubMed]
- Hasty P, Rivera-Perez J, Bradley A (1991) The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol 11:5586–5591 [DOI] [PMC free article] [PubMed]
- Helene C (1991) The anti-gene strategy: control of gene expression by triplex-forming-oligonucleotides. Anticancer Drug Des 6:569–584 [PubMed]
- Hirata R, Chamberlain J, Dong R, Russell DW (2002) Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol 20:735–738 [DOI] [PubMed]
- Hoess RH, Abremski K (1984) Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA 81:1026–1029 [DOI] [PMC free article] [PubMed]
- Kaufman CD, Zayed H, Miskey C, Walisko O, Izsvak Z (2004) Sleeping Beauty transposable element: evolution, regulation and genetic applications. Curr Issues Mol Biol 6:43–55 [PubMed]
- Kilby NJ, Snaith MR, Murray JA (1993) Site-specific recombinases: tools for genome engineering. Trends Genet 9:413–421 [DOI] [PubMed]
- Koduri RK, Miller JT, Thammana P (2001) An efficient homologous recombination vector pTV(I) contains a hot spot for increased recombinant protein expression in Chinese hamster ovary cells. Gene 280:87–95 [DOI] [PubMed]
- Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI (1990) Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA 87:2211–2215 [DOI] [PMC free article] [PubMed]
- Kramer BP, Fussenegger M (2005) Transgene control engineering in mammalian cells. Methods Mol Biol 308:123–143 [DOI] [PubMed]
- Lee G, Saito I (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216:55–65 [DOI] [PubMed]
- Liu XY, Gu JF (2006) Targeting gene-virotherapy of cancer. Cell Res 16:25–30 [DOI] [PubMed]
- Malphettes L, Fussenegger M (2006) Improved transgene expression fine-tuning in mammalian cells using a novel transcription-translation network. J Biotechnol 124:732–746 [DOI] [PubMed]
- Mandell JG, Barbas CF III (2006) Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res 34:W516–W523 [DOI] [PMC free article] [PubMed]
- May T, Hauser H, Wirth D (2006) Current status of transcriptional regulation systems. Cytotechnology 50:109–119 [DOI] [PMC free article] [PubMed]
- Mazur X, Fussenegger M, Renner WA et al (1998a) Higher productivity of growth-arrested Chinese hamster ovary cells expressing the cyclin-dependent kinase inhibitor p27. Biotechnol Progr 14:705–713 [DOI] [PubMed]
- Mazur X, Eppenberger HM, Bailey JE et al (1999b) A novel autoregulated proliferation-controlled production process using recombinant CHO cells. Biotechnol Bioengg 65:144–150 [DOI] [PubMed]
- Meents H, Enenkel B, Eppenberger HM, Werner RG, Fussenegger M (2002) Cell survival, and mitochondria number in CHO-DG44 grown in suspension and serum-free media. Biotechnol Bioeng 80 [DOI] [PubMed]
- Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS, Russell DW (2006) Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol 24:1022–10226 [DOI] [PubMed]
- Pavlou AK (2004) The market of therapeutic recombinant proteins to 2010. J Comm Biotechnol 10:363–367(5) [DOI]
- Perkins BD, Wilson JH, Wensel TG, Vasquez KM (1998) Triplex targets in the human. Biochemistry 37:11315–11322 [DOI] [PubMed]
- Rouet P, Smih F, Jasin M (1994) Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA 91:6064–6068 [DOI] [PMC free article] [PubMed]
- Sauer B (1998) Inducible gene targeting in mice using the Cre/lox system. Methods 14:381–392 [DOI] [PubMed]
- Seibler J, Bode J (1997) Double-reciprocal crossover mediated by FLP-recombinase: a concept and an assay. Biochemistry 36:1740–1747 [DOI] [PubMed]
- Smales CM, Dinnis DM, Stansfield SH, Alete D, Saga EA, Birch JR, Racher AJ, Marshall CT, James DC (2004) Comparative proteomic analysis of GS-NSO murine myeloma cell lines with varying recombinant monoclonal antibody production rate. Biotechnol Bioeng 88:474–488 [DOI] [PubMed]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317:230–234 [DOI] [PubMed]
- Soares MB, Schon E, Efstratiadis A (1985) Rat LINE1: the origin and evolution of a family of long interspersed middle repetitive DNA elements. J Mol Evol 22:117–133 [DOI] [PubMed]
- Sorrell DA, Kolb AF (2005) Targeted modification of mammalian genomes. Biotechnol Adv 23:431–469 [DOI] [PubMed]
- Sutter NB, Scalzo D, Fiering S, Groudine M, Martin DI (2003) Chromatin insulation by a transcriptional activator. Proc Natl Acad Sci USA 100:1105–1110 [DOI] [PMC free article] [PubMed]
- Te Riele H, Maandag ER, Berns A (1992) Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci USA 89:5128–5132 [DOI] [PMC free article] [PubMed]
- Thorpe HM, Smith MC (1998) In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA 95:5505–5510 [DOI] [PMC free article] [PubMed]
- Vasquez KM, Marburger K, Intody Z, Wilson JH (2001) Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA 98:8403–8410 [DOI] [PMC free article] [PubMed]
- Ward MA, Abramow-Newerly W, Roder JC (1993) Effect of vector topology on homologous recombination at the CHO aprt locus. Somat Cell Mol Genet 19:257–264 [DOI] [PubMed]
- Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M (2004) Gas-inducible transgene expression in mammalian cells and mice. Nat Biotechnol 22:1440–1444 [DOI] [PubMed]
- Wlaschin KF, Seth G, Hu W-S (2006) Hu WSToward genomic cell culture engineering. Cytotechnology 50:121–140 [DOI] [PMC free article] [PubMed]
- Wong NSC, Yap MGS, (2006) Wang DICEnhancing recombinant glycoprotein sialylation through CMP-sialic acid transporter over expression in chinese hamster ovary cells. Biotechnol Bioeng 93:1005–1016 [DOI] [PubMed]
- Wurm FM (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 22:1393–1398 [DOI] [PubMed]
- Wurm FM, Petropoulos CJ (1994) Plasmid integration, amplification and cytogenetics in CHO cells: questions and comments. Biologicals 22:95–102 [DOI] [PubMed]
- Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, Wakitani M, Niwa R, Sakurada M, Uchida K, Shitara K, Satoh M (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng 87:614–622 [DOI] [PubMed]