Abstract
CCNs are structurally related matricellular proteins that are highly expressed in many embryonic and adult tissues, including the skeletal system and tumors, where canonical cap-dependent translation is suppressed under hypoxic environments. CCNs are encoded by mRNAs containing long G/C rich 5′-untranslated regions (5′-UTRs). Given that they are expressed under conditions of cellular stress, it has been suggested that the long G/C-rich regions contain internal ribosomal entry sites (IRES) that allow these mRNAS to be translated under conditions where cap-dependent translation is suppressed. Previously published work supported this possibility. However, recent studies have shown that a number of previously reported cellular IRES elements do not in fact possess IRES activity. Here we aimed to reveal whether the 5′UTRs of CCNs harbor IRES activities. The 5′UTRs of CCN1, 2, and 4 were tested in this study. Our results showed that the 5′UTRs of these genes do not contain IRES elements, but instead appear to contain cryptic promoters. Both promoterless and hairpin-containing dicistronic tests showed that transcription was initiated by cryptic promoter elements in 5′UTRs of CCN1, 2, and 4. When dicistronic mRNAs were translated in vitro or in vivo, no IRES activities were detected in the 5′UTRs of CCN1, 2, and 4. Furthermore, these cryptic promoter activities from 5′UTRs of CCN1, 2, and 4 could be detected in various cell types, including chondrocytes, osteoblasts, and endothelial cells, where the cryptic promoter permitted varying degrees of activation. In addition, the core promoter element of the CCN2 5′UTR was identified. CCNs are expressed under conditions of cellular stress, and it has been suggested that some CCN family members utilize IRES-mediated translation initiation to facilitate this expression. We found no evidence for IRES activity, but rather found that the unusually long 5′UTRs of CCNs 1, 2, and 4 harbor cryptic promoters that showed varying degrees of activity in different cell types. These results suggest that these promoters may contribute to the regulation of CCN genes in vivo.
Keywords: CCN1 (Cyr61), CCN2 (CTGF), CCN4 (WISP-1), IRES, Cryptic promoter
Introduction
CCNs comprise a family of matricellular proteins that are conserved among vertebrates. Matricellular proteins are associated with the extracellular matrix, but rather than serving structural roles, they modulate signaling through their ability to engage integrins and to interact with growth factors (Bornstein and Sage 2002). There are six known CCNs: CCN1 (Cysteine-rich 61/Cyr61), CCN2 (Connective Tissue Growth Factor/CTGF), CCN3 (Nephroblastoma-overexpressed/Nov), CCN4 (Wnt-Induced Secreted Protein-1/WISP-1), CCN5 (WISP-2), and CCN6 (WISP-3). CCN proteins contain an N-terminal signal peptide followed by four conserved modules in the same order: an insulin-like growth factor binding protein domain, a von Willebrand type C domain, a thrombospondin domain, and a C-terminal cystine knot domain. CCN5 is the exception, in that it lacks the cystine knot domain. These domains allow CCNs to bind to growth factors and their receptors, to integrins, and to each other, providing opportunities for CCNs to regulate multiple types of signaling events within and between cells (Planque and Perbal 2003).
CCN-mediated signaling regulates diverse cellular processes, including cell adhesion, migration, proliferation, survival, and differentiation (Planque and Perbal 2003; Leask and Abraham 2006). However, how the diversity of potential binding partners such as specific ECM components, growth factors and their receptors, give individual CCN family members their different cellular activities remains an unanswered question. While the majority of their effects on cells can be explained by the ability of CCNs to stimulate expression of, and to act as ligands for, specific integrins (Lau and Lam 1999; Brigstock 2003), their interactions with multiple growth factors and their receptors are also significant (Abreu et al. 2002; Inoki et al. 2002; Mercurio et al. 2004).
CCNs are highly expressed in many embryonic and adult tissues, including the skeletal system. Most of the vertebrate skeleton is formed by endochondral ossification, in which cartilage templates are subsequently replaced by bone. This template eventually forms a growth plate, which is composed of stratified layers of chondrocytes at different stages of differentiation, including proliferative, pre-hypertrophic, and terminally differentiated hypertrophic chondrocytes. Cartilage is an avascular tissue that is hypoxic in nature (Schipani et al. 2001; Zelzer et al. 2004). In addition to the cellular stress imposed by hypoxia, hypertrophic chondrocytes face extra challenges, such as the requirement to produce extracellular matrix components and growth factors that support the invasion of blood vessels, osteoblasts, and other cell types that lead to the replacement of the cartilage with bone; these cells must achieve these goals while simultaneously undergoing apoptosis. Given the importance of hypertrophic chondrocytes in endochondral ossification and subsequent bone growth, understanding how these cells mediate the expression of key proteins at high levels in such a severe environment is an important, but poorly understood, issue.
CCNs are important regulators of skeletal growth and maintenance. Both CCN1 and CCN2 are highly expressed in hypertrophic chondrocytes of the growth plate (Wong et al. 1997; Friedrichsen et al. 2003; Ivkovic et al. 2003). CCN3 is expressed in pre-hypertrophic and early hypertrophic chondrocytes (Yu et al. 2003), and CCN4 is expressed in osteoblasts and perichondral mesenchyme (French et al. 2004). The loss of CCN2 in mice causes severe, lethal defects in chondrogenesis and growth plate angiogenesis (Ivkovic et al. 2003), while overexpression of CCN2 in cartilage results in decreased bone density, believed to be caused by precocious endochondral ossification (Nakanishi et al. 2001). Although loss of CCN1 leads to early embryonic lethality, precluding an analysis of its role in skeletal tissues (Mo et al. 2002), high levels of CCN1 expression in the growth plate and potent effects on chondrocytes in vitro strongly suggest essential roles in chondrogenesis. Finally, loss of CCN6 in humans leads to a severe form of childhood-onset arthritis characterized by cartilage destruction, progressive pseudorheumatoid dysplasia (Hurvitz et al. 1999).
Altered levels of CCN protein expression are also associated with tumorigenesis. In primary human breast carcinomas, elevated levels of CCN1, CCN2, and CCN4 have been associated with more advanced tumors (Xie et al. 2001). High levels of CCN1 expression are also seen in hepatic carcinomas, pancreatic cancer, and ovarian carcinomas (Zeng et al. 2004; Gery et al. 2005). Elevated CCN2 expression is found in many tumors, including hepatic carcinomas, fibrous mesenchymal tumors, and glioblastomas (Zeng et al. 2004; Kasaragod et al. 2001; Pan et al. 2002). CCN4 expression is significantly increased in most colon adenocarcinomas (Pennica et al. 1998). In many of these tumors, CCNs act as pro-proliferative, pro-tumorigenic factors, but there are also numerous examples where overexpression of CCN proteins in tumor cells may result in growth arrest and/or reduced tumourigenicity, which means that some CCN proteins also act as anti-proliferative agents (Planque and Perbal 2003). Thus, while the specific activities of CCNs in tumorigenesis are likely to be tissue- or context-dependent, there seems to be a general role for CCN proteins in regulating tumor growth.
The progression of tumor growth is influenced by environmental factors; one crucial environmental factor is reduced oxygen tension, or hypoxia, within the tumor. As a response to hypoxia, tumor cells initiate the expression of pro-angiogenic genes that promote neovascularization (Michiels 2003). In tumor cell lines, the expression of CCN1 and CCN2 is induced by hypoxia (Shimo et al. 2001; Kunz et al. 2003). Given the proangiogenic properties of these CCNs, these findings suggest that CCN1 and CCN2 may be important in the neovascularization response to hypoxia.
In both normal and tumourigenic cells, hypoxia has a profound effect on overall protein synthesis. Under conditions of hypoxia, levels of protein synthesis can be reduced to 30% of the level in normoxic cells (Kraggerud et al. 1995). One major mechanism through which hypoxia suppresses protein synthesis is by affecting the availability of the translation initiation factor eIF4E, a critical component of the cap-dependent translation initiation process (Tinton and Buc-Calderon 1999). In the canonical method of translation initiation, a ribosomal initiation complex scans 5′ capped mRNA, searching for an initiation codon. Translation initiation is inhibited if initiation factors become limited (as can happen under conditions such as hypoxia, apoptosis, or heat shock), or if the structure of the 5′mRNA impedes ribosomal scanning (Vagner et al. 2001; Komar and Hatzoglou 2005).
Under conditions in which canonical translation initiation is inhibited, the cell must utilize alternative mechanisms in order to translate essential proteins. Internal ribosome entry sites (IRESs) in the 5′ untranslated regions (UTRs) of certain cellular mRNAs have been proposed to allow translation initiation to precede when cap-dependent translation is inhibited (Vagner et al. 2001). Originally identified in picornaviruses, IRES activity permits direct interaction between mRNA and ribosomes in viral genes (Hellen and Sarnow 2001). To date, IRES elements have been described for relatively few cellular genes; among these are regulators of angiogenesis and apoptosis, such as HIF-1α, VEGF, c-myc, and XIAP (Bert et al. 2006; Baird et al. 2006). Because CCN1 and CCN2 are expressed in hypoxic tissues, in which cap-dependent translation may be compromised, we investigated the possibility that IRES elements are present in the 5′UTRs of these genes. Although no conserved structural characteristics of IRES-containing cellular mRNAs have been identified, features such as a GC-rich 5′UTR, a complex secondary RNA structure, and the presence of upstream AUG codons are frequently seen in cellular genes that are reputed to contain an IRES (Stoneley and Willis 2004). Ccn1 mRNA was previously identified in a microarray experiment as being efficiently translated under conditions of low levels of cap binding translation initiation factors (Johannes et al. 1999). Based on this property, it was concluded that Ccn1 mRNA possesses IRES activity (Johannes et al. 1999).
In order to assess whether the 5′UTRs of mRNAs encoding CCN family members might support IRES-mediated translation, we chose CCN1, 2, and 4 to be tested in a dicistronic luciferase reporter assay. We found that the 5′UTRs of CCN1, 2, and 4 appeared to have IRES activities that were functional in multiple cell lines using standard dicistronic luciferase reporter assays for IRES activity. However, these 5′UTRs do not display IRES activity when tested more rigorously in either in vitro or in vivo translation assays. We then used a promoterless luciferase assay to attempt to resolve this apparent discrepancy, and found that the 5′UTRs of CCN1, 2, and 4 exhibit evidence for the presence of cryptic promoters. Thus, IRES-mediated translation does not appear to be a significant mechanism for the regulation of CCN protein synthesis under hypoxic conditions. Furthermore, our data corroborates several recent reports indicating that the dicistronic luciferase reporter assay is not a sufficiently stringent method for assessing cellular IRES activity.
Materials and methods
Materials
Restriction Enzymes, pGL3-basic vector, pSP72 vector, Ribo m7G cap analog, and Dual-Luciferase reporter assay system were from Promega. Lipofectamine 2000, TaqPCRx DNA polymerase, and cell culture medium were purchased from Invitrogen. MEGAscript and Retic Lysate IVT (RRL) system were from Ambion. Midi plasmid purification kit, RNeasy Mini purification kit, and TransMessenger RNA transfection reagent were obtained from Qiagen.
Plasmid constructs
The luciferase reporter plasmids pR-F (formally pGL3-R), pR-Myc-F (formally pGL3-Rutr), and phpR-Mrt-F were kindly provided by A. Willis (University of Nottingham), and pR-HCV-F (formally pRL-Prom-HCV-FLuc) and pR-HCV-F(-p) (formally pRL-HCV-FLuc) by N. Elango.
CCN plasmid constructs pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F were generated as follows. The 5′UTRs of murine CCN1, CCN2, and CCN4 were amplified with following primer sets: Ccn1: 5′-CCCAGCTGGAATTCAGACCGTGAGCGAGA-3′ and 5′-GAGGACGCGCGGTGGTACCGG-3′; Ccn2: 5′-CCGAATTCTTCTCTCCAAGAAGACTCAGCC-3′ and 5′-TGGCGCAGGGCTAGGTACCGCCTAGGCC-3′; and Ccn4: 5′-CCGAATTCTGGAGGTGGGGACAGAGGAAAG-5′ and 5′-CGACTGAAGGTCCGGTACCGCCTAGGCC-3′. The PCR products were digested with EcoRI and NcoI and subcloned into pR-F.
In order to generate the hairpin-containing plasmids phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F, the plasmid phpR-Mrt-F, which contains a stable hairpin at the 5′end of Renilla luciferase, was digested with EcoRI and NcoI in order to remove the 5′UTR of Mrt. The resulting plasmid was designated phpR-F. The DNA sequence of this hairpin structure was originally reported by Stoneley et al. (1998). The 5′UTRs of CCN1, CCN2, and CCN4 were amplified as described above, digested with EcoRI and NcoI, and subcloned into phpR-F plasmid.
The dicistronic promoterless constructs were generated by removing the Simian virus 40 (SV40) promoter sequence between NheI and EcoRV digestion sites from pR-F, pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F, resulting in the plasmids pR-F(-p), pR-CCN1-F(-p), pR-CCN2-F(-p), and pR-CCN4-F(-p), respectively. The integrities of all plasmids were confirmed by restriction digest analysis and DNA sequencing.
To generate RNA expression constructs, the fragment of Renilla luciferase gene was first removed by EcoRV and XbaI digestion from pR-F and then was subcloned into pSP72 vector, designated pSP72-R. The fragments of firefly gene, CCN1-F, CCN2-F, and CCN4-F were removed by XbaI digestion from pR-F, pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F and were inserted into pSP72-R, designated pSP72-R-F, pSP72-R-CCN1-F, pSP72-R-CCN2-F, and pSP72-R-CCN4-F, respectively. The pSP72-R-HCV-F was generated by removing the R-HCV-F fragment from pR-HCV-F by Acc65I and BamHI digestion and by insertion of the fragment into pSP72 vector.
To assess cryptic promoter activity within the 5′UTR of CCN2, four smaller fragments of approximately 55 bp (designated CCN2-A, CCN2-B, CCN2-C, and CCN2-D) were cloned into the XhoI and HindIII sites of pGL3-basic vector. These 5′UTR fragments were generated with oligonucleotides as follows:
CCN2-A: 5′-TCGAGTTCTCTCCAAGAAGACTCAGCCAGATCCACTCCAGCTCCGACCCCAGGAGACCGA-3′ and 5′-AGCTTCGGTCTCCTGGGGTCGGAGCTGGAGTGGATCTGGCTGAGTCTTCTTGGAGAGAAC-3′; CCN2-B:5′-TCGAGACCTCCTCCAGACGGCAGCAGCCCCAGCCCAGCCGACAACCCCAGACGCCACCGA-3′ and 5′-AGCTTCGGTGGCGTCTGGGGTTGTCGGCTGGGCTGGGGCTGCTGCCGTCTGGAGGAGGTC-3′; CCN2-C: 5′-TCGAGCCTGGAGCGTCCAGACACCAACCTCCGCCCCTGTCCGAATCCAGGCTCCAGCCGA-3′ and 5′-AGCTTCGGCTGGAGCCTGGATTCGGACAGGGGCGGAGGTTGGTGTCTGGACGCTCCAGGC-5′; CCN2-D: 5′-TCGAGCGCCTCTCGTCGCCTCTGCACCCTGCTGTGCATCCTCCTACCGCGTCCCGATCA-3′ and 5′-AGCTTGATCGGGACGCGGTAGGAGGATGCACAGCAGGGTGCAGAGGCGACGAGAGGCGC-3′. These oligonucleotide sequences also include restriction enzyme recognition sites; annealed oligonucleotides were directly ligated into the XhoI and HindIII sites of pGL3-basic vector.
In vitro transcription and translation
The plasmids pSP72-R-F, pSP72-R-HCV-F, pSP72-R-CCN1-F, pSP72-R-CCN2-F, and pSP72-R-CCN4-F were linearized with NdeI. The capped and uncapped transcripts were synthesized with T7 MEGAscript high yield transcription kit according to the manufacturer’s protocol. The mRNAs were purified with Qiagen’s RNeasy Mini kit, quantified spectrophotometrically and their qualities were verified by Agilent 2100 Bioanalizer. Equal amounts of RNA (usually 2 μg of total RNA) were used in a 25 μl mixture in vitro translation system (RRL). The aliquot of 5 μl of each reaction mixtures were used in a luciferase activity assay (described below).
Cell culture, DNA and RNA transfection
HeLa (human cervical carcinoma) cells were provided by S. Smale (UCLA). ATDC5 (mouse chondrosarcoma) cells were obtained from the Riken Cell Bank. MC3T3-E1 (mouse preosteoblast) cells were obtained from American Type Culture Collection (ATCC). HUVEC (human umbilical vein endothelial) cells were obtained from Cascade Biologics. HUVEC cells were grown in Medium 200 supplemented with low serum growth supplement (Cascade Biologics). All other cells types were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin. All cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2. For dicistronic luciferase plasmid transient transfection, cells were grown to 90–95% confluence in 24-well plates and then transfected with 0.5 μg of luciferase plasmid. For monocistronic plasmid transient transfection, cells were co-transfected with 0.5 μg of luciferase plasmid and 0.25 μg of Renilla luciferase plasmid. Cells from both above transfection experiments were harvested after 48 h after transfection for luciferase activity assay.
RNA transfection was performed with TransMessanger Transfection Reagent according to manufacturer’s instruction. An aliquot of 2 μg of capped or uncapped mRNA were used to transfect 80–90% confluent cells grown in 24-well plate. Cells were harvested and lysed for luciferase activity assay after 4 h transfection of uncapped RNA or after 16 h transfection of capped RNA.
Luciferase assay
Transfected cells were washed twice with 1× PBS and then lysed in 100 μl of 1× passive lysis buffer. Firefly and Renilla luciferase activities were measured using a Dual luciferase reporter assay system according to the manufacturer’s instruction with the exception that only 50 μl of each assay reagent was used.
Results
The 5′UTRs of CCNs enhance expression of the second cistron in dicistronic constructs
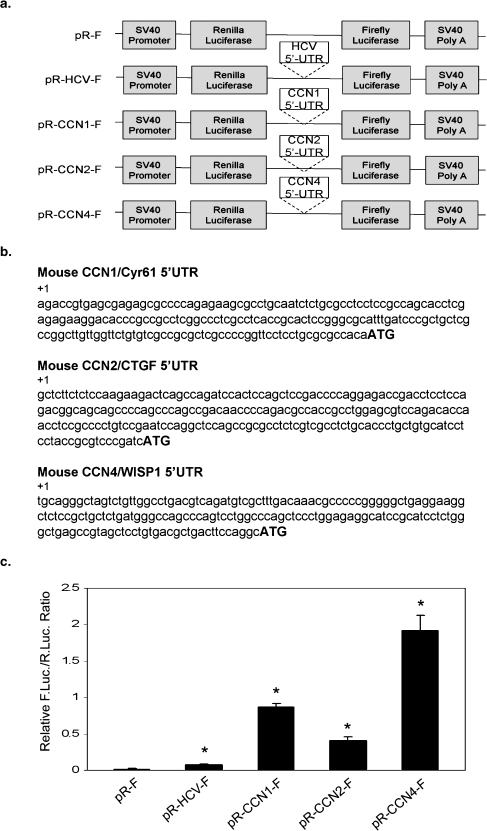
To characterize the function of the 5′UTRs of CCN transcripts in gene expression, we engineered 5′UTR-controlled dicistronic constructs by cloning the 5′UTR sequences of several members of the CCN family into the intergenic region of a dicistronic vector pRF, to obtain pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F (Fig. 1a). The IRES containing 5′UTR of HCV was used as a positive control. The pRF-based constructs contain a SV40 promoter that directs the transcription of Renilla luciferase as the first cistron. The firefly luciferase is the second cistron, and is regulated by the inserted 5′UTR sequence. These plasmids were transfected into HeLa cells, and both Renilla and firefly luciferase activities were measured. The sequences of the 5′UTRs are shown in Fig. 1b. As shown in Fig. 1c, although firefly luciferase activity was low for pR-HCV-F, the positive control, the activity was statistically significant compared to the negative control, pRF, where no firefly activity was detected. The 5′UTRs of CCN1, 2, and 4 stimulated much higher levels of firefly luciferase activity. The activities are ∼47, 22, and 105 fold in pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F, respectively, over the negative control pRF, and ∼12 fold, 10 fold, and 40 fold over the positive control, pR-HCV-F. These results suggest that (a) the 5′UTRs of these CCNs may enhance read-through ability of the ribosome through the intergenic region, (b) the 5′UTRs of these CCNs may contain IRES activities that enhance the translation of firefly luciferase from the dicistronic mRNA by internal initiation, as suggested previously for the HCV 5′UTR and for CCN1 (Johannes et al. 1999; Tsukiyama-Kohara et al. 1992), or (c) the 5′UTRs of these CCNs contain either cryptic promoters which direct the transcription of firefly luciferase or (d) splicing acceptor sites that create monocistronic transcripts of firefly luciferase. Indeed, some publications have indicated that the commonly used pRF vector can, in a gene-specific manner, lead to spurious splicing events that delete Renilla luciferase, generating monocistronic firefly luciferase transcripts (Sherrill et al. 2004; Van Eden et al. 2004a; Holcik et al. 2005).
Fig. 1.
The 5′UTRs of CCN1, 2, and 4 are capable of driving expression of firefly luciferase in the dicistronic reporter assay. a Schematic diagrams of the reporter plasmids. All constructs contain Renilla and firefly luciferase cassettes encoded by a single dicistronic mRNA, transcribed under the control of the SV40 promoter. pRF, a negative control, contains no insert between the Renilla and firefly cassettes. Hence, translation initiates at the AUG upstream of Renilla, and terminates after the stop codon in the Renilla sequence. pR-HCV-F is a positive control, as it contains a documented viral IRES sequence from hepatitis C virus (Tsukiyama-Kohara et al. 1992). The 5′ UTRs of CCN1, 2, and 4 were identified as described in Materials and Methods. b Sequences of the 5′UTR CCN1, 2, and 4 inserted into the plasmids. Sequences were derived from GeneBank NM_010516, NM_010217, NM_018865, respectively. c Relative firefly luciferase activity generated from dicistronic reporter assays. HeLa cells were transfected with dicistronic plasmid and lysates were prepared from 48 h post-transfected cells. The Renilla and firefly luciferase activities were measured and values are normalized as the ratio of firefly to Renilla activity. Transfection in triplicate was repeated 3 times and values are expressed as mean ± 1 S.D. *, P = 0.005 compared with empty vector (two-tailed Student’s t test)
The 5′UTR does not promote ribosome read-through in the dicistronic constructs
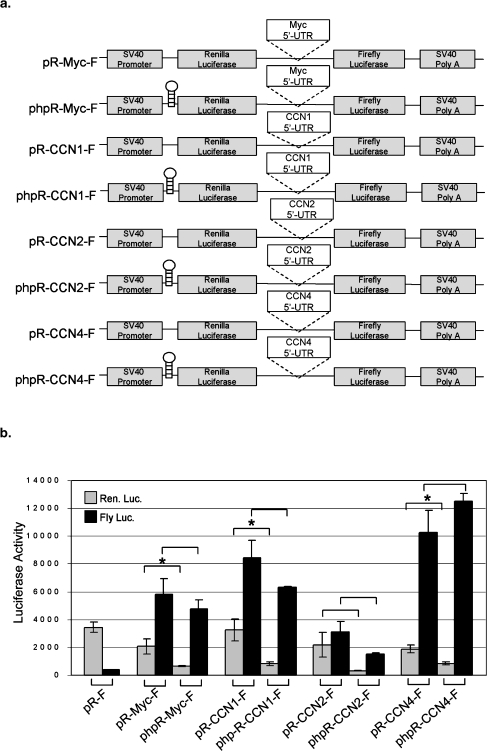
To determine whether the ability of the 5′UTRs of CCNs to drive high levels of expression of firefly luciferase is due to translational read-through of the intergenic regions, we engineered a 60 bp hairpin insertion upstream of Renilla luciferase in each construct, designed phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F (Fig. 2a). This hairpin is known to prevent read-through in dicistronic constructs (Stoneley et al. 1998). A 5′UTR containing reported IRES activity from c-myc (Stoneley et al. 1998) was used as a positive control. As shown in Fig. 2b, the inserted stable hairpin could reduce Renilla luciferase activity (first cistron) by ∼75–85% in phpR-Myc-F, phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F, as compared with the corresponding non-haripin-containing plasmids pR-Myc-F, pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F, respectively, indicating that stable hairpin structures prevent cap-dependent translation of Renilla luciferase through a ribosomal scanning mechanism, as expected. However, firefly luciferase activity (second cistron) produced by phpR-Myc-F, phpR-CCN1-F, and phpR-CCN2-F was reduced by less than ∼40% or even increased (∼20% in phpR-CCN4-F) as compared with pR-Myc-F, pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F, respectively. These results indicate that firefly luciferase activity produced by plasmids containing the 5′UTRs of CCN1, 2, and 4 are independent of cap-dependent translational read-through from the first cistron (Renilla luciferase), but support the possibilities that the expression of firefly luciferase might be promoted by IRES elements in the 5′UTR, or by cryptic promoters.
Fig. 2.
The expression of firefly luciferase in dicistronic constructs containing 5′UTRs from CCN1, 2, and 4 is not due to translational readthrough. a Schematic diagrams of the reporter plasmids used to test translational readthrough. The hairpin sequence inserted between SV40 promoter and Renilla luciferase cassette (−55 kcal/mol) has been shown previously to prevent passage of ribosomes, and hence blocks translation (Stoneley et al. 1998). b The 5′UTR driven luciferase activity is not reduced by stable hairpin insertion. As expected, pRF directs expression of Renilla, but not firefly luciferase. The positive control, pR-Myc-F, directs expression of both Renilla and firefly luciferase. However, as shown previously (Stoneley et al. 1998), insertion of a hairpin sequence upstream of the Renilla coding region in phpR-Myc-F blocks translation of Renilla luciferase in this plasmid. On the other hand, firefly luciferase activity is not blocked, reputedly due to the ability of the c-myc 5′UTR to direct expression of firefly luciferase. Similar results were obtained for phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F. Transfection in triplicate was repeated three times and values are expressed as mean ± 1 S.D. *P ≤ 0.04 compared with non-hairpin inserted vector; NS not significant (two-tailed Student’s t test)
5′UTRs of CCNs do not display an IRES activity in dicistronic mRNA assay
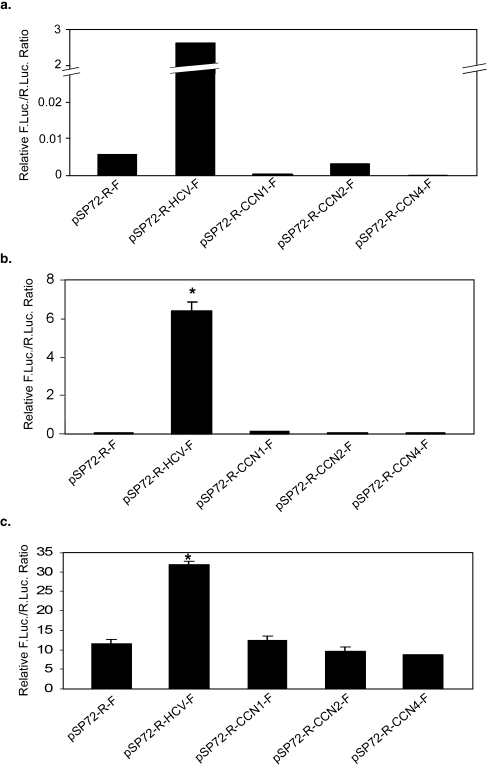
To discriminate whether the effect of the 5′UTRs of CCN1, 2, and 4 on the expression of firefly luciferase as a second cistron is due to the presence of IRES activity or cryptic promoter activity, we generated capped and uncapped dicistronic mRNAs in vitro and used them to program translation both in vitro and in vivo. Because bacteriophage RNA polymerases such as SP6 or T7 RNA polymerase initiate transcription from a highly specific promoter sequence, mRNA transcribed in vitro using these enzymes is essentially devoid of any monocistronic downstream luciferase transcripts. RNA transfection is thus one of the most stringent methods for characterizing translation efficiency and identifying whether eukaryotic regulatory sites possess IRES activity. This method avoids the requirement for transcription. Capped transcripts were first used in a rabbit reticulocyte lysate (RRL) in vitro translational system. As shown in Fig. 3a, there was strong IRES activity from the 5′UTR of HCV in the in vitro translation assay, but there was no enhancement of firefly luciferase activity conferred by transcripts from the 5′UTRs of CCN1, 2, and 4 as compared to pRF negative control.
Fig. 3.
In vitro and in vivo translation assays do not support the presence of IRES activities in the 5′UTRs of CCN1, 2, and 4. a In vitro translation assay. The 5′UTR contained on the pSP72-R-HCV-F positive control permits expression of firefly luciferase, indicative of IRES activity. However, levels of firefly luciferase activity directed by pSP72-R-CCN1-F, pSP72-R-CCN2-F, and pSP72-R-CCN4-F are not significantly different from that directed by the pSP72-R-F negative control. Values shown are from one of three repeated assays. b and c In vivo translation of uncapped and capped mRNAs, respectively, in HeLa cells. The pSP72-R-HCV-F mRNAs direct expression of firefly luciferase, consistent with the existence of an IRES element in the 5′UTR of HCV. However, expression of firefly luciferase was not above that directed by the pSP72-R-F negative control for pSP72-R-CCN1-F, pSP72-R-CCN2-F, and pSP72-R-CCN4-F, ruling out IRES activity for the 5′UTRs of these genes. Values are expressed as mean ± 1 S.D from triplicate transfection assay and assay was repeated three times. *p < 0.01 compared with non-insert vector mRNA (two-tailed Student’s t-test)
It has been shown that RRL may lack factors required for IRES-dependent translation initiation for some cellular and viral IRESs (Stoneley et al. 2000). Therefore, as an additional test for IRES activity, uncapped mRNA produced in vitro was purified and transfected into HeLa cells, and Renilla and firefly luciferase activities were measured 4 h after transfection, as uncapped mRNAs have been known to be stable for at least this long (Van Eden et al. 2004b). As shown in Fig. 3b, firefly luciferase activities in RNAs containing 5′UTRs from CCNs1, 2, and 4 were very low, comparable to that conferred by the negative control pRF transcript. In contrast, the HCV containing transcript expressed ∼90 fold higher firefly luciferase activity than the pRF control, confirming that the function of the HCV IRES is independent of the cap-structure of the dicistronic mRNA.
As a further test, capped dicistronic mRNAs were introduced into HeLa cells to program translation in vivo. Cell lysates were collected at 16 h post-transfection and luciferase activities were measured. As shown in Fig. 3c, as expected, firefly luciferase was poorly translated in control pRF mRNAs, and in any of mRNAs containing 5′UTRs of CCNs. However, there was ∼2.7 fold higher firefly luciferase activity directed by pR-HCV-F mRNA compared to pRF. Therefore, these data further indicate that the 5′UTRs of CCNs 1, 2 and 4 do not contain IRES elements to mediate internal ribosome entry.
5′UTR of CCNs contain functional promoters
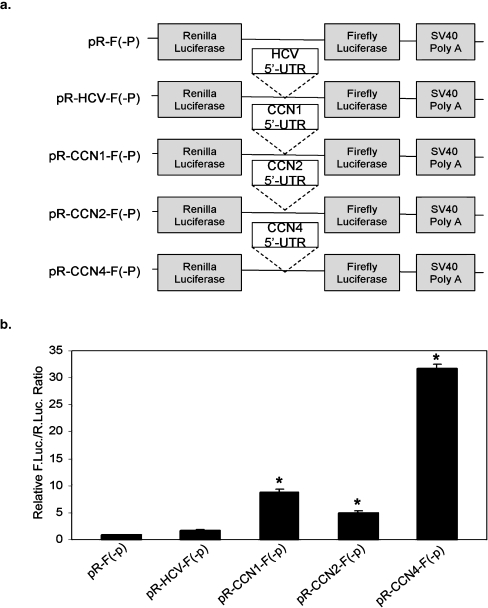
The results described above argue against the previously suggested IRES activity for the 5′UTR of CCN1 (Johannes et al. 1999), and indicate that the 5′UTRs of CCN2 and 4 do not contain IRES elements, either. Given that these UTRs nonetheless directed firefly luciferase expression in dicistronic assays (Figs. 1 and 2), we tested whether these UTRs contain cryptic promoters. To do so, we generated promoterless dicistronic constructs by removing the SV40 promoter from pR-F, pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F, designated pR-F(-p), pR-CCN1-F(-p), pR-CCN2-F(-p), and pR-CCN4-F(-p), respectively (Fig. 4a). This approach has been shown previously to provide a sensitive method to distinguish between IRES elements and cryptic promoters (Han and Zhang 2002). These constructs were transfected into HeLa cells and the activities of Renilla and firefly luciferase were measured. As expected, Renilla luciferase activity in all promoterless constructs was reduced to minimal values as compared to the corresponding promoter-containing constructs (data not shown). However, the firefly luciferase activities in pR-CCN1-F(-p), pR-CCN2-F(-p), and pR-CCN4-F(-p) were enhanced by ∼10, ∼6, and ∼35 fold, respectively, compared to pRF(-p) (Fig. 4b). These enhancements are similar to those generated by the pR-CCN1-F, pR-CCN2-F, and pR-CCN4-F promoter-containing constructs in Fig. 1a. These significant increases in firefly luciferase activity by the 5′UTRs of CCNs in this promoterless assay suggest the presence of a strong cryptic promoter in these sequences.
Fig. 4.
The 5′UTRs of CCN1, 2 and 4 contain cryptic promoters. a Schematic diagram of the promoterless constructs used to assay for cryptic promoter activity in HeLa cells. pR-F(-p), pR-CCN1-F(-p), pR-CCN2-F(-p), and pR-CCN4-F(-p) are identical to pR-F, pR-F-CCN1-F, p-R-CCN2-F, and pR-CCN4-F, but lack the SV40 promoter as described in Materials and Methods. b Firefly luciferase activity relative to Renilla activity from the same construct. As expected, neither firefly nor Renilla activity is conferred by the pR-F(-p) negative control. Similarly, the pR-HCV-F(-p)construct does not direct expression of either luciferase. This is consistent with the presence of an IRES (see Fig. 3), but not a cryptic promoter, in the 5′UTR of HCV. However, the 5′UTRs of CCN1, 2, and 4 are all able to direct much higher levels of expression of firefly luciferase than either pR-F(-p) or pR-HCV-F(-p), consistent with the presence of cryptic promoters, but not IRES elements, in the 5′UTRS of the CCNs. Transfection was performed in triplicate for two repeated individual assays and values are expressed as mean ± 1 S.D. *P ≤ 0.002 compared with empty vector (two-tailed Student’s t test)
The 5′UTRs of CCNs promote expression of second cistron firefly luciferase in multiple cell types
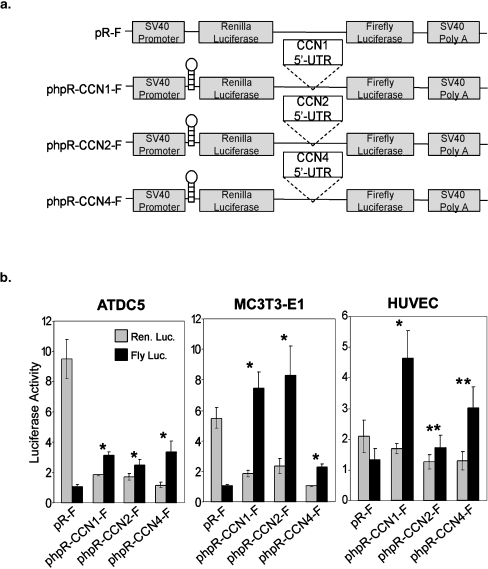
The functional promoter in the 5′UTRs of CCN1, 2 and 4 prompted us to explore whether this element can be utilized ubiquitously in endothelial cells, chondrocytes, and osteoblasts. We chose to examine these cell types because CCNs 1, 2, and 4 have been shown previously to play important roles in blood vessels, cartilage, and bone (Brigstock 2003; Ivkovic et al. 2003; Mo et al. 2002; Takigawa 2003). We used the well-characterized cell types ATDC5 (chondrocytes), MC3T3-E1 (pre-osteoblasts), and HUVEC (human umbilical vein endothelial cells). We transfected phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F (Fig. 5a) into each of these cell lines and measured Renilla and firefly luciferase activities. In each line, Renilla luciferase activity was repressed by the presence of the hairpin, as expected (see Fig. 2) (Fig. 5b). However, firefly luciferase activity was not silenced by the insertion of the hairpin sequence before the first cistron, consistent with the presence of a cryptic promoter in the 5′UTRs of CCN1, 2 and 4. Interestingly, firefly luciferase activity varied for the different CCNs in the tested cell lines. For example, the 5′UTRs of CCN1, 2 and 4 were all moderately active in ATDC5 cells, but only the 5′UTRs of CCN1 and 2 were highly active in MC3T3-E1 cells, and the 5′UTRs of CCN1 and CCN4 were much more active in HUVEC cells than was the 5′UTR of CCN2. Thus, these observations suggest that the promoters located in the 5′UTR of CCN1, 2, and 4 are active in endothelial cells, chondrocytes, and osteoblasts, but to different extents.
Fig. 5.
The 5′UTRs of CCN1, 2, and 4 promote firefly luciferase translation in multiple cell types. a Schematic diagrams of the hairpin containing reporter plasmids used in multiple cell types. b The 5′UTRs of CCN1, 2, and 4 exhibit cell-type variability in cryptic promoter in transiently transfected cells. In all cell lines, transfection with the control pR-F did not induce firefly luciferase activity as expected. However, the firefly luciferase activity was induced significantly in hairpin containing plasmids as shown in Fig. 2b in ATDC5, MC3T3-E1, and HUVEC cells. The relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity in pR-F, phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F (data not shown). In ATDC5 cells, transfected with phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F results in a 3-, 2-, and 3-fold, respectively, induction of firefly luciferase activity. In MC3T3-E1 cells, transfected with phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F results in a 7-, 7-, and 2-fold, respectively, induction of firefly luciferase activity. In HUVEC cells, transfected with phpR-CCN1-F, phpR-CCN2-F, and phpR-CCN4-F results in a 3-, 1-, and 3-fold, respectively, induction of firefly luciferase activity. Transfection was performed in triplicate assays and values are expressed as mean ± 1 S.D. *p < 0.01 and **p < 0.04, relative luciferase activity of each construct compared with empty vector (two-tailed Student's t-test)
Mapping of the CCN2 promoter in the 5′UTR of CCN2
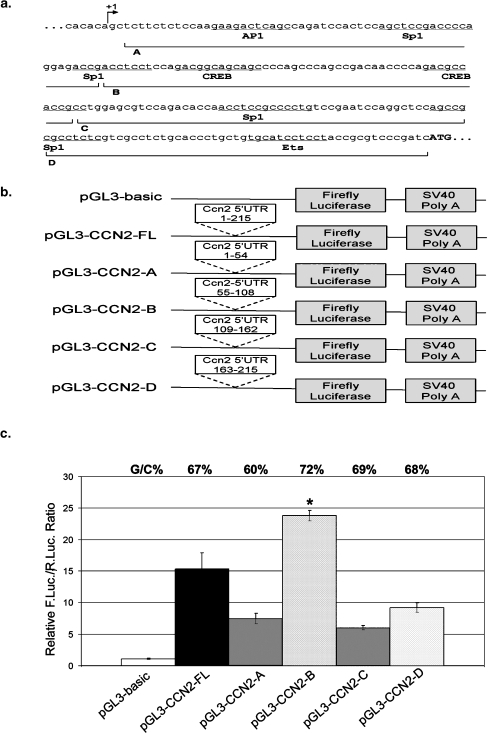
The above results strongly suggested the presence of cryptic promoters in the 5′UTR sequences of CCN family members. To further validate this observation, initial mapping of the cryptic promoter region in the CCN2 5′UTR was carried out by dissecting the sequence into four regions (each about 55 bp, Fig. 6a) and each fragment was subcloned into the pGL3-basic promoterless reporter vector (Fig. 6b). These constructs were transfected into HeLa cells and resulting induction of firefly luciferase activity was measured and compared with that conferred by the entire 5′UTR of CCN2. As shown in Fig. 6c, each CCN2 5′UTR fragment was able to induce firefly luciferase activity at least fivefold over that induced by the pGL3-basic control vector. However, one segment of CCN2 5′UTR, segment B, containing nucleotide 55–108 of the 5′UTR, has significant promoter activity, nearly 25 fold over the control vector, 3–4 fold higher than any other segment, and about 1.6 fold over the entire 5′UTR containing construct, suggesting that region B, from +55 to +108, contains a critical element of the cryptic promoter in the 5′UTR sequence of CCN2.
Fig. 6.
The 5′UTR of the mouse CCN2 transcript contains several putative binding sites for transcription factors. a Sequence of the 5′ UTR of CCN2. Putative binding sites identified by CISTER (Frith et al. 2001) and ConSite (Sandelin et al. 2004) are indicated. The putative transcription start site was identified by BLAST analysis of the mouse EST database, and by comparison to the defined start of transcription start site of human CCN2 gene. The sequences of the individual segments tested in reporter assays are delineated by solid lines. b Schematic diagram of constructs used to test the activities of segments of the 5′UTR of CCN2 in Fig. 6c. c Reporter assays for promoter activity of segments of the 5′UTR of CCN2. HeLa cells were co-transfected with pGL3-basic, pGL3-CCN2-FL, pGL3-CCN2-A, pGL3-CCN2-B, pGL3-CCN2-C, or pGL3-CCN2-D in combination with plasmid pSV-RenillaLuc. Cell lysates were harvested 48 h after transfection and Renilla and firefly luciferase activities were measured. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity. The segment in block B region exhibits an increase of relative luciferase activity, but blocks A, C, and D all reveal reduction of activities. The percent GC content for each UTR segment is indicated. Transfection was performed in triplicate for two repeated individual assays and values are expressed as mean ± 1 S.D. *P < 0.03 compared with pGL3-CCN2-FL vector (two-tailed Student's t-test)
Discussion
Since the initial suggestion that cellular genes may possess IRES activity, a large number of reports have provided evidence that a number of cellular genes, particularly those involved in stress responses, possess 5′UTRs with IRES activity (Bert et al. 2006; Baird et al. 2006). However, the validity of these conclusions has been questioned recently, because much of the evidence for the internal initiation hypothesis for cellular genes is incomplete (Kozak 2005). The extent to which cellular genes utilize IRES-mediated mechanisms is now a topic of considerable debate (Kozak 2005; Schneider et al. 2001). Given the proposed significance of this mode of regulation in hypoxic conditions, such as occurs in tumor growth, resolution of this issue is of scientific and clinical importance. It has been shown that the proangiogenic genes CCN1 and CCN2 are induced in tumor cells (Shimo et al. 2001; Kunz et al. 2003), and in hypoxic conditions (Higgins et al. 2004; Hong et al. 2006; Olbryt et al. 2006), suggesting that CCN1 and CCN2 expression may be important in the neovascularization response to hypoxia. As canonical eIF4E-initiated translation is suppressed under hypoxia (Kraggerud et al. 1995), many cellular genes associated with hypoxic and proangiogenic responses have been proposed to utilize IRES-mediated translation initiation (Higgins et al. 2004). Indeed, based on the observation of abundant CCN1 transcripts that remained associated with polysomes in viral infected cells, the 5′UTR of CCN1 was implied as possessing IRES activity (Johannes et al. 1999). Therefore, we examined the 5′UTRs of CCN1, 2, and 4, to verify the existence of IRES activity in 5′UTR regions. However, we found no evidence for IRES activity, but rather discovered cryptic promoters residing in these 5′UTRs.
In dicistronic DNA assays conventionally used to detect IRES activity, the 5′UTRs of CCN1, 2 and 4 triggered significant firefly luciferase activity (Fig. 1). This was also seen when we introduced a stable hairpin structure into the dicistronic construct, which successfully blocked the expression of the Renilla luciferase gene (first cistron), but could not block the expression of firefly luciferase (second cistron) (Fig. 2). This result ruled out the possibility that translational machinery reads through the dicistronic transcripts. Based on recent debates in the literature about the validity of the dicistronic assay (Komar and Hatzoglou 2005; Baird et al. 2006; Kozak 2005), direct transfection of dicistronic RNA transcripts has become a gold standard to test the existence of cellular IRES activity. When we tested the dicistronic RNAs in theses assays, the 5′UTRs of CCN1, 2 and 4 failed to enhance firefly luciferase translation either in vitro or in vivo. In contrast, the 5′UTR of HCV, a well-documented IRES containing element (Tsukiyama-Kohara et al. 1992), induced a relatively strong firefly luciferase activity when uncapped or capped dicistronic transcripts were transfected in cells (Fig. 3). This observation strongly suggests that there is no IRES activity in the 5′UTRs of CCN1, 2, and 4. Although we were unable to detect monocistronic transcripts by Northern analysis or by RT-PCR (data not shown), we cannot formally rule out the possibility that moncistronic transcripts are produced through alternative splicing at levels undetectable by the above methods. Nonetheless, our data suggest that the enhanced activity of firefly luciferase is due to the existence of cryptic promoter activity in the 5′UTRs of CCN1, 2, and 4 (Fig. 4). In addition, the promoter activity of the 5′UTRs of CCNs was measured in different cell types, including chondrocytes, osteoblasts, and endothelial cells, all of which are cell types displaying high levels of CCN1, 2, and 4 activity in vivo (Fig. 5). While the 5′UTR promoter activity was seen in all cell types, there were some differences (Fig. 5). For example, all three 5′UTRs drove moderate expression in ATDC5 chondrocytes to similar extents. However, the 5′UTRs of CCN1 and CCN2 were far more active in osteoblasts than was the 5′UTR of CCN4. Similarly, the 5′UTRs of CCN1 and CCN4 were more active than that of CCN2 in endothelial cells. These preliminary findings suggest that the promoter elements in the 5′UTRs may be involved in the tissue-specific expression of CCN genes in vivo.
Using transcription binding motif prediction software (http://forkhead.cgr.ki.se/cgi-bin/consite and http://zlab.bu.edu/~mfrith/cister.shtml), we found potential Sp1, Ets, AP, and CREB binding sites, all of which have been reported to function in TATA-less promoters (Smale and Kadonaga 2003), within the 5′UTRs of CCN1, 2, and 4 (Fig. 6a and data not shown). In the core element mapping assay, region 55–108 bp of the CCN2 5′UTR had the strongest promoter activity (Fig. 6c). This region contains a higher GC content (72%) and a higher density of predicted transcription factors than other regions. It contains a potential ATF/CREB binding site, which is known to regulate transcription in TATA-less promoters (Wu et al. 2004; Englander et al. 1991; Kim et al. 2002). CREB has been reported to have a major role in β-catenin-mediated regulation of CCN4/Wisp1 transcription (Xu et al. 2000) and CCN2/CTGF has been reported to be regulated by CREB through the MEK/ERK and JNK signaling pathways (Liu et al. 2006; Tullai et al. 2004). Consensus sites for SP1, which has long been known to exhibit TATA-less activity, are also predicted in regions B and C of the 5′UTR of CCN2. Sp7, which can potentially bind to this site, is required for osteoblast differentiation and bone formation (Nakashima et al. 2002). In addition, the Ets2 transcription factor inhibits mineralization and affects target gene expression during osteoblast maturation (Li et al. 2004). Ets1 and Ets2 are important in osteoblast differentiation and bone formation (Raouf and Seth 2000). Thus it is conceivable that these sites are functionally relevant to the regulation of CCN gene expression in vivo.
The existence of cryptic promoters in the 5′UTR of a gene is not common, although recent database analyses indicate that sequences located as far as 80 bp downstream of the transcription start site (corresponding to region B in the 5′UTR of CCN2) regulate transcription (Holmes et al. 2003; Yang et al. 2007). However, the usage of alternative promoters has been found in many genes (Kimura et al. 2006; Kim et al. 2005; Carninci et al. 2006). The usage of alternative promoters is important in regulating the expression level of genes and often is involved in tissue-specific or developmental stage-specific gene expression (Schibler and Sierra 1987). The possibility of alternative promoter usage has not previously been described for CCN family members. BLAST searches of the human and mouse EST databases utilizing the CCN2 5′ UTR reveal that the majority of transcripts start at the position indicated in Fig. 6a, but several transcripts were found with potential start sites in region B (data not shown), raising the possibility that the cryptic promoter may be active in vivo. Experiments to test whether this promoter element is utilized in vivo, and under what conditions, are warranted in the future. Interestingly, in our study, we found that the 5′UTRs of CCN1, 2, and 4 all enhanced firefly luciferase gene expression in transient transfection assays in HeLa cells under hypoxic conditions (1% of oxygen) as compared to normoxia (data not shown). There is a putative HIF-1α binding site predicted in the 5′UTR of CCN1 (data not shown) and some predicted AP binding sites in the 5′UTRs of CCN2 and 4. HIF-1α has been reported to interact with AP-1 in regulating the expression of CCN1 (Kunz et al. 2003). CCN2 has been shown to be a target gene of HIF-1α during hypoxia (Shimo et al. 2001; Higgins et al. 2004; Hong et al. 2006; Olbryt et al. 2006). HIF-1α is a major mediator enabling cellular adaptation to hypoxia. Additional studies to determine whether 5′UTRs of CCN family members function during hypoxia warrants further investigation.
Conclusions
Taken together, in this report, we show that, contrary to a previous report (Johannes et al. 1999), there are no IRES activities in the 5′UTRs of CCN1, 2, and 4, but there are cryptic promoters in these regions. Thus, IRES-mediated mechanisms do not account for the demonstrated ability of these genes to be expressed in conditions of cellular stress (e.g. under hypoxic conditions and in apoptotic hypertrophic chondrocytes). It is known that CCN family members are required for angiogenesis, chondrogenesis, and wound healing (Lau and Lam 1999; Takigawa 2003; Rachfal and Brigstock 2005). Their aberrant expression patterns are also associated with tumourigenesis (Planque and Perbal 2003). The likelihood that CCN gene expression is regulated by elements in untranslated regions is substantiated by a series of experiments that have demonstrated that the 3′UTR of CCN2 plays an important role in regulating the expression of this gene. The 3′UTR of CCN2 is unusually long (approximately 1 kb in length) and contains a cis-acting negative regulatory element that has a repressive effect on CCN2 expression (Kubota et al. 2005). This cis-repressive element is functional in multiple cell types and under conditions of hypoxia. However, under conditions of hypoxia, the 3′UTR element functions to increase the stability of CCN2 mRNA, thus increasing CCN2 expression rather than repressing it (Kondo et al. 2006). Therefore, regulation of CCN expression by elements in untranslated regions, in both the 5′ and 3′UTRs, may represent important mechanisms for fine-tuning the levels of CCN expression in a tissue-specific manner or under conditions of hypoxia.
Acknowledgements
The authors acknowledge A. Willis and N. Elango for the generous donation of plasmids and M. Hosseini for technical assistance. This work was supported by NIH (AR52686 to KML) and the Scleroderma Foundation.
Footnotes
Huang and Dornbach contributed equally to this work.
References
- Abreu JG, Ketpura NI, Reversade B, De Robertis EM (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol 4:599–604 [DOI] [PMC free article] [PubMed]
- Baird SD, Turcotte M, Korneluk RG, Holcik M (2006) Searching for IRES. RNA 12:1755–1785 [DOI] [PMC free article] [PubMed]
- Bert AG, Grepin R, Vadas MA, Goodall GJ (2006) Assessing IRES activity in the HIF-1alpha and other cellular 5′ UTRs. RNA 12:1074–1083 [DOI] [PMC free article] [PubMed]
- Bornstein P, Sage EH (2002) Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol 14:608–616 [DOI] [PubMed]
- Brigstock D (2003) The CCN family: a new stimulus package. J Endocrinol 178:169–175 [DOI] [PubMed]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC et al (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38:626–635 [DOI] [PubMed]
- Englander EW, Widen SG, Wilson SH (1991) Mammalian beta-polymerase promoter: phosphorylation of ATF/CRE-binding protein and regulation of DNA binding. Nucleic Acids Res 19:3369–3375 [DOI] [PMC free article] [PubMed]
- French DM, Kaul RJ, D’Souza AL, Crowley CW, Bao M, Frantz GD, Filvaroff EH, Desnoyers L (2004) WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am J Pathol 165:855–867 [DOI] [PMC free article] [PubMed]
- Friedrichsen S, Heuer H, Christ S, Winckler M, Brauer D, Bauer GR (2003) CTGF expression during mouse embryonic development. Exp Cell Res 312:175–188 [DOI] [PubMed]
- Frith MC, Hansen U, Weng Z (2001) Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics 17:878–889 [DOI] [PubMed]
- Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, Scott D, Yi WS, Popoviciu ML, Said JW, Koeffler HP (2005) Ovarian carcinomas: CCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res 11:7243–7254 [DOI] [PubMed]
- Han B, Zhang JT (2002) Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol Cell Biol 22:7372–7384 [DOI] [PMC free article] [PubMed]
- Hellen CU, Sarnow P (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15:1593–1612 [DOI] [PubMed]
- Higgins DF, Biju MP, Akai Y, Wutz A, Johnson RS, Haase VH (2004) Hypoxic induction of Ctgf is directly mediated by Hif-1. Am J Physiol Renal Physiol 287:F1223–F1232 [DOI] [PubMed]
- Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S (2005) Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA 11:1605–1609 [DOI] [PMC free article] [PubMed]
- Holmes A, Abraham DJ, Chen Y, Denton C, Shi-wen X, Black CM, Leask A (2003) Constitutive connective tissue growth factor expression in scleroderma fibroblasts is dependent on Sp1. J Biol Chem 278:41728–41733 [DOI] [PubMed]
- Hong KH, Yoo SA, Kang SS, Choi JJ, Kim WU, Cho CS (2006) Hypoxia induces expression of connective tissue growth factor in scleroderma skin fibroblasts. Clin Exp Immunol 146:362–370 [DOI] [PMC free article] [PubMed]
- Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, Roudier J, Holderbaum D, Pauli RM, Herd JK, Van Hul EV et al (1999) Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet 23:94–98 [DOI] [PubMed]
- Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y (2002) Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J 16:219–221 [DOI] [PubMed]
- Ivkovic S, Yoon B, Popoff S, Safadi F, Libuda D, Stephenson R, Daluiski A, Lyons K (2003) Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791 [DOI] [PMC free article] [PubMed]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci U S A 96:13118–13123 [DOI] [PMC free article] [PubMed]
- Kasaragod AB, Lucia MS, Cabirac G, Grotendorst GR, Stenmark KR (2001) Connective tissue growth factor expression in pediatric myofibroblastic tumors. Pediatr Dev Pathol 4:37–45 [DOI] [PubMed]
- Kim SW, Lee SH, Kim KS, Kim CH, Choo YK, Lee YC (2002) Isolation and characterization of the promoter region of the human GM3 synthase gene. Biochim Biophys Acta 1578:84–89 [DOI] [PubMed]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436:876–880 [DOI] [PMC free article] [PubMed]
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H et al (2006) Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res 16:55–65 [DOI] [PMC free article] [PubMed]
- Komar AA, Hatzoglou M (2005) Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem 280:23425–23428 [DOI] [PubMed]
- Kondo S, Kubota S, Mukudai Y, Moritani N, Nishida T, Matsushita H, Matsumoto S, Sugahara T, Takigawa M (2006) Hypoxic regulation of stability of connective tissue growth factor/CCN2 mRNA by 3′-untranslated region interacting with a cellular protein in human chondrosarcoma cells. Oncogene 25:1099–1110 [DOI] [PubMed]
- Kozak M (2005) A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res 33:6593–6602 [DOI] [PMC free article] [PubMed]
- Kraggerud SM, Sandvik JA, Pettersen EO (1995) Regulation of protein synthesis in human cells exposed to extreme hypoxia. Anticancer Res 15:683–686 [PubMed]
- Kubota S, Mukudai Y, Moritani NH, Nakao K, Kawata K, Takigawa M (2005) Translational repression by the cis-acting element of structure-anchored repression (CAESAR) of human ctgf/ccn2 mRNA. FEBS Lett 579:3751–3758 [DOI] [PubMed]
- Kunz M, Moeller S, Koczan D, Lorenz P, Wenger RH, Glocker MO, Thiesen HJ, Gross G, Ibrahim SM (2003) Mechanisms of hypoxic gene regulation of angiogenesis factor Cyr61 in melanoma cells. J Biol Chem 278:45651–45660 [DOI] [PubMed]
- Lau L, Lam S (1999) The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res 248:44–57 [DOI] [PubMed]
- Leask A, Abraham DJ (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810 [DOI] [PubMed]
- Li V, Raouf A, Kitching R, Seth A (2004) Ets2 transcription factor inhibits mineralization and affects target gene expression during osteoblast maturation. In Vivo 18:517–524 [PubMed]
- Liu B, Yu J, Taylor L, Zhou X, Polgar P (2006) Microarray and phosphokinase screenings leading to studies on ERK and JNK regulation of connective tissue growth factor expression by angiotensin II 1a and bradykinin B2 receptors in Rat1 fibroblasts. J Cell Biochem 97:1104–1120 [DOI] [PubMed]
- Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC (2004) Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development 131:2137–2147 [DOI] [PubMed]
- Michiels C (2003) Endothelial cell functions. J Cell Physiol 196:430–443 [DOI] [PubMed]
- Mo F-E, Muntean A, Chen C-C, Stolz D, Watkins S, Lau L (2002) CYR61 (CCN1) is essential from placental development and vascular integrity. Mol Cell Biol 22:8709–8720 [DOI] [PMC free article] [PubMed]
- Nakanishi T, Yamaai T, Asano M, Nawachi K, Suzuki M, Sugimoto T, Takigawa M (2001) Overexpression of connective tissue growth factor/hypertrophic chondrocyte-specific gene product 24 decreases bone density in adult mice and induces dwarfism. Biochem Biophys Res Commun 281:678–681 [DOI] [PubMed]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17–29 [DOI] [PubMed]
- Olbryt M, Jarzab M, Jazowiecka-Rakus J, Simek K, Szala S, Sochanik A (2006) Gene expression profile of B 16(F10) murine melanoma cells exposed to hypoxic conditions in vitro. Gene Expr 13:191–203 [DOI] [PMC free article] [PubMed]
- Pan LH, Beppu T, Kurose A, Yamauchi K, Sugawara A, Suzuki M, Ogawa A, Sawai T (2002) Neoplastic cells and proliferating endothelial cells express connective tissue growth factor (CTGF) in glioblastoma. Neurol Res 24:677–683 [DOI] [PubMed]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M et al (1998) WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci U S A 95:14717–14722 [DOI] [PMC free article] [PubMed]
- Planque N, Perbal B (2003) A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int 3:15 [DOI] [PMC free article] [PubMed]
- Rachfal AW, Brigstock DR (2005) Structural and functional properties of CCN proteins. Vitam Horm 70:69–103 [DOI] [PubMed]
- Raouf A, Seth A (2000) Ets transcription factors and targets in osteogenesis. Oncogene 19:6455–6463 [DOI] [PubMed]
- Sandelin A, Wasserman WW, Lenhard B (2004) ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res 32:W249–W252 [DOI] [PMC free article] [PubMed]
- Schibler U, Sierra F (1987) Alternative promoters in developmental gene expression. Annu Rev Genet 21:237–257 [DOI] [PubMed]
- Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS (2001) Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev 15:2865–2876 [DOI] [PMC free article] [PubMed]
- Schneider R, Agol VI, Andino R, Bayard F, Cavener DR, Chappell SA, Chen JJ, Darlix JL, Dasgupta A, Donze O et al (2001) New ways of initiating translation in eukaryotes. Mol Cell Biol 21:8238–8246 [DOI] [PMC free article] [PubMed]
- Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE (2004) BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 279:29066–29074 [DOI] [PubMed]
- Shimo T, Kubota S, Kondo S, Nakanishi T, Sasaki A, Mese H, Matsumura T, Takigawa M (2001) Connective tissue growth factor as a major angiogenic agent that is induced by hypoxia in a human breast cancer cell line. Cancer Lett 174:57–64 [DOI] [PubMed]
- Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72:449–479 [DOI] [PubMed]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE (1998) C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16:423–428 [DOI] [PubMed]
- Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE (2000) Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res 28:687–694 [DOI] [PMC free article] [PubMed]
- Stoneley M, Willis AE (2004) Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23:3200–3207 [DOI] [PubMed]
- Takigawa M (2003) CTGF/Hcs24 as a multifunctional growth factor for fibroblasts, chondrocytes and vascular endothelial cells. Drug News Perspect 16:11–21 [DOI] [PubMed]
- Tinton SA, Buc-Calderon PM (1999) Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett 446:55–59 [DOI] [PubMed]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A (1992) Internal ribosome entry site within hepatitis C virus RNA. J Virol 66:1476–1483 [DOI] [PMC free article] [PubMed]
- Tullai JW, Schaffer ME, Mullenbrock S, Kasif S, Cooper GM (2004) Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J Biol Chem 279:20167–20177 [DOI] [PubMed]
- Vagner S, Galy B, Pyronnet S (2001) Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep 2:893–898 [DOI] [PMC free article] [PubMed]
- Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE (2004a) Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10:720–730 [DOI] [PMC free article] [PubMed]
- Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE (2004b) Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA 10:469–481 [DOI] [PMC free article] [PubMed]
- Wong M, Kireeva ML, Kolesnikova TV, Lau LF (1997) Cyr61, product of a growth factor-inducible immediate-early gene, regulates chondrogenesis in mouse limb bud mesenchymal cells. Dev Biol 192:492–508 [DOI] [PubMed]
- Wu L, Kobayashi K, Sun T, Gao P, Liu J, Nakamura M, Weisberg E, Mukhopadhyay NK, Griffin JD (2004) Cloning and functional characterization of the murine mastermind-like 1 (Maml1) gene. Gene 328:153–165 [DOI] [PubMed]
- Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP (2001) Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res 61:8917–8923 [PubMed]
- Xu L, Corcoran R, Welsh J, Pennica D, Levine A (2000) WISP-1 is a Wnt-1- and β-catenin-responsive oncogene. Genes Dev 14:585–595 [PMC free article] [PubMed]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E (2007) Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389:52–65 [DOI] [PMC free article] [PubMed]
- Yu C, Le AT, Yeger H, Perbal B, Alman BA (2003) NOV (CCN3) regulation in the growth plate and CCN family member expression in cartilage neoplasia. J Pathol 201:609–615 [DOI] [PubMed]
- Zelzer E, Mamluk R, Ferrara N, Johnson RS, Schipani E, Olsen BR (2004) VEGFA is necessary for chondrocyte survival during bone development. Development 131:2161–2171 [DOI] [PubMed]
- Zeng ZJ, Yang LY, Ding X, Wang W (2004) Expressions of cysteine-rich61, connective tissue growth factor and Nov genes in hepatocellular carcinoma and their clinical significance. World J Gastroenterol 10:3414–3418 [DOI] [PMC free article] [PubMed]