Abstract
Microarray-based comparisons of long-lived and normal mouse strains represent a promising approach for dissecting the basis of lifespan extension in higher organisms. Recently, Boylston et al. (2006) generated a genome-wide data set that allowed expression levels of Snell (Pit1dw/dw) and Ames (Prop1df/df) long-lived mice to be compared with age-matched control mice across different ages (6–24 months). Longevity-associated genes were identified as those genes exhibiting differential expression between long-lived and normal mice at every age examined. In this communication, an alternative approach to identifying longevity-associated genes is suggested and applied to the data sets considered by Boylston et al. (2006). Longevity-associated genes are defined as those exhibiting significant genotype-by-age interaction with respect to expression levels of long-lived and normal mice, and a total of 63 longevity-associated genes are identified. This approach may lend greater confidence to the inference that expression of identified genes specifically underlies aging differences between long-lived and normal genotypes.
Keywords: Aging, Ames, Dwarf, Lifespan, Microarray, Pit1dw, Prop1df, Snell
Introduction
Long-lived strains of the laboratory mouse provide a valuable tool for dissecting and understanding factors associated with extended lifespan in mammals. In recent years, the number of known long-lived mutant mouse strains has increased appreciably (Miskin and Masos 1997; Flurkey et al. 2002; Bluher et al. 2003; Holzenberger et al. 2003; Kurosu et al. 2005). This growing collection will ultimately allow researchers to characterize differences between normal and long-lived strains at both the cellular and organismal levels. A key role in this progression will be occupied by DNA microarrays (Park and Prolla 2005; Spindler 2006). Microarrays allow transcript levels to be quantified on a genome-wide scale and are increasingly an affordable and standard means of investigating gene expression patterns. Microarrays have been used to identify expression-level differences between long-lived and normal mouse strains in several previous studies (Dozmorov et al. 2001, 2002; Boylston et al. 2004; Papaconstantinou et al. 2005; Boylston et al. 2006), and similar studies will be necessary in the future. It is therefore worthwhile to consider how these data should be analyzed in order to maximize the biological knowledge generated by this approach.
Boylston et al. (2006) recently provided a genome-wide analysis of expression differences between long-lived and normal mouse genotypes. This study utilized Affymetrix chips containing probe sets for 11,000–34,000 murine genes, representing a level of genome coverage considerably greater than that of earlier studies. Expression levels of both Snell (Pit1dw/dw) and Ames (Prop1df/df) long-lived dwarf mice were assayed at different ages (6–24 months), along with expression levels of corresponding age-matched control mice (Pit1+/? or Prop1+/?). These data represent a key step toward understanding mouse extended lifespan phenotypes at the gene expression level. In particular, based on these data, it is possible to identify key genes that may account for aging deceleration in Snell and Ames mouse genotypes. At the same time, however, given that Snell and Ames mice exhibit multiple endocrine abnormalities, many transcriptional changes associated with these models may be unrelated to aging (Carter et al. 2002).
Longevity-associated genes were defined by Boylston et al. (2006) as genes exhibiting differential expression between long-lived and control strains with respect to every age considered (6–24 months). Their analysis identified a total of 205 and 785 such genes from Pit1dw/dw and Prop1df/df mice, respectively, with 49 genes common to both of these gene sets. The present communication does not dispute that these identified genes are worthwhile candidates for further investigation. However, an alternative approach is suggested and applied, yielding a narrowed list of candidates. Genes identified by Boylston et al. (2006) exhibit an age-independent expression difference between long-lived and normal genotypes (see Table 1 from Boylston et al. 2006). Expression of such genes could be associated with any of several phenotypic characteristics distinguishing long-lived and normal genotypes, some of which are not associated with aging directly (e.g., body size). The suggestion that expression of identified genes impacts aging, rather than some other process or characteristic, may be strengthened for cases in which genotypic expression differences are age dependent. A natural statistical approach for identifying such genes is to evaluate genotype-by-age interaction effects. This approach is applied in the present communication, and it is suggested that identified genes are especially likely to underlie the aging processes that distinguish long-lived from normal mice.
Table 1.
Longevity-associated genes exhibiting significant genotype-by-age interaction with respect to expression levels of long-lived and normal mice
| Gene Symbol | Pattern | Description |
|---|---|---|
| Snell (dw/dw) | ||
| Pafah1b3b | A | Platelet-activating factor acetylhydrolase, isoform 1b, alpha 1 subunit |
| Pbx2a | A | Pre-B-cell leukemia transcription factor 2 |
| Ppyr1a,d | A | Pancreatic polypeptide receptor 1 |
| 1100001G20Rik a,d | A | RIKEN cDNA 1100001G20 gene |
| Aif1a | A | Allograft inflammatory factor 1 |
| 2610019E17Rika,d | A | RIKEN cDNA 2610019E17 gene |
| Cyb5r1b | A | Cytochrome b5 reductase 1 |
| Laptm5b | A | Lysosomal-associated protein transmembrane 5 |
| H2-Aaa | A | Histocompatibility 2, class II antigen A, alpha |
| Igh-6b,d | A | Immunoglobulin heavy chain 6 (heavy chain of IgM) |
| Syn1b | A | Synapsin I |
| Kifc2a,d | A | Kinesin family member C2 |
| Btg3a,d | A | B-cell translocation gene 3 |
| Cada | A | Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| Foxa3a,d | A | Forkhead box A3 |
| Mark2b | A | MAP/microtubule affinity-regulating kinase 2 |
| Lamp2b | B | Lysosomal membrane glycoprotein 2 |
| Amy1b | B | Amylase 1, salivary |
| Plscr2a | B | Phospholipid scramblase 2 |
| Aqp4a | B | Aquaporin 4 |
| Acot1a | B | Acyl-coenzyme A (CoA) thioesterase 1 |
| Aox1a | B | Aldehyde oxidase 1 |
| Gfm1b | B | G elongation factor, mitochondrial 1 |
| Thrspa | B | Thyroid hormone responsive SPOT14 homolog (Rattus) |
| BC031181b | B | cDNA sequence BC031181 |
| Esdb | B | Esterase D/formylglutathione hydrolase |
| Abcd3b | B | ATP-binding cassette, subfamily D (ALD), member 3 |
| Glo1b | B | Glyoxalase 1 |
| Chpt1a | B | Choline phosphotransferase 1 |
| Sdhab | B | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| Atg5a | B | Autophagy-related 5 (yeast) |
| Sar1bb | B | SAR1 gene homolog B (S. cerevisiae) |
| 4931406C07Rika,d | B | RIKEN cDNA 4931406C07 gene |
| Bphlb,d | B | Biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
| 1300002A08Rika | B | RIKEN cDNA 1300002A08 gene |
| Ehhadha | B | Enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| Hsd17b4a | B | Hydroxysteroid (17 beta) dehydrogenase 4 |
| Anxa8a | B | Annexin A8 |
| Gstm3a,d | B | Glutathione S-transferase, mu 3 |
| Dlstb | B | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| Fabp2b | B | Fatty-acid-binding protein 2, intestinal |
| Ghrb | B | Growth hormone receptor |
| Skp1ab | B | S-phase kinase-associated protein 1A |
| Chi3l1a | B | Chitinase 3-like 1 |
| Cdc2l1c | B | Cell division cycle 2-like 1 |
| Ames (df/df) | ||
| Limd2a,d | A | LIM domain containing 2 |
| Fasa | A | Fas [tumor necrosis factor (TNF) receptor superfamily member] |
| Adrb3a | A | Adrenergic receptor, beta 3 |
| Slc6a9b | B | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| 2610529C04Rikb | B | RIKEN cDNA 2610529C04 gene |
| Igfalsa,d | B | Insulin-like growth factor binding protein, acid labile subunit |
| Serpina3ca,d | B | Serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| Ces2a | B | Carboxylesterase 2 |
| Csada,d | B | Cysteine sulfinic acid decarboxylase |
| 2310047H23Rikb | B | RIKEN cDNA 2310047H23 gene |
| Mup1a,d | B | Major urinary protein 1 |
| Cdc34b | B | Cell division cycle 34 homolog (S. cerevisiae) |
| Cyp2f2b | B | Cytochrome P450, family 2, subfamily f, polypeptide 2 |
| Cadps2a,d | B | Ca2+-dependent activator protein for secretion 2 |
| Ara | B | Androgen receptor |
| Klra19b | A/B | Killer cell lectin-like receptor, subfamily A, member 19 |
| Aasdhpptb | A/B | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| Bcl9b | A/B | B-cell CLL/lymphoma 9 |
The type of pattern (A or B) indicates the specific type of genotype-by-age interaction associated with listed genes (see text and Figs. 1 and 2 for explanation)
XaInteraction significant (P < 0.05) based on two-factor analysis of variance (ANOVA) model and linear models for microarray data (LIMMA) analysis
XbInteraction significant (P < 0.05) based on two-factor ANOVA model, but not LIMMA analysis
XcInteraction significant (P < 0.05) based on LIMMA analysis, but not two-factor ANOVA model
XdDifferentially expressed at every age examined (criterion used by Boyleston et al. 2006)
Materials and methods
Detailed description of the experimental protocols followed in generating the data analyzed here is provided by Boylston et al. (2006). In brief, for dw/dw mice, four genotype-by-age treatment combinations were evaluated [dw/dw-6 months (n = 4), dw/dw-24 months (n = 3), control-6 months (n = 4), control-24 months (n = 3)]. For df/df mice, six genotype-by-age treatment combinations were evaluated [df/df-6 months (n = 5), df/df-12 months (n = 5), df/df-24 months (n = 6), control-6 months (n = 5), control-12 months (n = 5), control-24 months (n = 5)]. RNA was isolated from the liver tissue of experimental animals and hybridized to either MG U74Av2 (> 11,000 genes, dw/dw mice) or MG 430 2.0 (> 34,000 genes, df/df mice) oligonucleotide arrays.
Expression-level data sets processed using MAS 5.0 normalization were downloaded from Gene Expression Omnibus (series GSE3129 and GSE3150). Two approaches were used to identify genes exhibiting significant genotype-by-age interaction (at a significance level of 0.05). First, a two-factor analysis of variance (ANOVA) was applied to the expression data associated with each probe, where genotype and age were included as main-model effects along with their interaction. This approach is straightforward but requires estimation of multiple variance terms on a gene-by-gene basis, which could lead to poor performance of associated test statistics (Allison et al. 2006). A second approach, therefore, was also implemented in which genotype-by-age interactions were specified as contrasts using the linear models for microarray data (LIMMA) linear modeling package (Smyth 2004). For dw/dw mice, the design matrix specified only one contrast characterizing the genotype-by-age interaction between dw/dw and control mice at 6 and 24 months of age. For df/df mice, the design matrix specified three contrasts, which characterized the degree of genotype-by-age interaction between df/df and control mice at 6 and 12 months, 12 and 24 months, and 6 and 24 months, respectively. In both approaches described above, P values were adjusted across genes using the Benjamini–Hochberg correction (Benjamini and Hochberg 1995).
Results and discussion
A total of 63 genes were identified as exhibiting significant genotype-by-age interaction within one of the two long-lived strains. The majority of these genes (45/63) were identified on the basis of expression data generated from Snell (dw/dw) mice, whereas the remaining (18/63) genes were identified from Ames (df/df) mice. The complete list of 63 identified genes is provided in Table 1 along with annotations. No genes were found to exhibit genotype-by-age interaction with respect to both dw/dw and df/df genotypes. This lack of overlap does not reflect conservative statistical methodology. The Benjamini–Hochberg correction used to adjust P values, for instance, is generally regarded as nonconservative, as it assumes independence of expression among different genes (Allison et al. 2006).
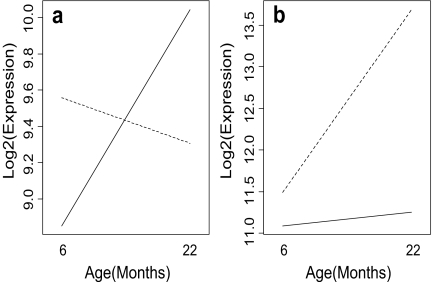
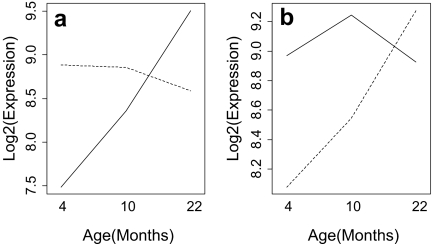
Two basic types of genotype-by-age interaction patterns were present among identified genes. These two patterns will be referred to as type A and B patterns, respectively, and are exemplified by select genes displayed in Figs. 1 and 2. Define M as the fold-change expression difference between long-lived and normal genotypes for a given gene (long-lived/normal). Type A patterns are those for which M declines with increased age (see Figs. 1a and 2a), whereas type B patterns are those for which M increases with increased age (see Figs. 1b and 2b). For each identified gene, the type of genotype-by-age interaction pattern present is indicated in Table 1. The majority of identified genes exhibited a type B pattern (29/45 for the dw/dw genotype, 12/18 for the df/df genotype).
Fig. 1.
Select longevity-associated genes exhibiting contrasting types of genotype-by-age interaction with respect to expression levels of Snell (dw/dw) long-lived mice and age-matched controls. Averaged expression profiles of dw/dw mice are represented by the dashed line, whereas those of control mice are represented by the solid line. In a, a type A expression pattern is illustrated by Foxa3 (forkhead transcription factor). In b, a type B expression pattern is illustrated by Gstm3 (glutathione S-transferase)
Fig. 2.
Select longevity-associated genes exhibiting contrasting types of genotype-by-age interaction with respect to expression levels of Ames (df/df) long-lived mice and age-matched controls. Averaged expression profiles of df/df mice are represented by the dashed line, whereas those of control mice are represented by the solid line. In a, a type A expression pattern is illustrated by Fas (fatty acid synthase). In b, a type B expression pattern is illustrated by Ar (androgen receptor)
Identified genes were analyzed to determine whether any gene ontology terms were significantly overrepresented (Beissbarth and Speed 2004). Three main trends emerged from this analysis. First, 27 of 63 identified genes were localized to the cytoplasm (P < 0.001), and interestingly, each of these 27 cytoplasmic genes were identified from the dw/dw long-lived strain. The second major trend was that many genes (18/63) were associated with transport biological processes (P = 0.034), the majority of which (14/18) were found to exhibit a type B pattern of genotype-by-age interaction. Lastly, eight of 63 identified genes were associated with organic acid metabolism (P = 0.008), and ten genes were associated with catalytic activity molecular functions (P < 0.036). Several lower-level ontologies connected to these main themes were also significantly overrepresented, including specific types of metabolism [acyl-coenzyme A (CoA), fatty-acid and carboxylic-acid metabolism], and catalytic enzyme activities (lyase, carboxylic ester hydrolase, oxidoreductase, and transferase activities).
Several genes listed in Table 1 have annotations supporting a role in lifespan determination. The growth hormone receptor (Ghr) and insulin-like growth factor binding protein (Igfals), for instance, are components of the IGF-I axis (Papaconstantinou et al. 2005), which has been widely implicated in lifespan determination within several model systems (e.g., Kimura et al. 1997; Bartke et al. 2003; Tatar et al. 2003). Table 1 also includes several other endocrine-related genes, including androgen receptor (AR), adrenergic receptor beta 3 (Adrb3), thyroid hormone responsive SPOT14 homolog (Thrsp), and hydroxysteroid dehydrogenase 4 (Hsd17b4). Whereas it is not surprising that expression levels of such genes differ between Pit1dw/dw and Prop1df/df mice and age-matched controls, the age dependence of this difference is suggestive of a role in aging.
A good illustration is provided by glutathione S-transferase (Gstm3), which exhibited a type B genotype-by-age interaction pattern (see Fig. 1b). Whereas normal mice exhibited declines or maintained steady levels of this transcript with age, Gstm3 transcript abundance increased with age in dw/dw dwarfs. Gstm3 protects against the deleterious effects of oxidative stress, which is thought to be a key factor underlying the deleterious effects of aging (Brown-Borg 2006). The extended longevity phenotypes associated with dw/dw dwarfs could therefore be due in part to enhanced oxidative stress resistance due to elevated expression of Gstm3 with age (which does not occur in normal mice) (Fig. 1b).
Acknowledgements
This work was supported by a research grant from the Michigan State University Quantitative Biology and Modeling Initiative and the Department of Statistics and Probability. The author thanks two anonymous reviewers for helpful comments on this manuscript.
References
- Allison DB, Xiangqui C, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7:55–65 [DOI] [PubMed]
- Bartke A, Chandrashekar V, Dominici F, Turyn D, Kinney B, Steger R, Kopchick JJ (2003) Insulin-like growth factor 1 (IGF-1) and aging: controversies and new insights. Biogerontology 4:1–8 [DOI] [PubMed]
- Beissbarth T, Speed TP (2004) Gostat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics 20:1464–1465 [DOI] [PubMed]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a powerful and practical approach to multiple testing. J Roy Stat Soc B 57:289–300
- Bluher M, Kahn B, Kahn CR (2003) Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299:572 [DOI] [PubMed]
- Boylston WH, Gerstner A, DeFord JH, Madsen M, Flurkey K, Harrison DE, Papaconstantinou J (2004) Altered cholesterologenic and lipogenic transcriptional profile in livers of aging Snell dwarf (Pitdw/dwj) mice. Aging Cell 3:283–296 [DOI] [PubMed]
- Boylston WH, DeFord JH, Papaconstantinou J (2006) Identification of longevity-associated genes in long-lived Snell and Ames dwarf mice. AGE 28:125–144 [DOI] [PMC free article] [PubMed]
- Brown-Borg HM (2006) Longevity in mice: is stress resistance a common factor? AGE 28:145–162 [DOI] [PMC free article] [PubMed]
- Carter CS, Ramsey MM, Sonntag WE (2002) A critical analysis of the role of growth hormone and IGF-1 in aging and lifespan. Trends Genet 18:295–301 [DOI] [PubMed]
- Dozmorov I, Bartke A, Miller RA (2001) Array-based expression analysis of mouse liver genes: effect of age and of the longevity mutant Prop1df. J Gerontol A Biol Sci Med Sci 56:B72–B80 [DOI] [PubMed]
- Dozmorov I, Galecki A, Chang Y, Krzesiecki R, Vergara M, Miller RA (2002) Gene expression profile of long-lived Snell dwarf mice. J Gerontol A Biol Sci Med Sci 57:B99–B108 [DOI] [PubMed]
- Flurkey K, Papaconstantinou J, Harrison DE (2002) The Snell dwarf mutation Pit1(dw) can increase lifespan in mice. Mech Ageing Dev 123:121–130 [DOI] [PubMed]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervara P, Le Bouc Y (2003) IGH-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421:182–187 [DOI] [PubMed]
- Kimura KD, Tissenbaum HG, Liu Y, Ruvkun G (1997) Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277:942–946 [DOI] [PubMed]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomur I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M (2005) Suppression of aging in mice by the hormone klotho. Science 309:1829–1833 [DOI] [PMC free article] [PubMed]
- Miskin R, Masos T (1997) Transgenic mice overexpressing urokinase-type plasminogen activator in the brain exhibit reduced food consumption, body weight and size and increased longevity. J Gerontol Ser A Biol Sci Med Sci 52(2):B118–B124 [DOI] [PubMed]
- Papaconstantinou J, Deford JH, Gerstner A, Hsieh C-C, Boylston WH, Guigneaux MM, Flurkey K, Harrison DE (2005) Hepatic gene and protein expression of primary components of the IGF-1 axis in long lived Snell dwarf mice. Mech Ageing Dev 126:692–704 [DOI] [PubMed]
- Park S-K, Prolla TA (2005) Lessons learned from gene expression profile studies of aging and caloric restriction. Ageing Res Rev 4:55–65 [DOI] [PubMed]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article 3 [DOI] [PubMed]
- Spindler SR (2006) Use of microarray biomarkers to identify longevity therapeutics. Aging Cell 5:39–50 [DOI] [PubMed]
- Tatar M, Bartke A, Antebi A (2003) The endocrine regulation of aging by insulin-like signals. Science 299:1346–1351 [DOI] [PubMed]