Abstract
The Pseudomonas syringae pv. tomato protein AvrPtoB is translocated into plant cells via the bacterial type III secretion system. In resistant tomato leaves, AvrPtoB acts as an avirulence protein by interacting with the host Pto kinase and eliciting the host immune response. Pto-mediated immunity requires Prf, a Pto-interacting protein with a putative nucleotide-binding site and a region of leucine-rich repeats. In susceptible tomato plants, which lack either Pto or Prf, AvrPtoB acts as a virulence protein by promoting P. syringae pv. tomato growth and enhancing symptoms associated with bacterial speck disease. The N-terminal 307 amino acids of AvrPtoB (designated AvrPtoB1–307) are sufficient for these virulence activities and for Pto-mediated avirulence. We report that AvrPtoB is phosphorylated by a Pto- and Prf-independent kinase activity that is conserved in several plant species, including tomato (Solanum lycopersicum), Nicotiana benthamiana, and Arabidopsis thaliana. AvrPtoB1–307 was phosphorylated in tomato protoplasts, and mass spectrometry identified serine 258 as the major in vivo phosphorylation site of this protein. An alanine substitution of Ser258 resulted in the loss of virulence and the diminution of avirulence activity of AvrPtoB1–307, whereas a phosphomimetic S258D mutant had activities similar to wild type AvrPtoB1–307. These observations suggest that AvrPtoB has evolved to mimic a substrate of a conserved plant kinase, leading to enhancement of its virulence and avirulence activities in the host cell.
Pathogens of plants have evolved strategies to evade or suppress both basal and R (resistance) gene-mediated host defenses (1). For example, Pseudomonas syringae pv. tomato (Pst),2 the causative organism of bacterial speck disease in tomato, uses its type III secretion system to deliver ~30 “effector” proteins into the plant cell (1–3). These sequence-diverse proteins are collectively essential for pathogenesis, and many appear to have functionally redundant roles in promoting bacterial virulence (4). Mounting evidence suggests that a major function of type III effectors is the suppression of the plant basal defenses that are induced following detection of pathogen-associated molecular patterns by host pattern recognition receptors (5–8). Pathogen-associated molecular patterns are structurally conserved and functionally indispensable in both pathogenic and nonpathogenic bacteria and reveal the presence of a potential danger to the plant upon recognition by pattern recognition receptors (9).
To overcome the suppression of basal defense by type III effectors, plants have evolved R genes that encode proteins that directly or indirectly recognize certain effectors thereby activating a strong plant immune response (10). R gene-mediated defense is usually associated with rapid localized cell death known as the hypersensitive response (11). Effectors which are detected by plant R proteins are referred to as avirulence (Avr) proteins. In response to this host surveillance mechanism, bacteria have evolved additional effectors to interfere with hypersensitive response-associated plant immunity (12–15). Type III effector proteins therefore play a central role in plant-pathogen interactions, and it is important to understand the molecular mechanisms they employ to manipulate host defense responses (1, 3, 16).
Type III effector proteins function within the plant cell, and interestingly, many of them appear to be post-translationally modified by host enzymes. For example, effectors have been reported to be acylated, phosphorylated, or proteolytically cleaved after their delivery into plant cells (8, 16, 17). In some cases, these post-translational modifications have been shown to be important for the function of effectors in the host cell. For example, effectors AvrPto, AvrRpm1, and AvrB are myristylated, and this modification promotes their localization to the plasma membrane, which is critical for the avirulence activity of all three effectors and for the virulence activity of AvrPto and AvrRpm1 (18, 19). Pseudomonas effectors AvrPto and AvrB are both phosphorylated by host kinases, and this alteration is required for their full avirulence and virulence activities (20, 21). Recently, two type III effectors from Rhizobium strain NGR234, NopL and NopP, have been shown to be phosphorylated by plant extracts (22–24). Both NopL and NopP are required for the nodulation ability of NGR234 on certain legumes, but it is unknown yet whether phosphorylation is involved in this function.
In the interaction between Pst strain DC3000 and tomato plants, resistance is conferred by the host R protein Pto, a serine/threonine kinase (25, 26). Although molecular details of the recognition mechanism are unknown, Pto physically interacts with either of two Pst DC3000 effectors, AvrPto or AvrPtoB, and activates immunity by interacting with the host protein Prf, an NB-ARC LRR protein (26–29). When either Pto or Prf is absent, the plant is susceptible to Pst DC3000. Moreover, in this case AvrPto and AvrPtoB act as virulence factors promoting bacterial growth and inducing ethylene biosynthesis, which promotes disease-associated host cell death (4, 30–33).
We have found previously that AvrPto is both phosphorylated and myristylated after it is delivered into the plant cell by Pst (21). The N terminus-dependent myristylation is critical for plasma membrane localization of AvrPto and indispensable for virulence activity and for eliciting Pto/Prf-mediated immunity (18, 21). Phosphorylation of AvrPto occurs at serine 149 located in the C-terminal region of AvrPto (21). An alanine substitution at Ser149 significantly decreased AvrPto virulence activity and partially compromised its ability to elicit Pto/Prf-mediated immunity, suggesting that phosphorylation of this residue plays an important role in AvrPto function (21).
AvrPtoB was originally identified as an avirulence protein based on its ability to trigger immunity on resistant tomato plants expressing Pto and Prf (34). AvrPtoB was later found to have two distinct avirulence determinants, both of which are in the N-terminal region of AvrPtoB. One, contained within the first 307 amino acids of the protein (AvrPtoB1–307), is recognized by the Pto kinase (13). The other requires the additional amino acids 308–387 (AvrPtoB1–387) and is recognized by the Fen kinase (13, 15).
AvrPtoB is a modular protein with both pathogenicity and virulence activities. The C-terminal region is an E3 ligase that acts as a pathogenicity factor by ubiquitinating the host R protein Fen, thereby promoting its degradation and causing disease susceptibility (15, 35, 36). The N-terminal region has two distinct virulence activities (33). In susceptible tomato plants (lacking either Pto or Prf), AvrPtoB1–307 is sufficient for promoting bacterial growth and enhancing disease symptoms associated with increased ethylene production (33). This ethylene-associated virulence is dependent on phenylalanine 173 in AvrPtoB. Substitution of an alanine for this residue abolishes the virulence activity of AvrPtoB1–307 (33). AvrPtoB residues 308–387 are required for a second virulence activity that involves the suppression of pathogen-associated molecular pattern-triggered immunity in Arabidopsis (33).
In susceptible tomato plants, the virulence activities of AvrPtoB1–307, AvrPtoB1–387, or the full-length AvrPtoB are indistinguishable (33), indicating that amino acids 308–553 either lack virulence activity in tomato or have an activity that is phenotypically redundant. Thus, AvrPtoB1–307 is sufficient both for eliciting Pto/Prf-dependent immunity in resistant tomato plants and for promoting bacterial virulence in susceptible tomato plants. Our primary interest in this work was the AvrPtoB virulence activity in tomato, and we therefore focused initially on AvrPtoB1–307.
Here we report that AvrPtoB is phosphorylated by a Pto- and Prf-independent protein kinase activity that is conserved in a diverse array of plant species. We used mass spectrometry to identify the in vivo phosphorylated residue and demonstrate that this residue plays an important role in AvrPtoB1–307 virulence activity. Our data support the emerging theme that type III effector proteins have evolved to mimic substrates of certain host enzymes to manipulate host defense signaling.
EXPERIMENTAL PROCEDURES
AvrPtoB Cloning and Mutagenesis
AvrPtoB1–307 tagged at the C terminus with a hemagglutinin (HA) epitope was generated previously (33). AvrPtoB1–307 S258A and S258D mutants were generated using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: 5′-AGACGCCGGTCGACAGGGCCCCGCCACGCGTCAACCAAAG-3′; 5′-CTTTGGTTGACGCGTGGCGGGGCCCTGTCGACCGGCGTCT, and 5′-AGACGCCGGTCGACAGGGACCCGCCACGCGTCAACCAAAG-3′; and 5′-CTTTGGTTGACGCGTGGCGGGTCCCTGTCGACCGGCGTCT-3′.
In Vitro Phosphorylation Assay of AvrPtoB
AvrPtoB was expressed in bacteria as a fusion protein in-frame with the C terminus of glutathione S-transferase (GST) and purified as described previously using a glutathione-Sepharose resin (21). Five leaf discs (~100 mg) from each plant species were ground in 1 ml of lysis buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 10 mM EDTA, 0.1% Triton X-100, and 10 μl/ml plant protease inhibitor mixture (Product 9599; Sigma-Aldrich)). Plant extracts were centrifuged at 8000 × g for 3 min, and 10 μg of the protein extract was used for an in vitro phosphorylation assay, which was performed with 10 μg of AvrPtoB-GST protein in the presence of 10 μCi of [γ-32P]ATP for 15 min at room temperature, followed by SDS-PAGE separation and autoradiography.
In Vivo 32P Labeling of AvrPtoB
HA-tagged AvrPtoB1–307 or its derivatives were transiently expressed in RG-prf3 tomato protoplasts by polyethylene glycol-mediated transformation (33). [32P]Orthophosphate (0.1mCi/ml) was added to the protoplasts 1 h after polyethylene glycol treatment and incubation continued for another 7 h in the dark. Protoplasts were collected and lysed with 1 ml of protoplast lysis buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 10 mM EDTA, 1% Triton X-100, and 10 μl/ml plant protease and phosphatase inhibitor I cocktails (Product 2850; Sigma-Aldrich)). AvrPtoB1–307-HA and derivatives were immunoprecipitated with anti-HA affinity matrix (Roche Applied Science) and subjected to SDS-PAGE, followed by vacuum drying of the gel and autoradiography.
Mass Spectrometry Analysis
100 μg of plasmid expressing HA-tagged AvrPtoB1–307 was transformed into 1 ml of RG-prf3 protoplasts (about 2 × 105 cells) and incubated for 9 h in the dark. Transformed protoplasts were collected and lysed with 1 ml of protoplast lysis buffer. The lysates were centrifuged at 12,000 × g for 20 min, and the supernatant was incubated with 100 μl of anti-HA affinity matrix at 4 °C for 4 h. The immunoprecipitation-complex was separated on 10% SDS-PAGE and stained with Coomassie Brilliant Blue G250. AvrPtoB1–307-HA was excised from the gel and sent for mass spectrometry analysis at the Proteomics and Mass Spectrometry Facility (Donald Danforth Plant Science Center, St. Louis, MO). The nano-ESI-MS/MS analysis was performed exactly as described previously (21).
Pseudomonas Protein Secretion and Western Blotting Assays
pCPP45 plasmids harboring avrPtoB1–307 or its derivatives were transformed into the DC3000ΔavrPtoΔavrPtoB strain by electroporation. The resulting strains were grown in minimal medium for 24 h at 18 °C, and secreted proteins were detected by standard Western blotting as described previously (37). The antibodies used for Western blotting were anti-HA (Roche Applied Science), anti-NptII (U.S. Biological Corp.), or anti-AvrPtoB (37). Detection of proteins was carried out using horseradish peroxidase-conjugated secondary antibodies and the ECL Plus detection system (Amersham Biosciences).
Measurement of Bacterial Populations in Tomato Leaves
Two genotypes of Rio Grande (RG) tomato lines were used: RG-PtoR (Pto/Pto, Prf/Prf) and RG-prf3 (Pto/Pto, prf/prf) (28). The method used to prepare P. syringae pv. tomato inoculum has been described previously (21). Five- to six-week-old greenhouse grown plants were vacuum-infiltrated with different P. syringae pv. tomato strains at an inoculum level of 104 colony-forming units/ml and maintained in a climate-controlled growth chamber under optimized conditions (see Ref. 21 for details). Bacterial populations in tomato leaves were measured at 2 or 4 days after infiltration. For consistency, day 2 samples were collected from the third and fourth leaves from the bottom of the plants. To assess the effect of Ser258 substitutions on the virulence and avirulence activities of AvrPtoB1–307 and mutants, analysis of variance of bacterial populations on plants was based on the pooled data from three experiments using the general linear model procedure of a statistical analysis system (SAS Institute Inc., Cary, NC). The least significance difference at a 0.05 probability level was used to test the differences between means.
RESULTS
To examine whether AvrPtoB is phosphorylated, we first adopted the in vitro phosphorylation assay, previously used for AvrPto, to test whether AvrPtoB could be phosphorylated by tomato leaf extracts (21). Recombinant AvrPtoB fused to GST was expressed in Escherichia coli and purified with a glutathione-Sepharose resin. The in vitro phosphorylation assay of AvrPtoB was performed in the presence of leaf extracts from resistant tomato RG-PtoR leaves (Pto/Pto, Prf/Prf) that express a functional Pto kinase. We observed incorporation of 32P into GST-AvrPtoB and not GST alone in the presence of leaf extracts (Fig. 1 and supplemental Fig. S1). GST-AvrPtoB without leaf extracts or RG-PtoR leaf extracts without AvrPtoB showed no evidence of 32P incorporation in this assay (Fig. 1). These observations indicated that AvrPtoB can be phosphorylated by a kinase activity present in RG-PtoR tomato leaves.
FIGURE 1. In vitro phosphorylation of AvrPtoB by leaf extracts from different tomato lines and other plant species.
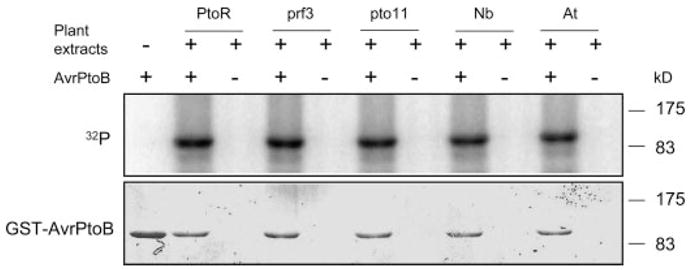
Recombinant GST-AvrPtoB protein was expressed in and purified from E. coli with glutathione-Sepharose resin. GST-AvrPtoB was incubated with leaf extracts in the presence of [γ-32P]ATP and separated by 10% SDS-PAGE, followed by autoradiography. Incorporation of 32P into GST-AvrPtoB (top panel) occurred in the presence of leaf extracts from tomato lines RG-PtoR (PtoR), RG-prf3 (prf3), RG-pto11 (pto11), N. benthamiana (Nb), and A. thaliana Columbia-0 (At). Equal loading of GST-AvrPtoB protein was verified by Coomassie Blue staining (bottom panel).
To assess whether phosphorylation of AvrPtoB is dependent on the Pto or Prf proteins that function to recognize the effector, we used leaf extracts from RG-pto11 (pto/pto Prf/Prf) or RG-prf3 (Pto/Pto prf/prf) plants for the in vitro phosphorylation analysis of AvrPtoB. RG-pto11 plants lack a functional Pto kinase because of a point mutation in the Pto gene, whereas RG-prf3 plants have a 1-kb deletion in the Prf gene rendering the plants susceptible to avirulent Pst strains despite their having a functional Pto gene (28). Leaf extracts from both RG-pto11 and RG-prf3 plants phosphorylated AvrPtoB as well as RG-PtoR leaf extracts did (Fig. 1). We further tested whether the kinase activity responsible for AvrPtoB phosphorylation is conserved in other plant species. Leaf extracts from the wild tobacco species Nicotiana benthamiana or Arabidopsis thaliana were tested, and both were found able to phosphorylate AvrPtoB (Fig. 1). Thus, AvrPtoB is phosphorylated by a Pto- or Prf-independent kinase activity that is conserved in several plant species.
We narrowed our initial focus to AvrPtoB1–307 because this region is recognized by Pto/Prf and is the minimal fragment in tomato with virulence activity. To test whether this region is phosphorylated in vivo, we developed a construct expressing AvrPtoB1–307 with a C-terminal HA tag and transiently transformed it into RG-prf3 protoplasts in the presence of [32P]orthophosphate. AvrPtoB1–307-HA was then immunoprecipitated with an anti-HA antibody and subjected to SDS-PAGE to monitor the phosphorylation of AvrPtoB1–307 as indicated by incorporation of 32P. AvrPtoB1–307 appeared as single 32P-labeled band (Fig. 2), indicating that the N-terminal domain of AvrPtoB is phosphorylated in plant cells.
FIGURE 2. In vivo phosphorylation of AvrPtoB1–307 in plant cells.
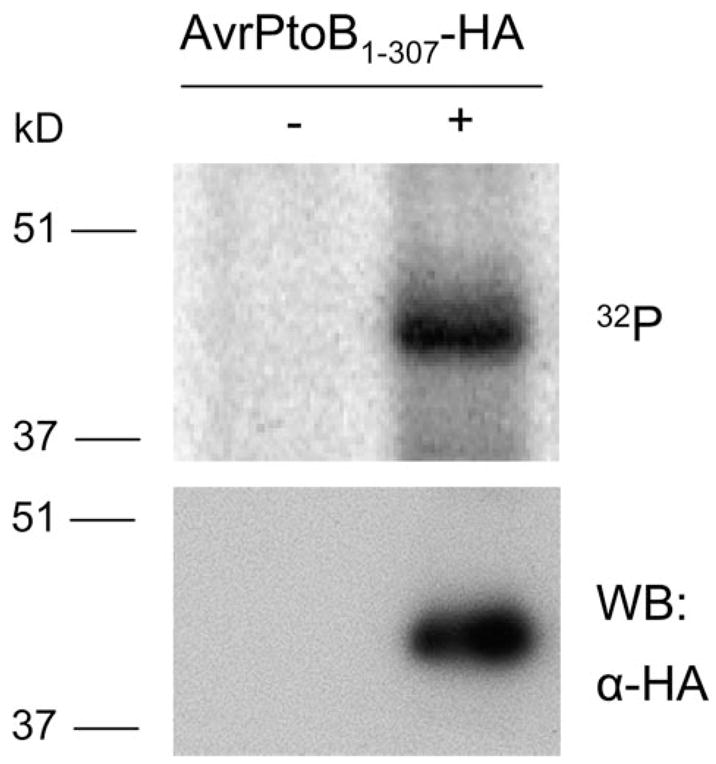
AvrPtoB1–307 with a C-terminal HA epitope tag was expressed in tomato RG-prf3 protoplasts by polyethylene glycol-mediated transformation in the presence of [32P]orthophosphate. AvrPtoB1–307-HA was immunoprecipitated with anti-HA affinity matrix, followed by SDS-PAGE separation and autoradiography. AvrPtoB1–307-HA incorporated [32P]phosphate (top panel). The identity of AvrPtoB1–307-HA band was confirmed by Western blotting (WB) with an anti-HA antibody using a small amount of lysate before immunoprecipitation (bottom panel).
To characterize the in vivo phosphorylation site of AvrPtoB1–307, we performed nano-ESI-MS/MS on the phosphorylated protein. AvrPtoB1–307-HA or an empty vector control were expressed in RG-prf3 tomato protoplasts, the proteins were immunoprecipitated with an anti-HA antibody and resolved by SDS-PAGE. Staining of this gel revealed a putative AvrPtoB1–307-HA band that was absent from the empty vector control (Fig. 3A). This band was excised from the gel for tryptic in-gel digestion and subjected to nano-ESI-MS/MS analysis. The resulting MS and MS/MS spectra were searched against the NCBInr data base using the Mascot software tool and revealed a clear identification of AvrPtoB as the protein present in this gel band (Fig. 3B). The protein was represented by several peptides providing an overall sequence coverage of ~65% (Fig. 3B). Among the identified peptides one was selected for MS/MS because it represented a single triply charged precursor with a mass-to-charge (m/z) ratio of 617.57, which matches the m/z of a singly phosphorylated peptide from AvrPtoB (namely amino acids 245–261; SSNTAASQTPVDRSPPR). A series of y-ions and several b-ion fragments revealed serine 258 to be the phosphorylated amino acid residue in this peptide (Fig. 3, C and D).
FIGURE 3. Identification of the in vivo phosphorylation site of AvrPtoB1–307 by mass spectrometry.
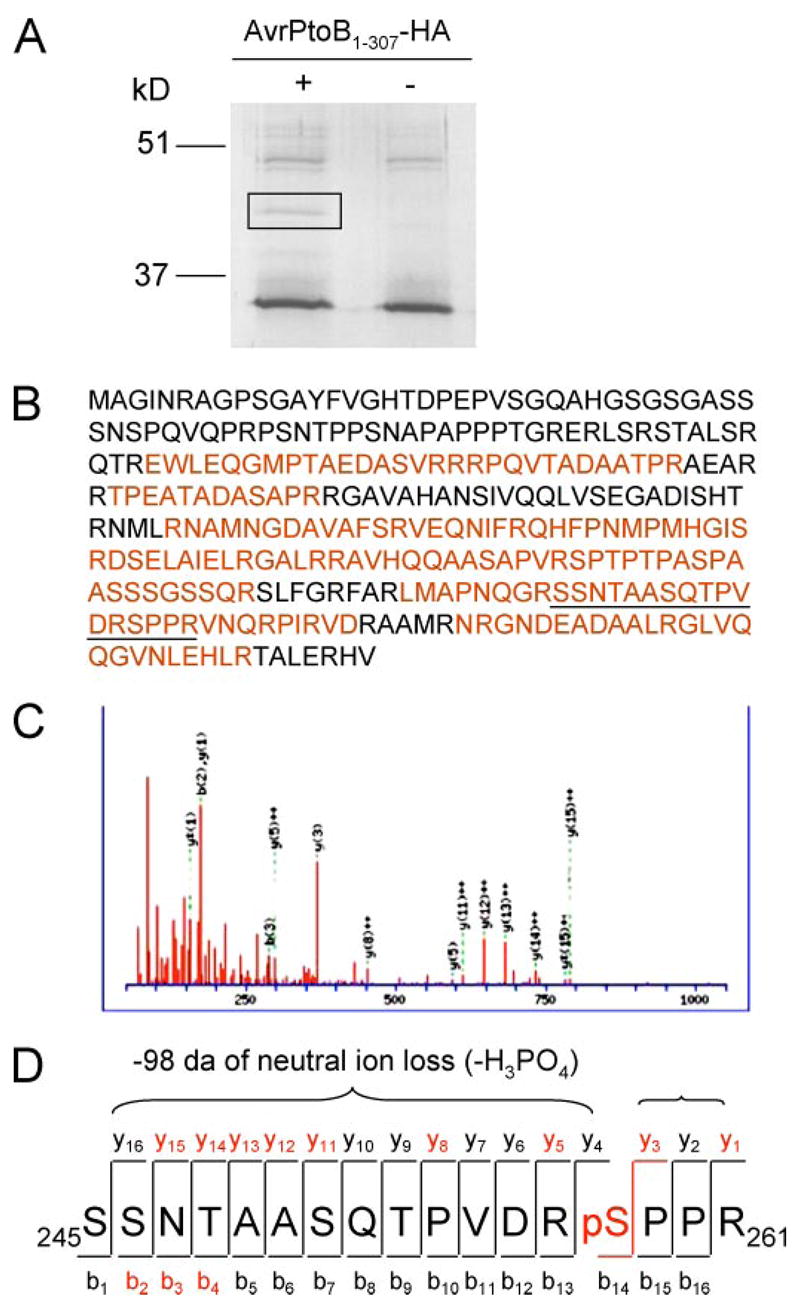
A, purification of AvrPtoB1–307 protein after expression in plant cells. AvrPtoB1–307-HA was transiently expressed in tomato RG-prf3 protoplasts, immunoprecipitated with anti-HA affinity matrix, and separated by SDS-PAGE. AvrPtoB1–307-HA appeared as a unique band in the Coomassie Blue-stained gel (marked by rectangle), and this band was excised for trypsin digestion. B, coverage of AvrPtoB1–307 sequence by MS/MS. Sequence recovered from ESI-MS/MS is highlighted in red. The peptide 245SSNTAASQTPVDRSPPR261 (underlined) with mass-to-charge (m/z) value of 617.57 was selected for collision-induced dissociation fragmentation. C, MS/MS spectrum of precursor 245SSNTAASQTPVDRSPPR261 indicates the phosphorylation on Ser258. D, diagram of the series of y fragment ions and several b fragment ions observed in C. Several y-ions featured loss of 98 Da because of neutral loss of phosphoric acid (−H3PO4). Red letters indicate ions present in the MS/MS spectrum.
We next sought to confirm that Ser258 is a phosphorylation site of AvrPtoB1–307 in the plant cell. We generated a construct expressing an AvrPtoB1–307(S258A) mutant and repeated the in vivo labeling assay to evaluate the level of [32P]orthophosphate incorporation of the mutant compared with the wild type AvrPtoB1–307 protein. The level of phosphate incorporation by each protein was evaluated by autoradiography after immunoprecipitation with anti-HA antibody and SDS-PAGE. The alanine substitution at Ser258 reduced the level of 32P incorporation by ~80% compared with wild type AvrPtoB1–307 (Fig. 4). Western blotting indicated that both AvrPtoB1–307 and the AvrPtoB1–307(S258A) mutant proteins were equally expressed in tomato protoplasts (Fig. 4A). Therefore, we conclude that Ser258 is the major in vivo phosphorylation site of AvrPtoB1–307. Because a low level of 32P incorporation of AvrPtoB1–307(S258A) mutant was detected, it is possible there are other weakly phosphorylated residues in AvrPtoB1–307 that were not identified by mass spectrometry. Alternatively, blocking of Ser258 might result in other residues being phosphorylated.
FIGURE 4. Serine 258 is the major in vivo phosphorylation site of AvrPtoB1–307.
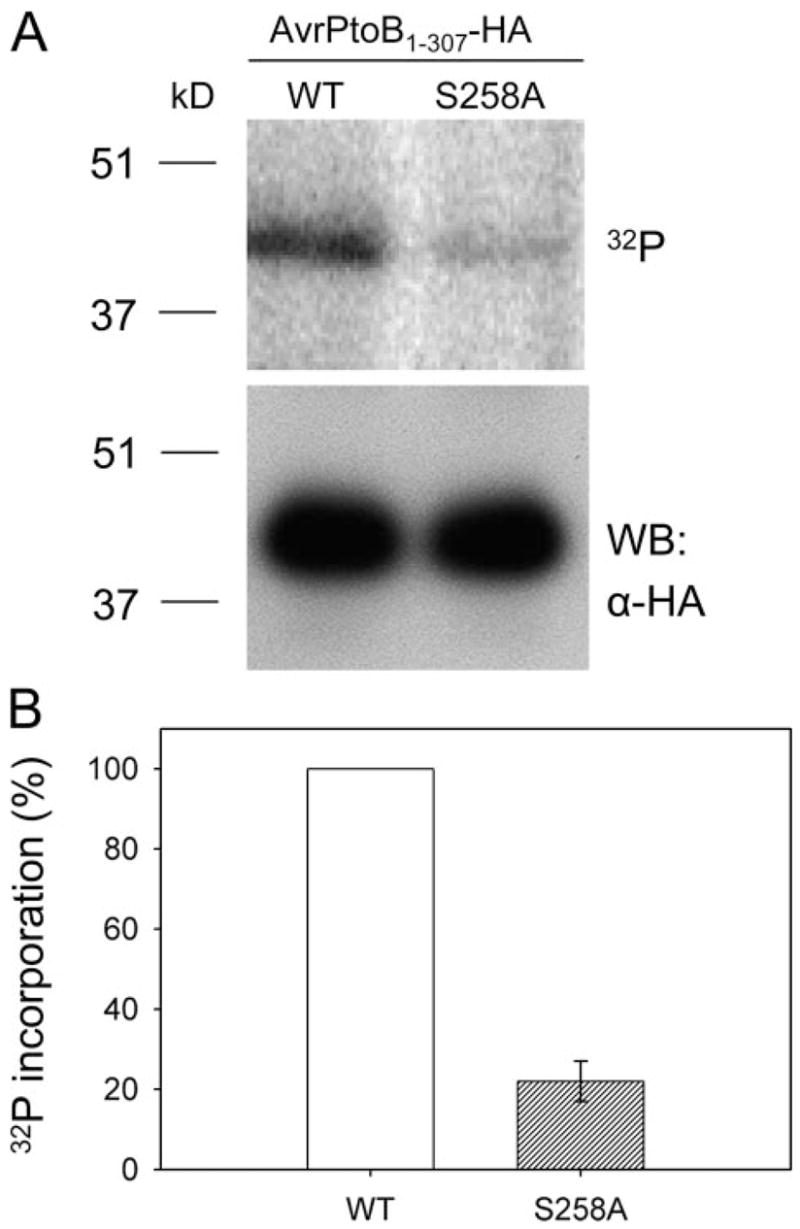
A, phosphorylation assay of wild type (WT) AvrPtoB1–307 and the AvrPtoB1–307(S258A) mutant in RG-prf3 tomato protoplasts. In vivo 32P labeling was performed as described in the legend to Fig. 2. Incorporation of 32 P into AvrPtoB1–307 and AvrPtoB1–307(S258A) mutant is shown by autoradiography after SDS-PAGE separation (top panel). Equal expression of WT AvrPtoB1–307 and AvrPtoB1–307(S258A) mutant in plant cells was confirmed by Western blotting with an anti-HA antibody using a small amount of lysate before immunoprecipitation (bottom panel). B, relative 32P incorporation level of WT AvrPtoB1–307 and AvrPtoB1–307(S258A) mutant. The data are shown as percentages of 32P incorporation with WT set to 100%. The bar represents standard error from three biological replicates.
To gain insight into a possible role of Ser258 phosphorylation in the virulence activity of AvrPtoB1–307, we used the DC3000ΔavrPtoΔavrPtoB strain, which has deletions in both avrPto and avrPtoB genes and is less virulent on susceptible tomato than the wild type DC3000 strain and no longer avirulent on resistant tomato plants (4). We transformed into this strain a broad host range plasmid expressing either wild type AvrPtoB1–307, AvrPtoB1–307(S258A), AvrPtoB1–307(S258D), or an empty vector control. The S258D substitution is a potential “phosphomimetic” mutation, mimicking the negative charge of a phosphorylated residue. The virulence activity of each of these strains was first evaluated by vacuum infiltrating them into susceptible RG-prf3 tomato plants and assessing their ability to promote more severe disease symptoms (Fig. 5A). At 4 days after infiltration, we observed that the strain expressing wild type AvrPtoB1–307 caused severe necrosis of the lower leaves, whereas the empty vector control strain did not cause this phenotype (Fig. 5A, red arrows). Significantly, the S258A substitution abolished this disease promoting activity of AvrPtoB1–307, whereas the possible phosphomimetic S258D variant retained its ability to cause enhanced disease of the lower leaves (Fig. 5A).
FIGURE 5. Contribution of phosphorylation of Ser258 to the virulence activity of AvrPtoB1–307.
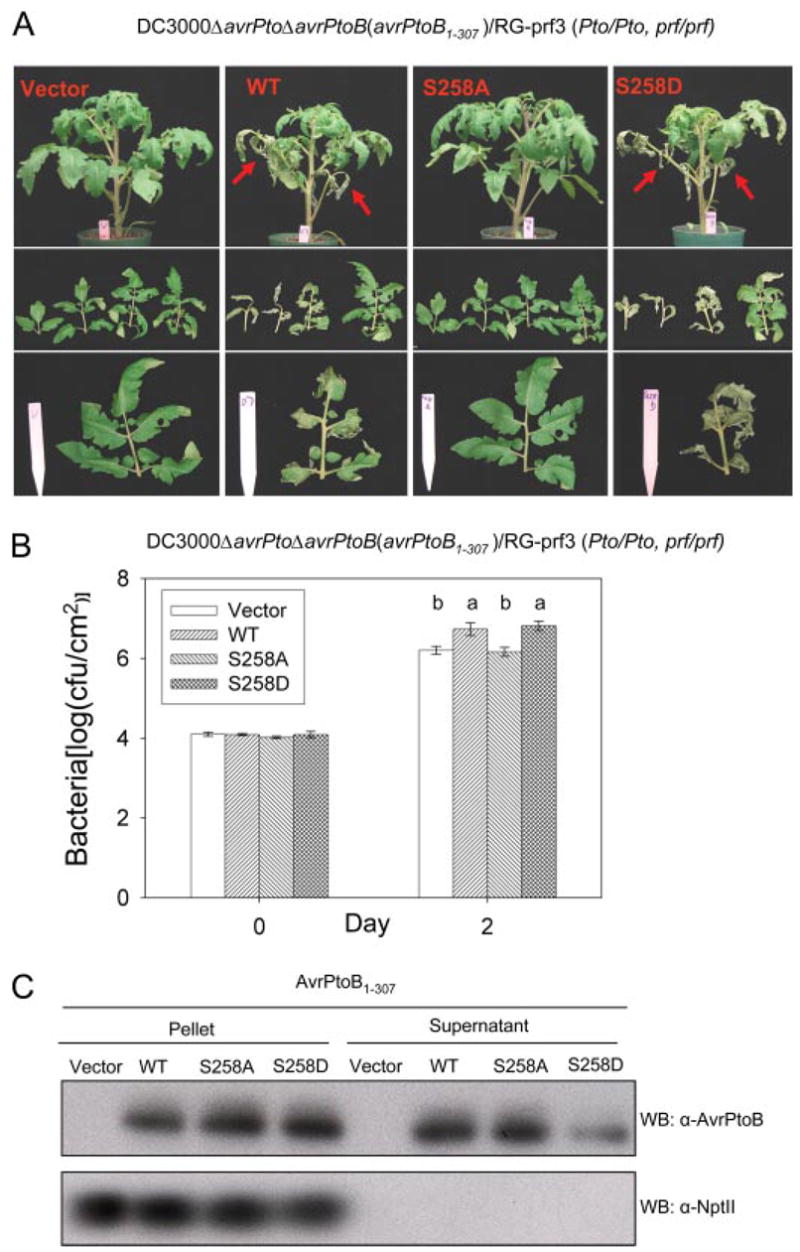
Pst DC3000ΔavrPtoΔavrPtoB strains carrying either empty vector, or plasmids expressing wild type (WT) AvrPtoB1–307, AvrPtoB1–307(S258A), or AvrPtoB1–307(S258D) mutants were vacuum-infiltrated into susceptible RG-prf3 tomato plants using an inoculum level of 104 cfu/ml. A, disease symptoms of plants 4 days after infiltration. Severe necrosis observed on lower leaves is indicated by red arrows in the top panel. The middle panel shows four diseased bottom branch leaves, and the bottom panel shows comparison of the third branch leaves among plants infiltrated with the different bacterial strains. B, Pst populations in RG-prf3 leaves shown in A at 2 days after inoculation. The data from three independent experiments were used for statistical analysis using the general linear model procedure of a statistical analysis system. The means shown with the same letters were not significantly different based on least significant difference test (p = 0.05). The bars indicate standard errors. C, expression and secretion of and derivatives from AvrPtoB1–307 DC3000ΔavrPtoΔavrPtoB grown in minimal medium under hrp induction conditions (37). Western blotting was performed using anti-AvrPtoB or anti-NptII antibody. Cytoplasmically localized NptII was used as a control for cell lysis. No NptII protein was detected in the culture medium with an anti-NptII antibody.
As another measure of virulence activity, we assessed bacterial growth of each strain in susceptible tomato leaves 2 days after infiltration (leaf necrosis at later time points interfered with growth measurements). The DC3000ΔavrPtoΔavrPtoB strains expressing wild type AvrPtoB1–307 or the AvrPtoB1–307(S258D) variant grew to a statistically significantly higher level than the strains carrying AvrPtoB1–307(S258A) or the empty vector control (Fig. 5B). To exclude the possibility that the disease and growth phenotypes we observed are due to unequal expression and/or secretion of the proteins from Pst, type III secretion assays were performed. Each of the three AvrPtoB1–307 proteins was expressed and secreted by Pst (Fig. 5C). Taken together, these assays indicate that the phosphorylation of Ser258 is required for the full virulence activity of AvrPtoB1–307.
We recently identified phenylalanine 173 as a key virulence determinant of AvrPtoB1–307 (33). To investigate the relationship between Phe173 and the phosphorylated residue Ser258, we generated F173A/S258A and F173A/S258D mutants in AvrPtoB1–307 and tested the virulence activities of these proteins (Fig. 6). If Phe173 and Ser258 act independently, they would be expected to contribute additively to the virulence of AvrPtoB1–307. Specifically, F173A/S258A and F173A/S258D may exhibit distinct degrees of virulence compared with F173A or S258A single mutants. However, we found that both F173A/S258A and F173A/S258D double mutants had virulence activity similar to that of the F173A or S258A single mutants, all of which led to less severe disease and lower bacterial populations than wild type AvrPtoB1–307 or, significantly, than the AvrPtoB1–307(S258D) protein (Fig. 6).
FIGURE 6. AvrPtoB virulence activity associated with Ser258 phosphorylation is dependent on Phe173-mediated virulence activity.
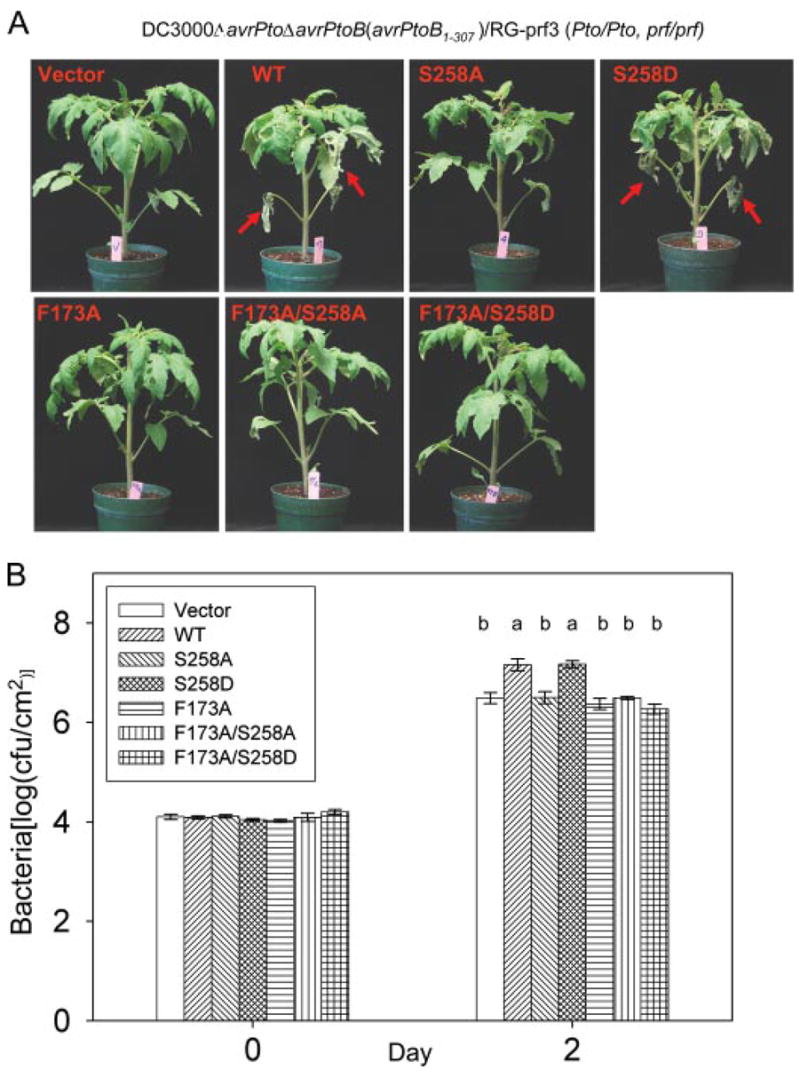
Disease assay with DC3000ΔavrPtoΔavrPtoB on susceptible RG-prf3 tomato plants was performed as described in the legend to Fig. 5. A, disease symptoms of plants 4 days after infiltration. Severe necrosis observed on lower leaves is indicated by red arrows. B, bacterial populations in RG-prf3 leaves at 2 days post-inoculation. The data from two independent experiments were used for statistical analysis using the general linear model procedure of a statistical analysis system. The means shown with the same letters were not significantly different from each other based on least significant difference test (p = 0.05). The bars indicate standard errors. WT, wild type.
Finally, we investigated whether the phosphorylation of Ser258 might affect the avirulence activity of AvrPtoB1–307. The same DC3000ΔavrPtoΔavrPtoB strains used above were vacuum-infiltrated into resistant RG-PtoR plants. Avirulence activity was evaluated 4 days after inoculation using the presence of visible specks and the level of bacterial growth. As expected, Pst strains carrying an empty vector caused speck disease on RG-PtoR tomato plants, whereas Pst expressing AvrPtoB1–307 did not exhibit any speck disease (data not shown). Interestingly, two to three specks/leaflet were observed on plants infiltrated with Pst expressing AvrPtoB1–307(S258A), whereas no specks were detected on the plants infiltrated with the Pst strain expressing AvrPtoB1–307(S258D) mutant (data not shown). Bacterial populations in leaves were measured at 4 days after infiltration. Pst strains expressing wild type AvrPtoB1–307 or AvrPtoB1–307(S258D) attained a population of ~105cfu/cm2, whereas the Pst strain carrying empty vector reached 108cfu/cm2 (Fig. 7). Notably, the strain expressing AvrPtoB1–307(S258A) reached a population about 10-fold greater than Pst strains expressing either AvrPtoB1–307 or AvrPtoB1–307(S258D) (Fig. 7). These results indicate that the S258A substitution partially compromises the avirulence activity of AvrPtoB1–307 and suggest that phosphorylation of Ser258 contributes to the full avirulence activity of AvrPtoB.
FIGURE 7. Requirement of phosphorylation of Ser258 for the full avirulence activity of AvrPtoB1–307.
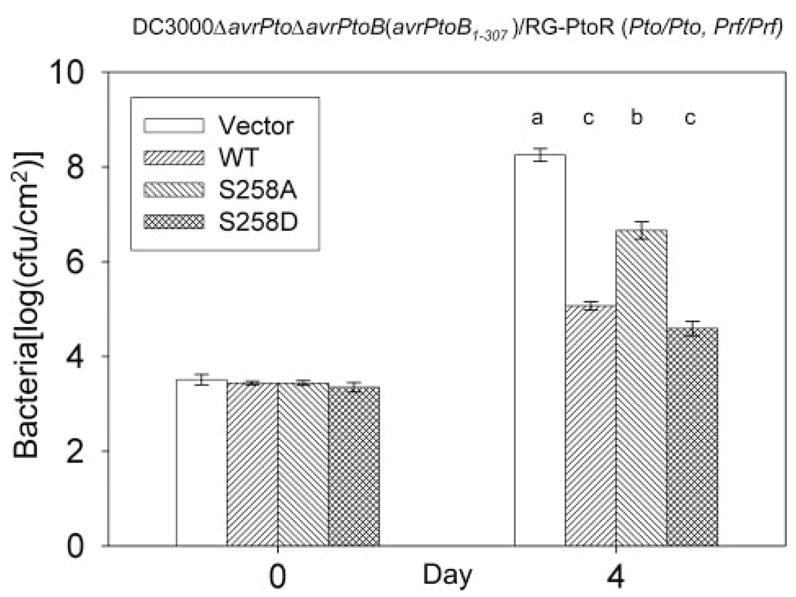
The experiments were performed as described in the legend to Fig. 5 except the resistant tomato line, RG-PtoR, was used. Bacterial populations at 4 days after inoculation of leaves. The means shown with the same letters were not significantly different based on least significant difference test (p = 0.05). The bars indicate standard errors. WT, wild type.
DISCUSSION
We have identified a phosphorylation site in the type III effector AvrPtoB that enhances the ability of the protein to promote bacterial growth and enhance disease on tomato. The identity of the kinase(s) responsible for this phosphorylation is unknown, but it acts independently of both Pto and Prf, host proteins involved in recognition of AvrPtoB and subsequent immune signaling. Two other P. syringae effectors, AvrPto and AvrB, have been reported to be phosphorylated by host proteins (20, 21). In both of those cases, the kinase identity is also unknown but was found to be independent of host proteins that are involved in effector recognition (Pto/Prf for AvrPto and RIN4/RPM1 for AvrB (20, 21)). Several type III effectors of bacterial pathogens of plants have been shown previously to be acylated after delivery into plant cells, and our present data firmly establish that phosphorylation is another common post-translational mechanism involved in activation of type III effectors.
Substitution of a negatively charged amino acid into a protein can often mimic a phosphorylated residue. This appeared to be the case for S258D as AvrPtoB1–307(S258D) exhibited full virulence and avirulence activity (in contrast to AvrPtoB1–307(S258A), which was compromised for both of these activities). We took advantage of the phosphomimetic S258D substitution to examine the relationship between a previously identified virulence determinant, Phe173, and the phosphorylated residue Ser258. Significantly, introduction of F173A into the AvrPtoB1–307(S258D) protein completely abolished the ability of the protein to enhance disease symptoms and increase Pst growth. This result suggests that Phe173 may be the “primary” virulence determinant of AvrPtoB1–307 and that phosphorylation of Ser258 facilitates the function of Phe173. This facilitation could be based on either an intramolecular interaction between the two regions carrying these residues or possibly on their interaction with a host protein. These observations differ from what we have found previously for AvrPto. AvrPto has two virulence determinants, the CD loop and a phosphorylated residue Ser149, and they appear to act additively and not synergistically (21).
In earlier work, we found that an alanine substitution at Phe173 of AvrPtoB1–307 also abolishes Pto-mediated recognition of this protein (i.e. avirulence activity of AvrPtoB1–307 is disrupted (33)). Here we found that the S258A substitution partially compromised AvrPtoB1–307 avirulence activity. This is again consistent with the possibility that the regions containing these amino acids are involved in either an intramolecular interaction or that they are both involved in an interaction with a host protein (in this case possibly Pto or Prf). Interestingly, this type of cooperativity between distinct parts of an effector for avirulence was also observed with AvrPto (21). In that case, we found that an alanine substitution of phosphorylated residue Ser149 compromised avirulence mediated by the CD loop. A fuller understanding of possible avirulence and virulence-associated interactions between Phe173 and Ser258 will probably require a crystal structure of AvrPtoB1–307. Although a three-dimensional structure for AvrPto exists (38), it unfortunately does not encompass Ser149 and therefore has shed no light on the interaction between that residue and the CD loop.
It remains unknown whether the same kinase activity is involved in the phosphorylation of both AvrPto and AvrPtoB. For both proteins, the phosphorylation involves a Pto- and Prf-independent kinase activity that is conserved in diverse plant species (note that even protein extracts from a monocot, rice, are able to specifically phosphorylate AvrPto).3 However, there does not appear to be any sequence similarity between the phosphorylation sites of AvrPto (145NPSGpSIRMS153) and AvrPtoB (254PVDRpSPPRV262). It is possible, of course, that these two regions are located in a similar three-dimensional structure. We are currently using genomics and biochemical approaches to isolate the AvrPto kinase, and if this is successful, we will be able to directly test whether that kinase can phosphorylate AvrPtoB.
Although we do not yet have an estimate of the mass of the AvrPtoB kinase, it is intriguing that the Ser258 phosphorylation site bears some similarity to a consensus MAPK site (PX(pS/T)P (39)). It was reported recently that phosphorylation of the Rhizobium type III effector NopL was partially suppressed by the MAPK kinase inhibitor PD 98059, possibly suggesting involvement of a MAPK in that phosphorylation event (24). We have directly tested one MAPK, wound-induced protein kinase, with a well characterized role in defense signaling for the ability to phosphorylate AvrPtoB1–307 in vitro, but no phosphorylation was observed.4 However, plants have a large number of MAPKs (~20 in Arabidopsis), and a more systematic effort will be required to thoroughly examine this possibility.
Supplementary Material
Acknowledgments
We thank Dr. Kathy Munkvold and Tracy Rosebrock for critical reading of the manuscript, Dr. Sixue Chen (Proteomics and Mass Spectrometry Facility, Donald Danforth Plant Science Center, St. Louis, MO) for mass spectrometry analysis, and the Boyce Thompson Institute greenhouse staff for plant care.
Footnotes
This work was supported by National Institutes of Health Grant R01GM078021 and United States Department of Agriculture-National Research Institute Grant 2005-35301-15675).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The abbreviations used are: Pst, P. syringae pv. tomato; HA, hemagglutinin, MAPK, mitogen-activated protein kinase, E3, ubiquitin-protein isopeptide ligase, GST, glutathione S-transferase, ESI, electrospray ionization, MS, mass spectrometry, MS/MS, tandem MS, RG, Rio Grande.
J. Anderson and G. B. Martin, unpublished observations.
F. Xiao and G. B. Martin, unpublished observations.
References
- 1.Abramovitch RB, Anderson JC, Martin GB. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petnicki-Ocwieja T, Schneider DJ, Tam VC, Chancey ST, Shan L, Jamir Y, Schechter LM, Janes MD, Buell CR, Tang X, Collmer A, Alfano JR. Proc Natl Acad Sci U S A. 2002;99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 4.Lin NC, Martin GB. Mol Plant-Microbe Interact. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 6.He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. Proc Natl Acad Sci U S A. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alfano JR, Collmer A. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 9.Zipfel C, Felix G. Curr Opin Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Martin GB, Bogdanove AJ, Sessa G. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg JT, Yao N. Cell Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 12.Jamir Y, Guo M, Oh HS, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR. Plant J. 2004;37:554–565. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- 13.Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. EMBO J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abramovitch RB, Martin GB. Curr Opin Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angot A, Vergunst A, Genin S, Peeters N. PLoS Pathog. 2007;3:1–13. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mudgett MB. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 18.Shan L, Thara VK, Martin GB, Zhou JM, Tang X. Plant Cell. 2000;12:2323–2338. doi: 10.1105/tpc.12.12.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 20.Desveaux D, Singer AU, Wu AJ, McNulty BC, Musselwhite L, Nimchuk Z, Sondek J, Dangl JL. PLoS Pathog. 2007;3:456–469. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB. Plant Cell. 2006;18:502–514. doi: 10.1105/tpc.105.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skorpil P, Saad MM, Boukli NM, Kobayashi H, Ares-Orpel F, Broughton WJ, Deakin WJ. Mol Microbiol. 2005;57:1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 23.Bartsev AV, Deakin WJ, Boukli NM, McAlvin CB, Stacey G, Malnoe P, Broughton WJ, Staehelin C. Plant Physiol. 2004;134:871–879. doi: 10.1104/pp.103.031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartsev AV, Boukli NM, Deakin WJ, Staehelin C, Broughton WJ. FEBS Lett. 2003;554:271–274. doi: 10.1016/s0014-5793(03)01145-1. [DOI] [PubMed] [Google Scholar]
- 25.Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 26.Pedley KF, Martin GB. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 27.Xiao F, Lu M, Li J, Zhao T, Yi SY, Thara VK, Tang X, Zhou JM. Plant Physiol. 2003;131:1239–1249. doi: 10.1104/pp.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 29.Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang JH, Rathjen JP, Bernal AJ, Staskawicz BJ, Michelmore RW. Mol Plant-Microbe Interact. 2000;13:568–571. doi: 10.1094/MPMI.2000.13.5.568. [DOI] [PubMed] [Google Scholar]
- 31.Shan L, He P, Zhou JM, Tang X. Mol Plant-Microbe Interact. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 32.Cohn JR, Martin GB. Plant J. 2005;44:139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- 33.Xiao F, He P, Abramovitch RB, Dawson JE, Nicholson LK, Sheen J, Martin GB. Plant J. 2007 doi: 10.111/j.1365–313x.2007.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YJ, Lin NC, Martin GB. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 35.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 37.Lin NC, Abramovitch RB, Kim YJ, Martin GB. Appl Environ Microbiol. 2006;72:702–712. doi: 10.1128/AEM.72.1.702-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wulf J, Pascuzzi PE, Fahmy A, Martin GB, Nicholson LK. Structure. 2004;12:1257–1268. doi: 10.1016/j.str.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez FA, Raden DL, Davis RJ. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


