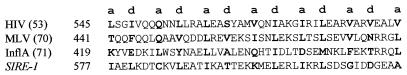Abstract
The soybean genome hosts a family of several hundred, relatively homogeneous copies of a large, copia/Ty1-like retroelement designated SIRE-1. A copy of this element has been recovered from a Glycine max genomic library. DNA sequence analysis of two SIRE-1 subclones revealed that SIRE-1 contains a long, uninterrupted, ORF between the 3′ end of the pol ORF and the 3′ long terminal repeat (LTR), a region that harbors the env gene in retroviral genomes. Conceptual translation of this second ORF produces a 70-kDa protein. Computer analyses of the amino acid sequence predicted patterns of transmembrane domains, α-helices, and coiled coils strikingly similar to those found in mammalian retroviral envelope proteins. In addition, a 65-residue, proline-rich domain is characterized by a strong amino acid compositional bias virtually identical to that of the 60-amino acid, proline-rich neutralization domain of the feline leukemia virus surface protein. The assignment of SIRE-1 to the copia/Ty1 family was confirmed by comparison of the conceptual translation of its reverse transcriptase-like domain with those of other retroelements. This finding suggests the presence of a proretrovirus in a plant genome and is the strongest evidence to date for the existence of a retrovirus-like genome closely related to copia/Ty1 retrotransposons.
Retroelements are ubiquitous components of bacterial and eukaryotic genomes that employ reverse transcriptase to sponsor their proliferation (1–3). They encompass a diverse collection of genetic elements that include DNA and RNA viruses, fungal mitochondrial plasmids, bacterial retrons, group II introns, and retrotransposons (1, 3). Infectious retroviruses and related, noninfectious retrotransposons are distinguished from other retroelements, including LINE retroposons, by their possession of long terminal repeats (LTR) (1, 3). Although retroviruses and integrated, endogenous retroviruses are primarily associated with mammalian genomes (2, 4), mammalian LTR retrotransposons have yet to be reported. LTR retrotransposons have been identified in the genomes of other vertebrates (5–8) and are routinely found in the genomes of lower animals, plants, and fungi (1, 3, 9–13). Based on sequence comparisons, some endogenous retroviruses have been shown to be closely related to known infectious retroviruses, whereas others are clearly retrovirus-like but do not correspond to any known infectious viruses (4). In addition to LTR, these retroelements are characterized by genes coding for structural core proteins (gag) and four enzymes: protease (prot), reverse transcriptase (rt), ribonuclease H (rh), and integrase (int) (1–3). Retroviral genomes encode an envelope protein that mediates both virion export from and entry into susceptible host cells (2, 14).
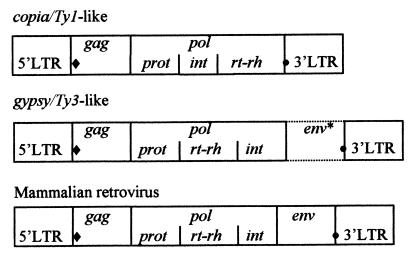
Most of the characterized LTR retrotransposons belong to either the copia/Ty1 or gypsy/Ty3 group (1). The two classes can be phylogenetically distinguished by amino acid comparisons of the catalytic proteins (1, 15, 16) and by the order of the loci in pol (Fig. 1). In all copia/Ty1 elements, int precedes rt and rh, whereas in gypsy/Ty3 group members, int resides at the 3′ end of pol. All vertebrate retroviruses and endogenous retroviruses conform to the latter configuration (1, 3), and phylogenetic analyses of the conserved regions within the reverse transcriptase suggest that retroviruses and gypsy/Ty3 retroelements are monophyletic (1).
Figure 1.
Organization of LTR retroelement genes. ♦, tRNA primer binding site; •, polypurine tract. ∗, Some gypsy/Ty3 group members possess an env-like ORF.
The coding sequences of many characterized plant retrotransposons and endogenous retroviruses are cluttered with disabling stop codons, frameshifts, and deletions, and appear to be nonfunctional (4, 12). Vestiges of ancient copia/Ty1-like sequences have been identified adjacent to several plant genes (17), and the maize genome apparently contains large clusters of nested retrotransposons (18).
Besides vertebrate retroviruses, five invertebrate gypsy/Ty3 class retroelements (19–23) and a sixth retroelement from a parasitic nematode (24) encode an envelope-like protein. Of these, gypsy from Drosophila melanogaster (25, 26), Tom from Drosophila ananassae (22), and TED from the nocturnal moth, Trichoplusia ni (27) produced the encoded protein. Horizontal, infectious-like transfer has been reported for gypsy particles (25, 26).
SIRE-1 is a relatively homogeneous family of copia/Ty1-related retroelements in the soybean genome (28, 29). Each of the several hundred copies is about 11 kb in length (ref. 28; H.M.L., unpublished data), making SIRE-1 one of the largest retroelements. The family was initially identified after a short segment was fortuitously amplified by the PCR and sequenced (28). We subsequently recovered and sequenced a 2.4-kb cDNA that encompassed the 3′ end of the 5′ LTR, a primer binding site complementary to Glycine max tRNAi-met, and an uninterrupted ORF whose conceptual translation produced a retroelement-like, gag-prot polyprotein (29). We now report the characterization of part of a genomic clone that confirms SIRE-1’s assignment to the copia/Ty1 family and contains an unprecedented ORF between the 3′ end of the pol ORF and the 3′ LTR. The full SIRE-1 sequence will be published elsewhere.
MATERIALS AND METHODS
Cloning and Sequencing.
A λFIXII soybean genomic library (Stratagene) was probed with radiolabeled copies of the SIRE-1 gag region as described (28, 29). Positive plaques were purified (30), and DNA from clones carrying the largest inserts were digested with several restriction enzymes. The DNAs were separated by agarose gel electrophoresis and a Southern blot was then probed with an end-labeled, LTR-specific oligonucleotide (30). To isolate possible full-size SIRE-1 inserts, clones in which the probe hybridized to two fragments were selected. DNA was isolated (31) from one phage candidate and digested with XbaI and HindIII. The fragments were then subcloned (30) into pSPORT1 (Life Technologies, Gaithersburg, MD) for automated DNA sequencing. Some longer subclones were unstable, most probably because of rearrangements sponsored by the long direct repeats. Two contiguous, stable subclones, one of which hybridized to the LTR probe, were sequenced on Applied Biosystems Prism 377 DNA sequencers by using pUC/M13 primers and internal primers synthesized at the Loyola University Macromolecular Analysis Facility.
Sequence Analysis and Database Searches.
Sequence alignments and ORF determinations were made by using the Genetics Computer Group package (32). Multiple amino acid sequence alignments with seven conserved rt domains suggested by Xiong and Eickbush (33) were made by using pileup (32). Trees were constructed by maximum parsimony or neighbor-joining by using paup (34). Predictions of α-helices were made by using four programs (35–38). Predictions of coiled coils were generated by using two programs (39, 40), as were predictions of transmembrane domains (41, 42).
Southern Hybridization Analysis.
Cloned and genomic DNAs from G. max cv Williams 82 were digested to completion in separate reactions with BamHI and EcoRI. The digested DNAs were run on a 0.8% agarose gel, blotted, and hybridized (30) to a rt-specific probe. After exposure and film development, the membrane was stripped, reexposed to ensure loss of signal, then reprobed with an ORF2-specific probe. The probes were generated by random primer, 32P-labeling (Amersham) of a PCR-amplified segments derived from the two coding regions.
RESULTS
Isolation and Sequence Analysis of Subclones.
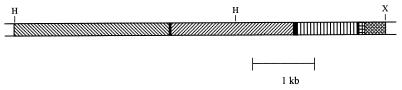
A genomic clone containing a possible full-size copy of SIRE-1 was isolated and subcloned as described above. DNAs from two contiguous subclones, the 3′ member of which hybridized to the LTR probe, were sequenced (GenBank accession nos. AF053008 and U96295).
To identify the LTR, the DNA sequence was aligned with that from the SIRE-1 cDNA clone (29) containing the last 178 bp of a 5′ LTR. The analysis fixed the location of the 3′ end of the LTR on the genomic clone, beyond which the two sequences were unrelated, indicating that the genomic sequence was a 3′ LTR. The genomic and cDNA sequences differed at only four positions (98% identity) over the 178 bp (see Fig. 2).
Figure 2.
Organization of SIRE-1 subclones. ▧, ORF1 (pol); ▨, ORF2; ▥, 3′ LTR; , cDNA overlap; ▩, flanking ORF. H, HindIII; X, XbaI.
An uninterrupted, 178-codon ORF adjacent to the 3′ end of the LTR extended to the 3′ end of the 3′ subclone (Fig. 2). This ORF was in the same orientation as the element. Database searches against this ORF by using either the DNA sequence (blastn) or the conceptual peptide sequence (blastp) did not retrieve any similar sequences. This ORF is presumably the downstream portion of an uncharacterized G. max gene split by the SIRE-1 insertion.
Translation of the remaining DNA produced two ORFs (Fig. 2). ORF1 extended 2,505 bp from the 5′ end of the 5′ subclone. blastp searches with the conceptual translation of this sequence retrieved the pol regions of several copia/Ty1-like retrotransposons. The alignments demonstrated that, in addition to the 3′ LTR, the subclones encompassed the int, rt, and rh domains of SIRE-1 (data not shown).
In all copia/Ty1-like retrotransposons, rh is at the 3′ end of pol and is closely followed by a polypurine tract and the 3′ LTR (see Fig. 1). However, the rh in SIRE-1 is followed by a long ORF in the region corresponding to retroviral envelope (env) genes (Fig. 2). ORF2 is immediately preceded by a TAA triplet and commences with a threonine codon 27 nt beyond the pol stop codon. ORF2 is therefore in the same reading frame as ORF1. Translation of ORF2 would require readthrough of the two stop codons or, alternatively, could be translated as the 3′ member of a spliced transcript (see below).
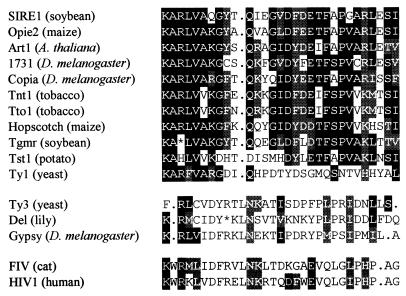
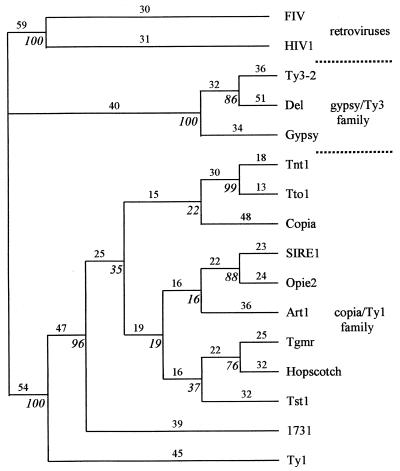
To confirm the assignment of SIRE-1 to the copia/Ty1 family, the conceptual translation of pol was aligned to seven conserved retroelement rt domains defined by Xiong and Eickbush (33). The alignments of the second domain are shown in Fig. 3. The aligned sequences were used to build phylogenetic trees by using maximum parsimony (Fig. 4) and neighbor joining (data not shown) (34). The tree building programs unambiguously placed SIRE-1 on the copia/Ty1 branch of the unrooted tree (Fig. 4).
Figure 3.
Multiple sequence alignment of the second conserved domain in rt by using pileup (32). copia/Ty1 consensus positions are highlighted. Amino acids identical to consensus are highlighted in black; amino acids similar to consensus are highlighted in gray. Opie-2 (18); Art1 (C. Herve, unpublished data, GenBank accession no. Y08010); 1731 (43); Copia (44); Tnt1 (45); Tto1 (46); Hopscotch (17); Tgmr (47); Tst1 (48); Ty1 (49); Ty3 (50); Del (51); Gypsy (21); FIV (52); HIV1 (53). A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr.
Figure 4.
Maximum parsimony tree based on the alignment of seven conserved domains in rt. Numbers above lines are the branch lengths; italicized numbers at nodes are bootstrap values (100 replicates). See Fig. 3 for references.
ORF2 is 648 codons in length. The derived theoretical protein has a molecular weight of 70 kDa. Despite its location immediately downstream of pol, the translated amino acid sequence (Fig. 5) does not exhibit significant sequence identity to any reported retroviral envelope proteins. This result is not entirely unexpected because known envelope sequences constitute a very heterogeneous collection, and only comparisons between those of closely related retroviruses (e.g., human and simian immunodeficiency viruses, but not human and feline immunodeficiency viruses) reveal recognizable, primary sequence similarities (data not shown). Alternatively, ORF2 could be a transduced cellular sequence. Bs1 from maize, a low copy-number LTR retrotransposon that lacks its own rt (54), contains segments derived from exons of a maize plasma membrane H-ATPase (55, 56).
Figure 5.
Conceptual translation of ORF2. Single underline, predicted transmembrane helix (41, 42); double underline, predicted coiled coil (39, 40); dotted underline, proline-rich region; bold, consensus of predicted α-helices (35–38); wavy underline, possible fusion peptide. ORFs were generated as described (32). See Fig. 3 for amino acid designations.
Identification of Envelope-Like Structural Elements.
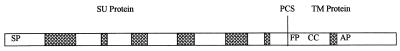
Retroviral env genes encode polypeptides that are cleaved by host proteases into two subunits—surface (SU) and transmembrane (TM) polypeptides—that are subsequently rejoined through disulfide linkages (14, 57). Although the primary sequences of these proteins may be diverse, all retroviral envelope proteins are glycosylated and share three, functionally conserved, hydrophobic transmembrane domains: a signal peptide near the amino terminal of SU (cleaved during processing), a membrane fusion peptide near the amino end of TM, and a distal anchor peptide (14, 57) (Fig. 6).
Figure 6.
Predicted and empirically deduced secondary structure features of retroviral envelope proteins (adapted from refs. 65 and 66). ▩, Predicted α-helices; SP, signal peptide; FP, fusion peptide; CC, coiled coil; AP, anchor peptide; PCS, peptide cleavage site.
Retroviral envelope glycoproteins contain between 4 and 30 N-glycosylated asparagines at Asn-Xaa-Ser/Thr motifs (57), with SU generally more heavily glycosylated than TM. The conceptual translation product of ORF2 from SIRE-1 has only two asparagines in this context. However, retroelement envelope proteins are also known to be O-glycosylated at serine and threonine residues (58, 59). O-glycosylation is correlated with clustering of hydroxy amino acids and elevated frequencies of proline (60). The amino half of the SIRE-1 theoretical env-like protein conforms to this pattern, and many of the serines and threonines are adjacent to proline. The amino acid composition of one extended, proline-rich region encompassing amino acids 60–127 is similar to the 60-amino acid proline-rich neutralization domain (61) of SU from mammalian leukemia viruses (Table 1). Proline, serine, and threonine are similarly elevated, and there is a nearly complete absence of aromatic amino acids. In SIRE-1, the spacing of many of the proline residues—(Xaa-Pro-Yaa)n or (Xaa-Pro)n—in this region, and from positions 188–197, is characteristic of many structural membrane proteins (62).
Table 1.
Comparison of 60-residue proline-rich regions from SIRE-1 and mammalian retroviruses
| Element | Amino
acid composition,
%
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | S+T | F+W+Y | I+L+V | H | N | Q | A+G | D+E | K+R | C+M | |
| SIRE-1 | 20 | 23 | 1 | 15 | 2 | 3 | 11 | 11 | 6 | 8 | 1 |
| FeLV | 18 | 22 | 0 | 20 | 3 | 5 | 8 | 13 | 3 | 5 | 2 |
| MLV | 28 | 18 | 0 | 20 | 0 | 2 | 6 | 12 | 4 | 10 | 0 |
FeLV, feline leukemia virus; MLV, murine leukemia virus. See Fig. 3 for amino acid abbreviations.
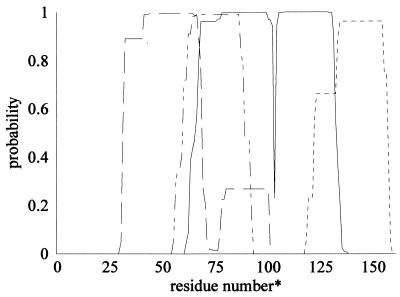
The putative env protein sequence was next evaluated for the presence of hydrophobic, membrane-spanning helices (41, 42). Both programs selected the same 13–20 amino acid region centered at residue 30 with high (70–82%) reliability (Fig. 5). The location of the predicted N-terminal, transmembrane helix is consistent with that expected for a signal peptide and is flanked by basic residues, a characteristic feature of most membrane-spanning peptides. Both programs (41, 42) recorded a second transmembrane helix centered at residue 519, but the reliabilities were considerably weaker and of questionable significance. There is, however, a hydrophobic region from residues 510–523 that could correspond to a fusion peptide (Fig. 6, see below).
Only two retroviral env peptides have been structurally characterized by x-ray crystallography (63, 64), but several env SU and TM sequences have been analyzed by structural prediction algorithms (57, 65, 66). Despite the considerable size and sequence diversity among retroviral envelope proteins, these analyses predict multiple α-helical regions similarly distributed throughout the sequence (Fig. 6). The SIRE-1 envelope-like sequence was evaluated by using several programs whose individual reliabilities ranged from 63% to 70% for predicting short helices in nonhomologous proteins (36–38), to as high as 89% for helices of length greater than eight residues (38). The accuracy of copredicted sites was significantly higher (35). The dispersal of the consensus α-helices predicted in SIRE-1 resembled that of retroviral proteins (Fig. 5). In addition, the sequence was evaluated for the possible presence of coiled coils (39, 40). Amino acids 580–611 were predicted to form a coiled coil with probabilities approaching 1.0, as were similarly located regions of retroviral TM proteins (Fig. 7). The sequence adheres well to the heptad repeat identified on the carboxyl side of several virus fusion peptides (67–71) (Fig. 8). The predicted coiled coil in the TM domains of HIV and Moloney murine leukemia virus have recently been confirmed by x-ray crystallography (63, 64). Because coiled coils are located near the N terminal in the TM proteins of HIV and the mouse virus, the location of the hydrophobic peptide beginning at residue 511 of the SIRE-1 ORF2 (Fig. 4) is appropriate for that of a fusion peptide.
Figure 7.
Output of coils program search for coiled coils (39) in SIRE-1 and selected retroviral transmembrane proteins. Similar results were obtained with ref. 40. Results shown are for a window of 21 amino acids. ——, SIRE-1; ⋅ ⋅ ⋅, HIV; - - -, murine leukemia virus; -··-··, influenza A virus. ∗, Numbering from the peptide cleavage site, except for SIRE-1, where residue 1 corresponds to the amino end of the putative hydrophobic fusion peptide.
Figure 8.
Heptad repeats in the coiled coil region of retroviral TM proteins. MLV, murine leukemia virus; InflA: influenza virus type A. See Fig. 3 for amino acid designations.
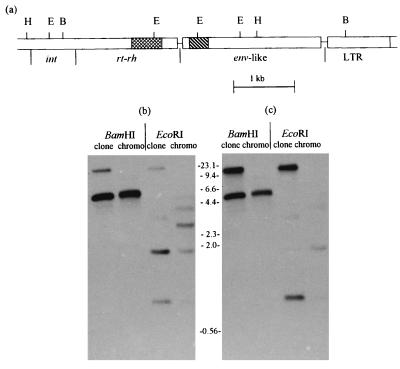
Comparison of Cloned and Chromosomal SIRE-1 Copies.
To confirm that the env-like gene was not a library or cloning artifact and that its relative location was representative of most, if not all, chromosomal copies of SIRE-1, genomic DNA was digested with restriction enzymes, and a Southern blot was sequentially probed with sequences from rt and ORF2. Fig. 9a shows the positions of the restriction sites relative to the SIRE-1 coding regions and the probes. As shown in Fig. 9 b and c, the rt and env-like probes annealed to the same 4.6-kb BamHI fragment in both the cloned and chromosomal DNAs, confirming that rt and the putative env are identically juxtaposed in the soybean genome and the clone. The EcoRI pattern is more complex. The rt probe (Fig. 9b), which spans the second EcoRI site (see Fig. 9a), hybridized with the expected fragments at 1.7 and 0.83 kb in both DNAs. However, there are additional bands in the genomic lane, suggesting that the EcoRI sites are polymorphic. The weak upper bands in the clone lanes of Fig. 9b are caused by the presence of low levels of vector DNA in the probe.
Figure 9.
Structural congruence of pol and env-like regions of SIRE-1 from a λ clone and chromosomal copies. (a) Restriction map showing HindIII (H), EcoRI (E), and BamHI (B) sites relative to rt- (▩) and env- (▧) specific probes. BamHI and EcoRI digests of clone and chromosomal (chromo) DNAs sequentially hybridized with rt (b) and ORF2 (c) probes
The env-like probe (Fig. 9c) hybridized with two unresolved bands in both cloned and genomic DNAs: the same 0.83-kb EcoRI fragment that hybridized with the rt probe and a 0.85-kb fragment representing the distal EcoRI fragment that overlaps the ORF2 probe. The env-like probe also hybridized weakly with some of the same putative polymorphic EcoRI bands observed with the rt probe. Unlike the upper bands in the clone lanes visualized with the rt probe, the env-like labeled bands in the same lanes are much more intense and are caused by a second copy of this sequence in the original λ clone. The λSIRE-1 clone actually contains one complete copy of SIRE-1 and part of a second copy (H.M.L., unpublished data). The presence of the truncated copy in the upper hybridizing bands was confirmed by a Southern blot of a BanII digest (data not shown). The two copies are not contiguous. We do not know whether the duplication is a cloning artifact or reflects a clustering of elements in the genome, as observed in maize (18).
DISCUSSION
Our data support the inference that SIRE-1 is an endogenous retrovirus closely related to copia/Ty1 retrotransposons. All previously characterized retroviruses and endogenous retroviruses are more closely related to gypsy/Ty3-like retroelements (Fig. 1). The possibility that in addition to the gypsy/Ty3 group, some copia/Ty1 members may actually be endogenous retroviruses suggests that retroviruses have evolved at least twice. The tree in Fig. 4 shows that SIRE-1 is unequivocally anchored in the copia/Ty1 family and is most closely related to opie-2 from maize. The bootstrap values for many of the nodes of the very similar tree generated by neighbor joining (data not shown) were 20–30% higher than the corresponding nodes of the more conservative maximum parsimony tree (Fig. 4). Internal nodes with less than 50% bootstrap support that were consistent with the consensus tree are shown, but their inclusion does not weaken the assignment of SIRE-1 to the copia/Ty1 family. SIRE-1 is not closely related to two other copia/Ty1-like soybean retroelements that have been fully (47) or partially (11) characterized. The former, Tgmr (47), is included on the tree in Fig. 4.
The predicted structural features of the ORF2 conceptual translation product are similar to those found in retroviral envelope proteins. The correspondence of conserved features is not perfect, however. The SIRE-1 envelope-like sequence has fewer glycosylation sites and appears to be missing a transmembrane anchor peptide. In addition, the SIRE-1 sequence has far fewer cysteine residues, some of which sponsor disulfide bridges within and between SU and TM (57). Retroviral envelope proteins are generated from spliced transcripts (2, 57). In the case of some avian retroviruses, splicing leads to an in-frame fusion of the gag start codon with the 5′ end of env (57), obviating the need for an initiation codon in env. An analogous splice in a SIRE-1 transcript would serve the same purpose, although no splice donor or acceptor consensus sequences were found in the expected regions. Cleavage of mammalian retroviral envelope precursors into SU and TM generally occurs at a conserved site near the amino terminal of the fusion peptide at the consensus (Arg/Lys)-Xaa-(Arg/Lys)-Arg (57). This sequence does not appear in the putative SIRE-1 envelope protein, and the only appropriately located tetrapeptide with at least two basic amino acids is at position 487. Complete adherence to the full catalog of generally conserved, retroviral envelope features, however, should not be expected because it is unlikely that the SIRE-1 and retroviral env genes are related by descent. Phylogenetic analyses suggest that the copia/Ty1 and gypsy/Ty3 groups diverged from each other prior to the emergence of enveloped retroviruses from the gypsy/Ty3 line of descent (1, 15, 33).
In addition to demonstrating the congruence of the cloned and chromosomal copies of SIRE-1, the comparable intensities of the hybridization signals between the clone and chromosomal lanes in Fig. 9 b and c, attest to the high copy number of “env”-containing SIRE-1 members. Although the possibility cannot be ruled out, we do not believe this env-like ORF is a transduced host gene. The presence in retrotransposons of apparently transduced host genes has been found only rarely (12, 55, 56). The maize Bs1 element appears to have sacrificed its rt gene to gain a cellular sequence and is apparently not capable of autonomous retrotransposition (54). The presence of the env-like ORF in most if not all of the several hundred copies of SIRE-1 suggests that this gene is an integral part of a retroelement genome that was, or is, functional, at least as a retrotransposon. Preliminary sequence analysis of regions upstream of int (H.M.L. and E. Gaucher, unpublished data) coupled with the previously characterized cDNA clone (29) indicate that ORF1 also encompasses gag and prot regions of appropriate length.
Neither retroviral genomes nor virions have been reported in plants, although both classes of retrotransposons are widespread. Plant caulimoviruses encode reverse transcriptase, but have DNA genomes and do not integrate into host chromosomes (72). Very few plant virus genomes encode an env gene. Those that do—rhabdoviruses and bunyaviruses (72)—also infect animal hosts, where envelope proteins sponsor viral-host cell membrane fusion. Intact plant cell walls may hinder this mode of virus transfer, and whether viral envelope proteins serve the same function in plant hosts as they do in animals is not known. This finding suggests that SIRE-1 may have originally been an infectious invertebrate retrovirus that was transferred to soybean by the invertebrate vector. In plants, intercellular virus spread is mediated by movement proteins (73), but there is no evidence for the existence of this property in any of the theoretical SIRE-1 gene products.
Most higher plant retrotransposons with copy numbers comparable to SIRE-1 are very heterogeneous and are composed of multiple subfamilies (10, 74–77) analogous to retroviral quasispecies (78). The absence of additional hybridizing bands in the chromosomal lanes in Fig. 9 a and b is therefore unusual. Genomic digests of soybean DNA generated by a dozen different restriction enzymes have now been probed with cloned copies of the gag-like, rt-like, and env-like regions of SIRE-1. Subfamilies were not detected in any of these digests, although a few low copy-number derivatives may be present (Fig. 9; also ref. 28 and H.M.L., unpublished data). This general, restriction-site homogeneity, the presence of long, uninterrupted ORFs within and adjacent to SIRE-1, and the near identities of the comparable 178 bp of the two LTRs suggest that the introduction and amplification of SIRE-1 in G. max and its wild progenitor, Glycine soja, is a relatively recent event. Functional copies of SIRE-1 may persist. Transcripts containing gag-like, rt-like, and env-like sequences have been detected by Northern blot hybridization and RT-PCR, and it appears that the 5′ end of some, if not all of these are located within the LTR (E. Lin and H.M.L., unpublished data).
The occurrence of genetically related retrotransposons among phylogenetically unrelated hosts has led to the assumption that these noninfectious elements can be transferred horizontally by some unknown mechanism (1). The observation that members of both major LTR retrotransposon families have env-like genes provides a foundation for the counter-intuitive proposal that the apparent horizontal transfer of LTR retrotransposons may be the result of transmission of closely related retroviral derivatives that subsequently lost their env gene. Although many dozens of presumed copia/Ty1-related retrotransposons have been detected by PCR amplification of conserved rt domains (5, 10, 11, 74–77), the number of fully sequenced elements is relatively small (1, 3, 12). It is conceivable that additional env regions may be encountered as more of these elements are fully sequenced.
Acknowledgments
We thank Z. Burki and C. Brown for their help with automated DNA sequencing, W. Ballard and J. Norman for their assistance with tree building, and S. Wessler for advice and comments on the manuscript. The assistance and contributions of Y.-A. Bi, M. Hughes, M. Grassi, A. Sverdlik, and S. Wakim are gratefully acknowledged. We are also grateful to the scientists and organizations that made their analytical resources available on the World Wide Web.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: LTR, long terminal repeat; rt, reverse transcriptase; int, integrase; rh, ribonuclease H; env, envelope; SU, surface protein; TM, transmembrane protein.
References
- 1.Eickbush T H. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 121–157. [Google Scholar]
- 2.Varmus H, Brown P. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 53–108. [Google Scholar]
- 3.Flavell A J. Comp Biochem Physiol B. 1995;110:3–15. doi: 10.1016/0305-0491(94)00122-b. [DOI] [PubMed] [Google Scholar]
- 4.Urnovitz H B, Murphy W H. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flavell A J, Jackson V, Iqbal M P, Riach I, Waddell S. Mol Gen Genet. 1995;246:65–71. doi: 10.1007/BF00290134. [DOI] [PubMed] [Google Scholar]
- 6.Flavell A J, Smith D B. Mol Gen Genet. 1992;233:322–326. doi: 10.1007/BF00587596. [DOI] [PubMed] [Google Scholar]
- 7.Tristem M, Kabat P, Herniou E, Karpas A, Hill F. Mol Gen Genet. 1995;249:229–236. doi: 10.1007/BF00290370. [DOI] [PubMed] [Google Scholar]
- 8.Greene J M, Otain H, Good P J, Dawid I B. Nucleic Acids Res. 1993;21:2375–2381. doi: 10.1093/nar/21.10.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britten R J, McCormack T J, Mears T L, Davidson E H. J Mol Evol. 1995;40:13–24. doi: 10.1007/BF00166592. [DOI] [PubMed] [Google Scholar]
- 10.Flavell A J, Smith D B, Kumar A. Mol Gen Genet. 1992;231:233–242. doi: 10.1007/BF00279796. [DOI] [PubMed] [Google Scholar]
- 11.Voytas D F, Cummings M P, Konieczny A, Ausubel F M, Rodermel S R. Proc Natl Acad Sci USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennetzen J L. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 13.Britten R J. Proc Natl Acad Sci USA. 1995;92:599–601. doi: 10.1073/pnas.92.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 15.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 16.McClure M A. Mol Biol Evol. 1991;8:835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- 17.White SE, Habera L F, Wessler S R. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 19.Saigo K, Kugiyama W, Matsuo Y, Inouye S, Yoshioka K, Yuki S. Nature (London) 1984;312:659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- 20.Inouye S, Yuki S, Saigo K. Eur J Biochem. 1986;154:417–425. doi: 10.1111/j.1432-1033.1986.tb09414.x. [DOI] [PubMed] [Google Scholar]
- 21.Marlor R L, Parkhurst S M, Corces V G. Mol Cell Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanda S, Mullor J L, Corces V G. Mol Cell Biol. 1994;14:5392–5401. doi: 10.1128/mcb.14.8.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friesen P D, Nissen M S. Mol Cell Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felder H, Herzceg A, deChastonay Y, Aeby P, Tobler H, Muller F. Gene. 1994;149:219–225. doi: 10.1016/0378-1119(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 25.Song S U, Gerasimova T, Kurkulos M, Boeke J D, Corces V G. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 26.Kim A, Terzian C, Santamaria P, Pelisson A, Prud’homme N, Bucheton A. Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozers M S, Friesen P D. Virology. 1996;226:252–259. doi: 10.1006/viro.1996.0653. [DOI] [PubMed] [Google Scholar]
- 28.Laten H M, Morris R O. Gene. 1993;133:153–159. doi: 10.1016/0378-1119(93)90089-l. [DOI] [PubMed] [Google Scholar]
- 29.Bi Y-A, Laten H M. Plant Mol Biol. 1996;30:1315–1319. doi: 10.1007/BF00019562. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Burmeister M, Lehrach H. Trends Genet. 1996;12:389. doi: 10.1016/s0168-9525(96)90092-x. [DOI] [PubMed] [Google Scholar]
- 32.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford D L. paup. Sunderland, MA: Sinauer Associates; 1997. [Google Scholar]
- 35.Geourjon C, Deleage G. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 36.Gibrat J F, Garnier J, Robson B. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 37.Levin J M, Robson B, Garnier J. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 38.Solamov A A, Solovyev V V. J Mol Biol. 1995;247:11–15. doi: 10.1006/jmbi.1994.0116. [DOI] [PubMed] [Google Scholar]
- 39.Lupas A, Dyke M, Van Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 40.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;374:166. doi: 10.1515/bchm3.1993.374.7-12.507. [DOI] [PubMed] [Google Scholar]
- 42.Rost B, Casadia R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fourcade-Peronnet F, d’Auriol L, Becker J, Galibert F, Best-Belpomme M. Nucleic Acids Res. 1988;16:6113–6125. doi: 10.1093/nar/16.13.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mount S M, Rubin G M. Mol Cell Biol. 1985;5:1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grandbastien M-A, Spielmann A, Cabouche M. Nature (London) 1989;347:376–380. doi: 10.1038/337376a0. [DOI] [PubMed] [Google Scholar]
- 46.Hirochika H, Otsuki H, Yoshikawa M, Otsuki Y, Sugimoto K. Plant Cell. 1996;8:725–734. doi: 10.1105/tpc.8.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharyya M K, Gonzales R A, Kraft M, Buzzell R I. Plant Mol Biol. 1997;34:255–264. doi: 10.1023/a:1005851623493. [DOI] [PubMed] [Google Scholar]
- 48.Camirand A, St-Pierre B, Marineau C, Brisson N. Mol Gen Genet. 1990;224:33–39. doi: 10.1007/BF00259448. [DOI] [PubMed] [Google Scholar]
- 49.Clare J, Farabaugh P. Proc Natl Acad Sci USA. 1985;82:2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen L J, Chalker D L, Sandmeyer S B. Mol Cell Biol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth D R, Kalitsis P, Joseph J L, Sentry J W. Proc Natl Acad Sci USA. 1989;86:5015–5019. doi: 10.1073/pnas.86.13.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olmsted R A, Hirsch V M, Purcell R H, Johnson P R. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratner L, Haseltine W, Patearca R, Livak K J, Starcich B R, et al. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 54.Jin Y-K, Bennetzen J L. Proc Natl Acad Sci USA. 1989;86:6235–6239. doi: 10.1073/pnas.86.16.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bureau T E, White SE, Wessler S R. Cell. 1994;77:479–480. doi: 10.1016/0092-8674(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 56.Palmgren M G. Plant Mol Biol. 1994;25:137–140. doi: 10.1007/BF00023232. [DOI] [PubMed] [Google Scholar]
- 57.Hunter E, Swanstrom R. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 58.Pinter A, Honnen W J. J Virol. 1988;62:1016–1021. doi: 10.1128/jvi.62.3.1016-1021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernstein H B, Tucker S P, Kar S R, McPherson S A, McPherson D T, Dubay J W, Lebowitz J, Compans R W, Hunter E. J Virol. 1995;69:2745–2750. doi: 10.1128/jvi.69.5.2745-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson I B H, Gavel Y, von Heijne G. Biochem J. 1991;275:529–534. doi: 10.1042/bj2750529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fontenot J D, Tjandra N, Ho C, Andrews P C, Montelaro R C. J Biomol Struct Dyn. 1994;11:821–836. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 62.Williamson M P. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 64.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 65.Gallaher W R, Ball J M, Garry R F, Martin-Amedee A M, Montelaro R C. AIDS Res Hum Retroviruses. 1995;11:191–202. doi: 10.1089/aid.1995.11.191. [DOI] [PubMed] [Google Scholar]
- 66.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 67.Chambers P, Pringle C R, Easton A J. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 68.Rabenstein M, Shin Y-K. Biochemistry. 1995;34:13390–13397. doi: 10.1021/bi00041a016. [DOI] [PubMed] [Google Scholar]
- 69.Hughson F M. Curr Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- 70.Shinnick T M, Lerner R A, Sutcliffe J G. Nature (London) 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 71.Gething M J, Bye J, Skehel J, Waterfield M. Nature (London) 1980;287:301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- 72.Matthews R E F. Plant Virology. New York: Academic; 1991. [Google Scholar]
- 73.Mushegian A R, Koonin E V. Arch Virol. 1993;133:239–257. doi: 10.1007/BF01313766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flavell A J, Dunbar E, Anderson R, Pearce S R, Hartley R, Kumar A. Nucleic Acids Res. 1992;14:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.VanderWiel P L, Voytas D F, Wendel J F. J Mol Evol. 1993;36:429–447. doi: 10.1007/BF02406720. [DOI] [PubMed] [Google Scholar]
- 76.Pearce S R, Harrison G, Li D, Heslop-Harrison J S, Kumar A, Flavell A J. Mol Gen Genet. 1996;250:305–315. doi: 10.1007/BF02174388. [DOI] [PubMed] [Google Scholar]
- 77.Wang S, Zhang Q, Maughan P J, Saghai Maroof M A. Plant Mol Biol. 1997;33:1051–1058. doi: 10.1023/a:1005715118851. [DOI] [PubMed] [Google Scholar]
- 78.Holland J J, de la Torre J C, Steinhauer D A. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]