Summary
Plant innate immunity relies on the recognition of pathogen effector molecules by nucleotide-binding-leucine-rich repeat (NB-LRR) immune receptor families. Previously we have shown the N immune receptor, a member of TIR-NB-LRR family, indirectly recognizes the 50-kDa helicase (p50) domain of Tobacco mosaic virus (TMV) through its TIR domain. We have identified an N receptor-interacting protein, NRIP1, that directly interacts with both N's TIR domain and p50. NRIP1 is a functional rhodanese sulfurtransferase and is required for N to provide complete resistance to TMV. Interestingly, NRIP1 that normally localizes to the chloroplasts is recruited to the cytoplasm and nucleus by the p50 effector. As a consequence, NRIP1 interacts with N only in the presence of the p50 effector. Our findings show that a chloroplastic protein is intimately involved in pathogen recognition. We propose that N's activation requires a pre-recognition complex containing the p50 effector and NRIP1.
Introduction
Plants have evolved a refined, two-branched system of innate immunity to prevent the ingress of would be phytopathogens. The first line of defense employs receptors that detect non-specific microbial-associated molecular patterns (MAMPs) such as flagellin (Ausubel, 2005). In response, pathogens have evolved effector molecules to evade MAMP-triggered immunity (MTI). If a pathogen evades this line of defense, it must overcome a second line of defense to become pathogenic. This defense system, recently termed “effector-triggered immunity” (ETI) (Jones and Dangl, 2006), employs specific plant-encoded immune receptors called resistance (R) proteins to recognize specific pathogen-encoded effectors. Although ETI relies solely on germ-line encoded molecules, it remarkably provides disease resistance that rivals both the specificity and the range of mammalian adaptive immunity.
Plant immune receptors contain domains that are also found in pattern recognition receptors (PRRs) required for mammalian innate immunity. The vast majority of plant immune receptors have a nucleotide binding (NB) and a leucine-rich repeat (LRR) domain (NB-LRR), which are also present in the animal CATERPILLER/NOD/NLR superfamily of intracellular PRRs (Ausubel, 2005; Soosaar et al., 2005). Plant NB-LRR proteins are subdivided into two sub-classes by their amino-terminal domain: CC-NB-LRRs have a coiled-coiled (CC) domain and TIR-NB-LRRs have a Toll-interleukin-1 (TIR) homology domain. The TIR domain is also found in important animal innate immunity proteins, such as Toll in Drosophila and Toll-like receptors (TLRs) in mammals (Ausubel, 2005; Soosaar et al., 2005). Despite their structural similarities with animal innate immunity molecules, plant immune receptors are functionally more similar to mammalian adaptive immunity in that they recognize specific pathogen effectors rather than non-specific PAMPs.
Historically, it was posited that one immune receptor recognizes one pathogen effector by a direct interaction. Indeed, the immune receptors Pi-ta, RRS1, N, and L alleles were shown to directly interact with their corresponding pathogen effectors (Deslandes et al., 2003; Dodds et al., 2006; Jia et al., 2000; Ueda et al., 2006). However, many attempts to observe such direct interactions between other receptor-effector pairs have been unsuccessful. Considering the limited repository of plant immune receptors compared to the vast number of pathogens, it was proposed that immune receptors may also recognize effectors indirectly by monitoring key host factors (Jones and Dangl, 2006). This model of recognition, eloquently termed the ‘guard hypothesis,’ proposes immune receptors ‘guard’ key host factors required for pathogen virulence (Van der Biezen and Jones, 1998). Pathogen effectors interact with or modify these host factors and immune receptors perceive the altered host factor to initiate a defense response.
An indirect recognition mechanism has been shown for multiple R proteins and their cognate pathogen effectors. RIN4 is the classic example of a host target that is guarded by CC-NB-LRRs and is modified by pathogen effectors (Mackey et al., 2002). The immune receptors RPM1 and RPS2 recognize modifications to RIN4 induced by three different pathogen effectors (Axtell and Staskawicz, 2003; Mackey et al., 2003). Similarly, RPS5 recognizes the cleavage of the host factor, PBS1, by the pathogen effector, AvrPphB (Ade et al., 2007; Shao et al., 2003). The Cf-2 immune receptor recognizes its pathogen effector by monitoring a host cysteine protease (Rooney et al., 2005). Finally, Pto, which was originally identified as an immune receptor, may actually be a host factor guarded by the NB-LRR protein, Prf (Mucyn et al., 2006).
Interestingly, indirect pathogen recognition has been described only for CC-NB-LRRs and the LRR receptor-like Cf-2 protein. TIR-NB-LRRs, however, comprise approximately 60% of the total NB-LRRs in the Arabidopsis genome, (Meyers et al., 2003), suggesting that TIR containing NB-LRRs are a significant class of immune receptors. Recently we have shown that N, an immune receptor belonging to the TIR-NB-LRR class, recognizes Tobacco mosaic virus (TMV) by an association of N's TIR domain with the 50-kDa helicase domain of TMV's replicase (p50) (Burch-Smith et al., 2007). N immune receptor is localized to the cytoplasm and the nucleus. While it was shown that nuclear N is required for a defense response, recognition of the p50 effector by N occurs within the cytoplasm (Burch-Smith et al., 2007). Interestingly, N's TIR domain fails to directly interact with p50 in yeast two-hybrid and in vitro assays, suggesting that N and p50 associate indirectly in planta (Burch-Smith et al., 2007).
Since the N(TIR) association with p50 is indirect, other factors must play a role in the N immune receptor recognition of the p50 effector protein. To search for such factors, we conducted a yeast-two hybrid screen with N's TIR domain. We identified a host protein called N receptor-interacting protein 1 (NRIP1) that associates with both N's TIR domain and the p50 effector in yeast-two hybrid assays. We show by co-immunoprecipitation and fluorescence microscopy in intact, living tissue that NRIP1 interacts with both N and p50. NRIP1 is required for N-mediated resistance to TMV and has in vitro sulfurtransferase activity. Interestingly, NRIP1 is normally localized to chloroplasts, but is recruited to the cytoplasm and nucleus by the p50 effector. We envision that after NRIP1 changes localization, it forms a mature p50-NRIP1 complex that is recognized through N's TIR domain to activate successful defense signaling. Our findings present a novel model for the involvement of host factors in effector recognition.
Results
NRIP1 interacts with the N immune receptor's TIR domain
To search for novel proteins that mediate the association between the N immune receptor with the p50 effector, we conducted a yeast two-hybrid screen with the TIR domain of N as bait. We identified N receptor-interacting protein 1 (NRIP1), which was represented by six identical, independent clones in the screen. Next, we isolated the full-length NRIP1 coding sequence from Nicotiana benthamiana (bankit1044291) for direct yeast two-hybrid analyses with the full-length or with individual domains of N. Yeast containing the N(TIR) domain as bait and NRIP1 as prey activated LEU2 expression and grew on media lacking leucine (Figure 1A, row 2). Therefore, NRIP1 and N(TIR) interact directly in a yeast two-hybrid assay. In contrast, NRIP1 does not interact with the TIR domain of another TIR-NB-LRR resistance protein, BS4 (Figure 1A, row 1) suggesting that NRIP1 specifically interacts with N's TIR domain. NRIP1 failed to interact with the NB, LRR, and TIR-NB domains of N or the full-length N protein (Figure 1A, row 3-6).
Figure 1. NRIP1 interacts with the TIR domain of N and is required for N-mediated resistance to TMV.
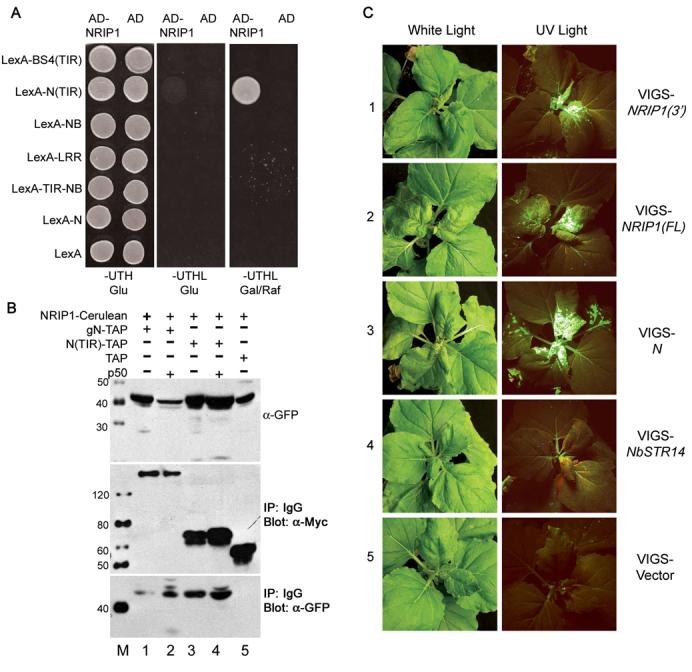
(A) NRIP1 interacts with N's TIR domain in a yeast two-hybrid assay (row 2) but not with other N domains or with full length N (row 3–6). NRIP1 does not interact with the TIR domain from the R protein, BS4 (row 1), or LexA alone (row 7). Interactions were determined by the ability to turn on the LEU2 reporter gene and subsequent growth on plates lacking leucine (Column 3).
(B) Coimmunoprecipitation of NRIP1-Cerulean with either gN-TAP or N(TIR)-TAP in the presence or absence of p50. All proteins were expressed transiently in N. benthamiana. The top panel shows the input of NRIP1-Cerulean detected with α-GFP. Middle panel shows gN-TAP, N(TIR)-TAP or TAP control immunoprecipitated with IgG beads and detected with α-Myc to the 9xMyc in the TAP tag. The bottom panel shows the NRIP1-Cerulean co-immunoprecipitated with gN-TAP, N(TIR)-TAP or TAP and detected with α-GFP. NRIP1-Cerulean co-immunoprecipitated with gN-TAP and N(TIR)-TAP in the absence (lanes 1 and 3) and presence (lanes 2 and 4) of p50. The TAP alone control was not pulled down (lane 5).
(C) N-containing N. benthamiana plants were infiltrated with VIGS vector control, VIGS-STR14, VIGS-N, VIGS-NRIP1(3') or VIGS-NRIP1(FL). Silenced plants were infected with TMV-GFP nine days after infiltration of silencing constructs. TMV-GFP appears green on a background of reddish-brown chlorophyll autofluorescence under UV light. TMV-GFP overcomes N-mediated resistance and moves from the inoculated leaves to upper, uninfected leaves in NRIP1(3')- and NRIP1(FL)-silenced plants (row 1 and 2 respectively) while in STR14-silenced plants (row 4) and the vector control plants (row 5) the virus does not spread to the upper uninfected parts of the plants. TMV-GFP moved robustly in N-silenced plants (Row 3).
The lack of an interaction between full-length N and NRIP1 in yeast two-hybrid assays may be caused by the limitations of studying interactions in yeast. Therefore, we conducted in vivo coimmunoprecipitations using transient Agrobacterium infiltration to investigate the association of N and NRIP1 in plants. We fused the full genomic clone of NRIP1, including its endogenous 5' and 3' regulatory sequences and introns (bankit1044280), with the Cerulean variant of enhanced cyan fluorescent protein (ECFP) to generate NRIP1-Cerulean. For N expression, we used the previously described full genomic clone of N fused to a tandem affinity purification (TAP) tag (gN-TAP) (Burch-Smith et al., 2007). NRIP1-Cerulean and gN-TAP were coexpressed in N. benthamiana with or without the p50 effector from the U1-eliciting strain of TMV, hereafter referred to as p50. Isolated gN-TAP immunocomplexes contained NRIP1-Cerulean (Figure 1B, bottom panel, lanes 1 and 2), confirming the in vivo association of N and NRIP1. Interestingly, this association was enhanced by p50 (Figure 1B, bottom panel, lane 2). We also confirmed the association of N's TIR domain and NRIP1-Cerulean. NRIP1-Cerulean coimmunoprecipitated with N(TIR)-TAP in the absence and presence of p50, but not with TAP alone (Figure 1B, bottom panel, lane 3-5). Thus, NRIP1 associates with full-length N in vivo, even though it did not interact in a yeast two-hybrid assay.
NRIP1 is required for N-mediated resistance to TMV
To examine the biological significance of NRIP1 in N-mediated defense against TMV, we used our well established virus-induced gene silencing (VIGS) system and TMV expressing GFP (TMV-GFP) movement assay (Liu et al., 2002). Briefly, if a gene is silenced that is dispensable for N-mediated resistance, inoculation with TMV-GFP does not result in systemic infection. Silencing a gene required for N function, however, results in TMV-GFP movement throughout the plant. To determine if NRIP1 is required by N, we knocked down NRIP1 expression using either VIGS-NRIP1(3') containing the 3' region of NRIP1 for high silencing specificity or VIGS-NRIP1(FL) containing the full-length NRIP1 mRNA sequence for robust silencing efficiency. As a negative control we used VIGS-NbSTR14 that silences an unrelated, chloroplastic sulfurtransferase with high homology to Arabidopsis STR14 (Bauer et al., 2004; Yang et al., 2003).
Nine days after the introduction of VIGS vectors, plants were infected with TMV-GFP and virus movement was monitored under ultra-violet (UV) light. In the VIGS STR14-silenced plants and vector control, TMV-GFP was restricted to the inoculated leaves and was unable to spread to the upper parts of the plant (Figure 1C, row 4 and 5). However, TMV-GFP spread to the upper parts of NRIP1(3')-silenced, NRIP1(FL)-silenced, and N-silenced control plants (Figure 1C, rows 1-3 and Figure S1D). The amount of TMV-GFP movement compared to N-silenced plants was ∼47% in NRIP1(3')- and ∼77% in NRIP1(FL)-silenced plants (Figure S1C). These data suggest that silencing NRIP1 partially abrogates N's function. Alternatively, the hypomorphic phenotype may be a result of incomplete silencing of NRIP1, since NRIP1 mRNA levels were reduced by only 86% ± 9.3% in NRIP1(FL)-silenced plants and 47% ± 4.6% in NRIP1(3')-silenced plants compared to VIGS vector controls (Figure S1A). Taken together, these results indicate that NRIP1 is partially required for an effective N-mediated resistance response to TMV.
NRIP1 is localized to the chloroplasts in the absence of p50 effector
Previously we have shown that the p50-N association occurs in the cytoplasm (Burch-Smith et al., 2007); therefore, we investigated if NRIP1 is in the same location. The subcellular localization prediction program, TargetP (Emanuelsson et al., 2000), predicts that NRIP1 contains a putative chloroplast-targeting sequence (Figure S2). Furthermore, homologs of NRIP1 from Nicotiana tobaccum and Arabidopsis thaliana localize to chloroplasts (Bauer et al., 2004; Yang et al., 2003). To determine NRIP1's subcellular localization, we generated homozygous transgenic plants containing NRIP1 tagged with Cerulean under the control of NRIP1's genomic promoter. Confocal microscopy of intact, living leaf tissue from these transgenic plants revealed NRIP1-Cerulean fluorescence in large, discrete structures that colocalized with chloroplast autofluorescence (Figure 2A). More specifically, NRIP1-Cerulean was observed in stromules, suggesting that NRIP1-Cerulean is localized to the soluble, stromal fraction of chloroplasts (Figure 2A).
Figure 2. NRIP1 localization is p50-dependent.
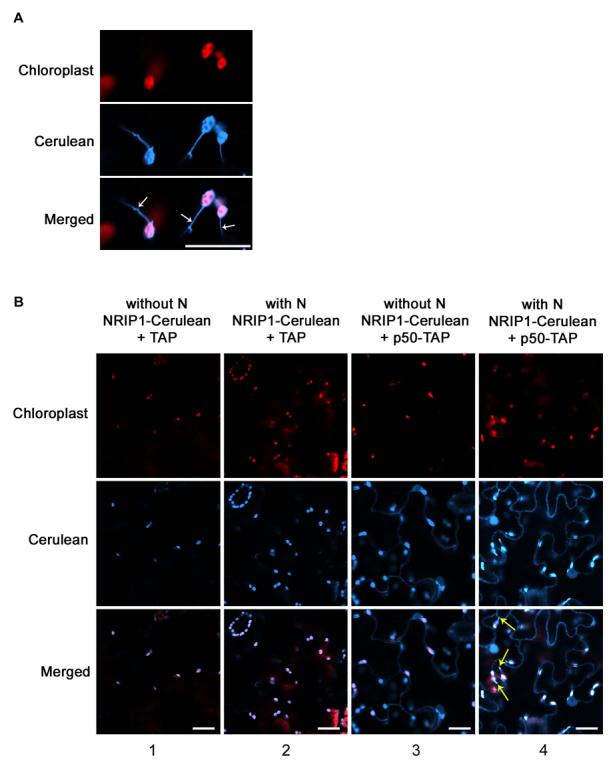
(A) In NRIP1-Cerulean transgenic plants, NRIP1-Cerulean colocalized with the red autofluorescence of chloroplasts. NRIP1-Cerulean was found in stromules (arrows). Scale bar is 20 μm.
(B) NRIP1-Cerulean localized to the chloroplasts in wild-type NRIP1-Cerulean (column 1) and N-containing NRIP1-Cerulean (Column 2) transgenic plants expressing TAP alone. NRIP1-Cerulean redistributed to chloroplasts, cytoplasm, and nucleus in the presence of p50-TAP in NRIP1-Cerulean transgenic plants without N (column 3) and with N (column 4). Red structures are chloroplasts. Yellow arrows mark stromules. Scale bars equal 20 μm.
It was unclear how chloroplast localized NRIP1 can interact with cytoplasmic and nuclear localized N. Therefore to test if N alters the subcellular localization of NRIP1, we also generated homozygous transgenic NRIP1-Cerulean plants with the N gene. Both NRIP1 and N were under the control of their native promoters. Our analyses indicated that NRIP1-Cerulean localized exclusively to the chloroplasts both with and without N (Figure S3A, column 1 and 2), suggesting that N does not affect NRIP1's subcellular localization.
The p50 effector alters the subcellular localization of NRIP1
Next, we tested whether p50 alters the localization of NRIP1 in NRIP1-Cerulean transgenic plants. Surprisingly, in non-N-containing plants transient expressing p50-TAP, NRIP1-Cerulean fluorescence was observed not only in the chloroplasts, but also in the cytoplasm and the nucleus. (Figure 2B, column 3). The altered localization was enhanced when p50-TAP was infiltrated into N-containing transgenic NRIP1-Cerulean plants (Figure 2B, column 4). Interestingly, we observed a strong induction of stromules in NRIP1-Cerulean N-containing transgenics expressing p50-TAP (Figure 2B, column 4 and Figure S3B, column 4). Expression of TAP alone had no effect on NRIP1's chloroplast localization in non-N and N-containing NRIP1 transgenic plants (Figure 2B, column 1 and 2). ). Interestingly, unlike NRIP1-Cerulean, the localization of the closest Arabidopsis homology AtSEN1 was not altered in the presence of p50, and remained solely in the chloroplasts (Figure S4). These results suggest that NRIP1 is a Solanaceae-specific component recruited by TMV's p50 effector.
Next, we tested if TMV infection will have a similar effect as the p50 effector on the localization of NRIP1-Cerulean. Localization of NRIP1-Cerulean was altered to include the nucleus and cytoplasm when TMV was expressed in non-N-containing NRIP1 transgenic plants and enhanced in N-containing NRIP1 transgenic plants (Figure S3A, column 3 and 4). Together these results indicate that NRIP1 subcellular localization is dependent on TMV's p50 effector.
NRIP1 interacts with the p50 effector
Given that p50 alters the localization of NRIP1, we tested if p50 and NRIP1 interact. We first examined this interaction in a yeast two-hybrid assay. We found that yeast carrying p50 as bait and NRIP1 as prey activated expression of the LEU2 reporter gene (Figure 3A), suggesting that p50 and NRIP1 interact directly in a yeast two-hybrid assay.
Figure 3. NRIP1 associates with p50.
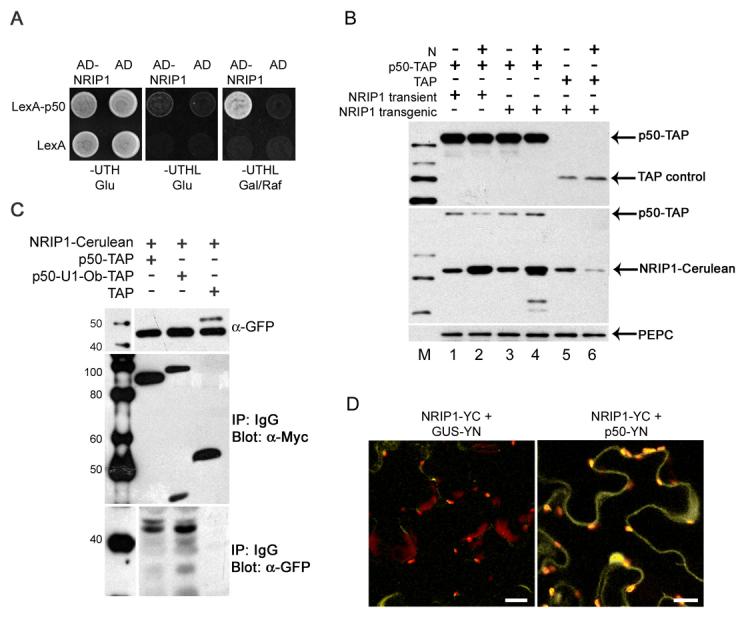
(A) NRIP1 interacts with LexA-p50 (top row) but not LexA alone (bottom row) in a yeast two-hybrid assay. The interaction was determined by the ability to turn on the LEU2 reporter gene and to grow on plates lacking leucine (column 3).
(B) TAP alone or p50-TAP was expressed in non-N-containing (lane 1 and 3) or N-containing (lane 2 and 4) NRIP1-Cerulean transgenic plants. Transient expression of NRIP1-Cerulean was coexpressed with p50-TAP in non-N-containing (lane 1) or N-containing (lane 2) N. benthamiana plants. The level of TAP-containing proteins was detected with α-Myc antibodies to the 9xMyc in the TAP tag (top panel). The level of NRIP1-Cerulean was detected with α-GFP antibodies (Middle panel). α-PEPC was used to detect largest PEPC band as a loading control (Bottom panel).
(C) Coimmunoprecipitation of NRIP1-Cerulean with p50. Top panel shows input of NRIP1-Cerulean detected with α-GFP. Middle panel shows p50-TAP, p50-U1-Ob-TAP or TAP control immunoprecipitated with IgG beads. α-Myc was used to detect the 9xMyc in the TAP tag. The bottom panel is NRIP1-Cerulean coimmunoprecipitated with p50-TAP, p50-U1-Ob-TAP, or TAP control and detected with α-GFP (TAP proteins detected by α-GFP are not shown). NRIP1-Cerulean was pulled down by both p50-TAP (lane 1) and p50-U1-Ob-TAP (lane 2) but not the TAP alone control (lane 3).
(D) BiFC assays to confirm the interaction of NRIP1 with p50. Coexpression of NRIP1-YC with p50-YN reconstituted BiFC (panel 2) but not with GUS-YN (panel 1). Scale bars equal 20 μm.
To determine if NRIP1 and p50 associate in vivo, genomic NRIP1 including its endogenous 5' and 3' regulated sequences fused to Cerulean was transiently coexpressed with p50-TAP or TAP-tag alone. We first determined if transiently expressed NRIP1-cerulean levels are comparable to that of NRIP1-cerulean levels expressed in transgenic plants. Western blot analysis reveals similar protein levels in the transient Agroinfiltration expression (Figure 3B, lane 1 and 2) compared to the transgenic NRIP1-cerulean lines (Figure 3B, lane 3 and 4). High levels of expression typically associated with transient expression by Agroinfiltration were kept in check by using a short 46h expression time, native promoters, and by omitting silencing suppressors. Therefore, transiently expressing NRIP1 is unlikely to result in overexpression.
NRIP1-Cerulean was transiently coexpressed with p50-TAP or TAP-tag alone (Figure 3C, middle panel). NRIP1-Cerulean coimmunoprecipitated with p50-TAP but did not with TAP alone (Figure 3C, bottom panel, lane 1 and 3). We also tested if NRIP1-Cerulean can associate with the non-eliciting p50-U1-Ob (Abbink et al., 2001). Interestingly, p50-U1-Ob-TAP can also associate with NRIP1-Cerulean (Figure 3C, lane 2). These results suggest that NRIP1 associates with p50 derived from both the eliciting and non-eliciting strains of TMV in vivo. Furthermore, NRIP1 is the first host protein reported to directly interact with both N and p50.
The change of NRIP1's subcellular localization in the presence of p50, prompted us to carefully look at NRIP1 interactions in intact, living tissue. To this end, we monitored the association of transiently expressed p50 and NRIP1 in living N. benthamiana leaves using Bimolecular Fluorescence Complementation (BiFC) assays (Hu et al., 2002). To conduct BiFC, we split the YFP variant, Citrine, at the 155th amino acid. NRIP1, in the context of its full genomic regulatory sequence, was tagged with the carboxy-terminal half of Citrine (YC155) to create NRIP1-YC. p50 was tagged with the amino-terminal half of Citrine (YN155) to create p50-YN. Rather than overexpressing p50 using the strong 35S viral promoter, which can result in non-specific BiFC (TBS and SPD-K, unpublished results), we placed p50 under the weaker 5' and 3' regulatory sequences of N. The widely used reporter gene, ß-glucoronidases (GUS), tagged with YN155, under the control of the 5' and 3' regulatory sequences of N (GUS-YN), was used as a control. NRIP1-YC and p50-YN were co-expressed in N. benthamiana. Reconstituted fluorescence was detected in the cytoplasm, nuclei and chloroplasts (Figure 3D, column 2). Co-expression of NRIP1-YC and GUS-YN did not reconstitute fluorescence (Figure 3D, column 1). As expected, NRIP1-YC, p50-YN, and GUS-YN did not produce fluorescence when expressed separately (Figure S5). These results confirm that p50 can interact with and alter the localization of NRIP1-Cerulean in intact tissue.
NRIP1 associates with N in intact, living tissue only in the presence of p50
The association of NRIP1 with N was strengthened by the presence of p50 in coimmunoprecipitation assays (Figure 1B, lane 2). To examine this association in the context of subcellular localization, we conducted BiFC assays by transiently expressing proteins in N. benthamiana leaves. NRIP1-YC was coexpressed with the previously characterized gN-YN, which expresses N fused to YN155 in N's full genomic context (Burch-Smith et al., 2007). Coexpression of NRIP1-YC and gN-YN with Cerulean alone did not result in fluorescence (Figure 4, column 1) suggesting that under normal conditions NRIP1 and N do not associate. This result was not surprising because NRIP1-Cerulean does not share N's subcellular localization. However, in the presence of p50-Cerulean, NRIP1-YC and gN-YN reconstituted fluorescence (Figure 4, column 3). As expected, NRIP1-YC and GUS-YN do not associate, even in the presence of p50-Cerulean (Figure 4, column 2). These results confirm that N and NRIP1 associate only in the presence of p50 in intact, living tissue.
Figure 4. NRIP1 associates with N only in the presence of p50.
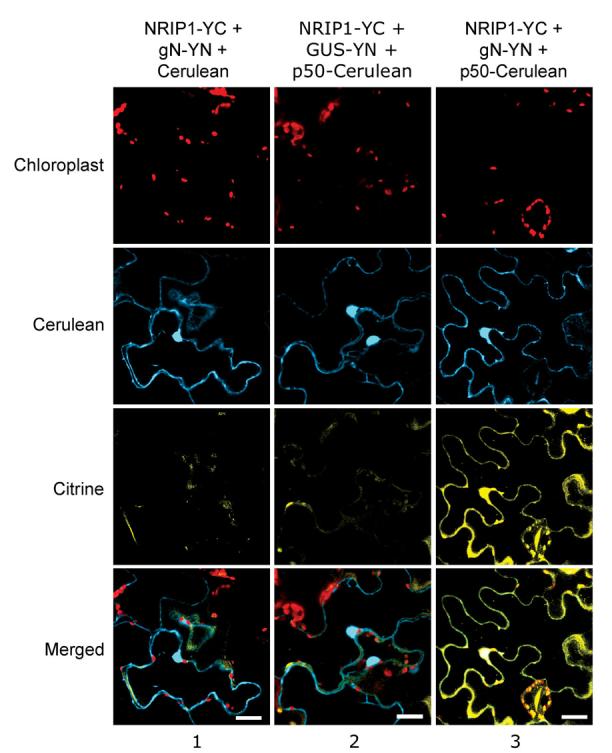
BiFC assays were used to study the association of NRIP1 and N in the context of subcellular compartmentalization. Coexpression of NRIP1-YC and N-YN did not result in Citrine BiFC in the presence of Cerulean alone (column 1). However, in the presence of p50-Cerulean, NRIP1-YC and N-YN resulted in Citrine BiFC (column 3). NRIP1-YC and the control GUS-YN did not reconstitute Citrine BiFC in the presence of p50-Cerulean (column 2). Scale bars equal 20 μm.
Ectopic expression of NRIP1 in the cytoplasm and nucleus without p50 does not induce N-mediated defense
Since p50 causes NRIP1 to partially move to the same location as N, we wanted to determine if N simply recognizes the change of NRIP1's localization to activate a defense response. To this end, we deleted the chloroplast targeting signal peptide (SP) of NRIP1 to form (-SP)NRIP1-Cerulean. Transiently expressed (-SP)NRIP1-Cerulean constitutively localizes to the cytoplasm and nucleus (Figure S6). When (-SP)NRIP1-Cerulean was co-expressed with TAP alone in N-containing plants, it did not induce hypersensitive response (HR) cell death (Figure 5A, top left). However, coexpression of (-SP)NRIP1-Cerulean with p50-TAP induces HR cell death (Figure 5A, top right). Thus, ectopically expressing NRIP1 and N in the cytoplasm and nucleus was not sufficient to trigger N-mediated defense responses. Hence, we hypothesize that N recognizes only a mature NRIP1-p50 pre-recognition complex, or alternatively, requires unknown components that are recruited by or contained within a NRIP1-p50 pre-recognition complex.
Figure 5. Constitutively localized cytoplasmic (-SP)NIP interacts with N only in the presence of p50.
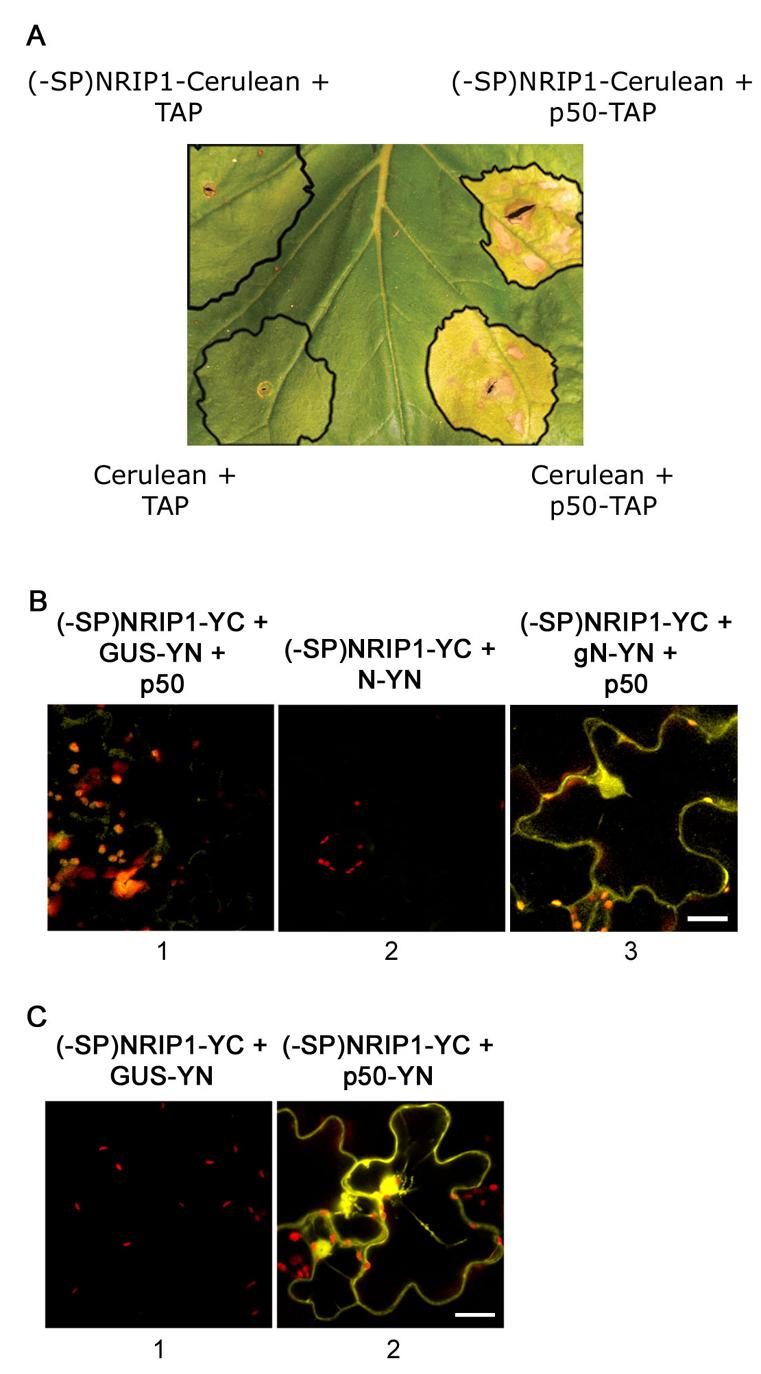
(A) (-SP)NRIP1-Cerulean coinfiltrated with p50-TAP (top right) induces HR cell death in N-containing N. benthamiana plants. (-SP)NRIP1-Cerulean coinfiltrated with TAP alone (top left) does not induce cell death. HR occurs normally when Cerulean alone is coinfiltrated with p50-TAP (bottom right). Coinfiltration of Cerulean alone and TAP alone does not induce HR (bottom left). The black outlines mark the site of infiltration.
(B) BiFC assays were implemented to study the association of (-SP)NRIP1 with N. Coexpression of (-SP)NRIP1 with N-YN in the presence of p50-Cerulean (not shown) reconstituted Citrine BiFC (panel 3). However, coexpression of (-SP)NRIP1 and GUSYN with p50-Cerulean (not shown) did not reconstitute Citrine BiFC (panel 1). Also, coexpression of (-SP)NRIP1-YC and N-YN without p50 did not result in Citrine BiFC (panel 2) Scale bars equal 20 μm.
(C) BiFC assays were implemented to study the association of (-SP)NRIP1 with p50. Coexpression of (-SP)NRIP1-YC with p50-YN reconstituted BiFC (panel 2) but not with GUS-YN (panel 1). Scale bars equal 20 μm.
To test if N recognizes a NRIP1-p50 complex, we used BiFC assays to determine if (-SP)NRIP1 and N can associate in the presence of p50. Coexpression of (-SP)NRIP1-YC and N-YN did not reconstitute fluorescence (Figure 5B, column 2). However, coexpression of (-SP)NRIP1-YC and N-YN in the presence of p50-Cerulean reconstituted fluorescence (Figure 5B, column 3), but the coexpression of (-SP)NRIP1-YC and GUS-YN in the presence of p50-Cerulean did not result in BiFC (Figure 5B, column 1). As expected, (-SP)NRIP1-YC can associate with p50-YN (Figure 5C, column 2). These findings support our hypothesis that a mature NRIP1-p50 complex (possibly containing other components) is required for NRIP1 to interact with N. Therefore, in intact tissue where subcellular localization is undisturbed, NRIP1 only associates with N when it is in a complex with p50.
NRIP1 is a functional sulfurtransferase and this activity is not required for its association with p50 and N
NRIP1 is homologous to rhodaneses that catalyze the in vitro transfer of a sulfur atom from suitable sulfur donors to nucleophilic sulfur acceptors. Thiosulfate:cyanide sulfurtransferases (TSTs) use thiosulfate as a sulfur donor and 3-mercaptopyruvate:cyanide sulfurtransferases (MSTs) use 3-mercaptopyruvate as a sulfur donor in vitro (Bordo and Bork, 2002). To characterize NRIP1's rhodanese activity, we tested whether purified GST-NRIP1 uses thiosulfate or 3-mercaptopyruvate as a sulfur donor, and transfers that sulfur atom to cyanide. NRIP1 was able to use thiosulfate as a substrate (Figure 6A, line 1) but not 3-mercaptopyruvate (Figure 6A, line 2). Furthermore, a mutant NRIP1(C145S) that lacks the catalytic cysteine in the predicted active site (Miller-Martini et al., 1994), exhibited no sulfurtransferase activity (Figure 6A, line 3) indicating that NRIP1 is a canonical TST rhodanese.
Figure 6. NRIP1's sulfurtransferase activity is not required for NRIP1 to associate with p50.
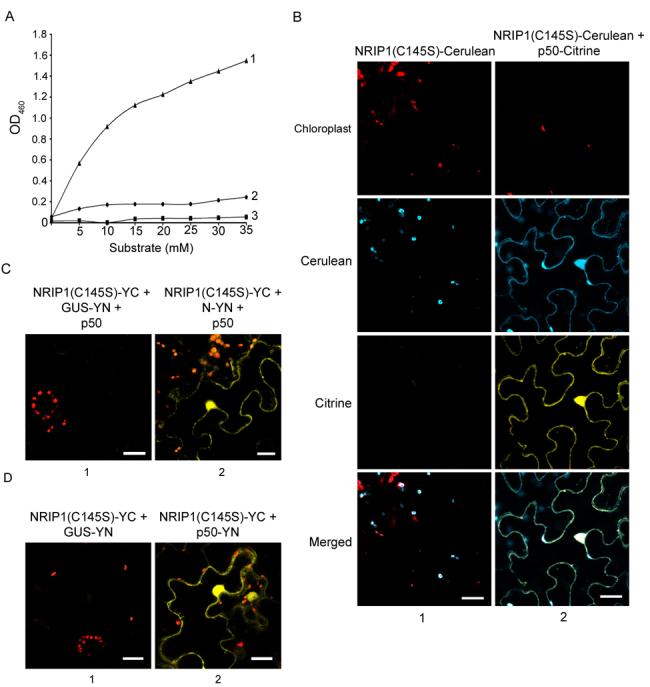
(A) NRIP1 exhibits thiosulfate sulfurtransferase activity in in vitro assays. GST-NRIP1 generates Fe(SCN)3 with sodium thiosulfate as a substrate (line 1) but not with 3-mercaptopyruvate (line 2). An inactive mutant (C145S) of NRIP1 was unable to use sodium thiosulfate as a substrate (line 3). Production of Fe(SCN)3 was quantified in a spectrophotometer at OD460.
(B) NRIP1(C145S)-Cerulean localized to the chloroplasts of N. benthamiana epidermal cells (Column 1). NRIP1(C145S)-Cerulean changed localization to the cytoplasm and nucleus in the presence of p50-Citrine (column 2). Scale bars equal 20 μm.
(C) Coexpression of NRIP1(C145S)-YC and N-YN in the presence of p50-Cerulean (not shown) produced BiFC (column 2) but not with GUS-YN (column 1). Scale bars equal 20 μm.
(D) Coexpression of NRIP1(C145S)-YC with p50-YN produced Citrine BiFC fluorescence (column 2) but not with GUS-YN (column 1). Scale bars equal 20 μm.
Next, we tested if NRIP1's sulfurtransferase activity is required for its association with p50 or N. To abolish the sulfurtransferase activity, we mutated the active site of NRIP1-Cerulean and NRIP1-YC to create NRIP1(C145S)-Cerulean and NRIP1(C145S)-YC respectively. Mutating its active site does not affect NRIP1's ability to localize to chloroplasts or to redistribute localization in the presence of p50 (Figure 6B, columns 1 and 2). To examine the association of NRIP1(C145S) with N and p50 we used BiFC assays. Coexpression of NRIP1(C145S)-YC and N-YN in the presence of p50-Cerulean (fluorescence not shown) results in BiFC (Figure 6D, column 2). The transient expression of NRIP1 constructs containing the C145S was lower (data not shown) and required the presence of the silencing suppressor, p19, for three days; presumably to allow accumulation of sufficient protein to observe the association. Furthermore, coexpression of NRIP1(C145S)-YC and GUS-YN expressed with p50-Cerulean (fluorescence not shown) did not reconstitute fluorescence, even in the presence of P19 (Figure 6D, panel 1). As expected, NRIP1(C145S)-YC did not associate with the GUS-YN control (Figure 6C, column 1); but, NRIP1(C145S)-YC and p50-YN associated, reconstituting Citrine fluorescence (Figure 6, column 2). These results suggest that NRIP1's ability to associate with N or p50 is independent of its sulfurtransferase activity.
Discussion
Recently, we showed that the TIR domain of the N immune receptor is both necessary and sufficient for the association with the p50 effector (Burch-Smith et al., 2007). Since N and p50 do not directly interact in a yeast two-hybrid or an in vitro pull down assay, other host components may be required for this association (Burch-Smith et al., 2007). Our investigation of N-interacting proteins has identified NRIP1 as a TIR domain interacting protein. NRIP1 interacts directly with N's TIR domain and TMV's p50 effector in yeast two-hybrid analyses and is the first protein identified that interacts with both a TIR-NB-LRR immune receptor and its cognate elicitor. In planta, NRIP1 is recruited by p50 to the same subcellular locations where N and p50 associate, but NRIP1 associates with N only in the presence of the p50 effector. We show that NRIP1 is a functional rhodanese sulfurtransferase, but that activity is not required for p50 or N to associate with NRIP1. Furthermore, NRIP1 is required for a complete N-mediated defense response against TMV. Taken together, our data suggest that p50 and NRIP1, possibly along with other host proteins, constitute a pre-recognition complex that is recognized by N (Figure 7).
Figure 7. Model of recognition by an N-NRIP1-p50 complex.
(A) In uninfected plants, NRIP1 localizes to the stroma of chloroplasts and N localizes to the cytoplasm and nucleus (not shown).
(B) When TMV infects the plant, NRIP1 is first retained in or recruited to the cytoplasm by TMV's p50 domain via protein-protein interactions.
(C) NRIP1 mediates the association of p50 and N's TIR domain to form an active immune complex in the cytoplasm. This complex may contain unknown (X) host components required for the activation that are only recruited by a NRIP1-p50 complex. Activated cytoplasmic N either enters the nucleus or sends a signal to nuclear-localized N to initiate a defense response.
A chloroplastic protein functions in effector-triggered immunity
The localization of R proteins, pathogen effectors, and key host factors during plant innate immunity has revealed important insights into recognition mechanisms. Chloroplastic proteins are often overlooked since no immune receptors have been predicted to be chloroplast localized. However, chloroplasts have an additional role in effector-triggered immunity during the regulation of HR-associated programmed cell death, particularly that resulting from viral infection (Seo et al., 2000). Chloroplasts synthesize salicylic acid (SA) (Wildermuth et al., 2001), an important local and systemic signaling molecule in the establishment and maintenance of defense (van Loon et al., 2006). Interestingly, carbonic anhydrase in the chloroplasts is an SA-binding protein (SABP3) that is required for an effective defense response (Slaymaker et al., 2002).
Here we show an important, novel role for a chloroplast-localized protein during plant innate immunity as part of a plant immune receptor complex. Confocal fluorescence microscopy allowed us to observe NRIP1 in its native state and subcellular location in the stroma of N. benthamiana chloroplasts. Remarkably, TMV's p50 effector drastically altered NRIP1's subcellular localization to include the cytoplasm and nucleus, in addition to the chloroplasts. An interesting question is how does p50 alter the localization of NRIP1? A low level of NRIP1 must exist in the cytoplasm since NRIP1 must travel through the cytoplasm before it is imported. Hence, one possibility is p50 intercepts NRIP1 on its way to the chloroplasts. The binding of p50 to NRIP1 may directly mask NRIP1's chloroplast targeting signal or p50 may indirectly disrupt global chloroplast import affecting the translocation of NRIP1 and possibly other chloroplastic proteins. Alternatively, chloroplast-localized NRIP1 may be released into the cytoplasm and nucleus. The export of NRIP1 may be similar to the release of pro-cell death signals, such as cytochrome c, from the mitochondria (Reviewed in (Gogvadze et al., 2006). One of the key initiation steps of apoptosis in mammalian cells is the permealization of mitochondrial membranes and subsequent release of pro-death signals. Released cytochrome c leads to activation of the apoptosome in the cytoplasm and this initiates a caspase cascade leading to cell death (Zou et al., 1999). Similarly, NRIP1 may be released from chloroplasts by a p50-induced permealization of the outer membrane by an unknown mechanism or using the machinery required for retrograde signaling (see below). Since the recognition of pathogens by plant immune receptors results in a type of programmed cell death (hypersensitive response), it is possible that NRIP1 or other chloroplastic factors act as pro-death signals that are recognized by immune receptors such as N.
The strong induction of thread-like stromules in plants undergoing a defense response (Figure 2 and Figure S3) suggests that there may be a general alteration of chloroplast structure following pathogen recognition. Stromules are associated with stress responses, such as high temperatures (Holzinger et al., 2007) and it is likely that the induction of stromules is a general response to abiotic and biotic stresses (this study). Although the exact role of stromules remains elusive, one putative function is to increase the surface area of chloroplasts to aid in the transport to and from the cytosol or organelles (Natesan et al., 2005). Stromules have been observed in close contact with nuclei in electron and confocal micrographs (Holzinger et al., 2007; Kwok and Hanson, 2004). We also observed the tips of numerous stromules in contact with nuclei (data not shown). Currently, there is no evidence suggesting stromules directly connect nuclei and chloroplasts; however, their close association may enhance the import of chloroplastic factors, such as NRIP1, to the nucleus. Stromules may have an important role during chloroplast-to-nucleus or chloroplast-to-cytoplasm retrograde signaling. Chloroplast-to-cytoplasm retrograde signaling that leads to release of NRIP1 or other chloroplastic proteins might activate the cell death process through immune receptors. Additionally, chloroplast-to-nucleus retrograde signaling might participate in the transcriptional reprogramming during pathogen defense. Furthermore, NRIP1 may have an unknown function within the nucleus. Increasing the size of NRIP1 with two copies of CFP does not disrupt NRIP1's nuclear localization (data not shown); therefore, NRIP1 does not passively diffuse into the nucleus, but may be actively imported by an unknown mechanism.
A second question related to NRIP1's localization is why does p50 alter the localization of NRIP1? The association of p50 (and p50-U1-Ob) with NRIP1 suggests NRIP1 may play a role in TMV's infection cycle. Several other host factors are known to function in basal immunity, in addition to their roles in effector-triggered immunity (Jones and Dangl, 2006). It is possible that NRIP1 also possesses an additional role in basal immunity. Interestingly, the expression of NRIP1's Arabidopsis homolog AtSEN1 is regulated in response to defense-related molecules such as SA and during basal defense (Schenk et al., 2005). NtDin1, the close tobacco homolog of NRIP1, is required for the function of molybedenum containing proteins such as nitrate reductase and xanthine dehydrogenase (Yang et al., 2003) that have been implicated in the formation of reactive oxygen species that function during defense (Apel and Hirt, 2004; Shapiro, 2005). Thus, the disruption of chloroplasts may have the desirable consequence of reducing the host's capacity for effective defense responses. Indeed, the bacterial effector HopI1 localizes to chloroplasts, disrupts their structure, and suppresses SA production, presumably to aid in the successful colonization of the host by the pathogen (Jelenska et al., 2007). In addition, silencing a component of photosystem II of chloroplasts results in a 10-fold increase in TMV accumulation (Abbink et al., 2002) and depletion of photosystem II core complex by TMV is the cause for yellow mosaic symptoms (Lehto et al., 2003). It is likely then that NRIP1 is not the only chloroplastic protein that will be affected by TMV and it is tempting to speculate that other chloroplastic proteins may function like NRIP1 during defense against other pathogens. Indeed, we found a strong putative chloroplast targeting signal within the recently discovered Tm-1 gene product that confers resistance to tomato mosaic virus (ToMV) (Ishibashi et al., 2007). Moreover, Tm-1 protein directly binds to the replicase proteins of ToMV and inhibits virus replication.
NRIP1 is a novel component of the N immune receptor complex
Mounting evidence suggests that the amino-terminal variable domain of NB-LRR immune receptors is required for indirect recognition of pathogens, and the carboxy-terminal LRR domain recognizes pathogens via direct interactions. The host factors, RIN4, PBS1, and Pto bind N-terminal domains of NB-LRR proteins (Ade et al., 2007; Mackey et al., 2002; Mucyn et al., 2006). In this study, we demonstrated that NRIP1 binds to the N-terminal TIR domain of N and the p50 effector using three independent methods. In yeast two-hybrid assays, NRIP1 associates with the TIR domain of N but not with full-length N; however, in co-immunoprecipitation assays, NRIP1 associates with both the TIR domain alone and full-length N. We were not surprised by this discrepancy between yeast two-hybrid and in vivo assays because a similar difference was observed for another NB-LRR immune receptor, RPM1, and the associated host factor, TIP49a (Holt et al., 2002). Although N and N(TIR) coimmunoprecipitate with NRIP1 in the absence of p50, in BiFC assays that maintained subcellular compartmentalization in living cells, NRIP1 did not interact with N unless p50 was present (Figure 4). Moreover, ectopically expressing NRIP1 in the cytoplasm by removing its chloroplast targeting peptide was not sufficient for NRIP1 and N to associate without p50 (Figure 5). In conclusion, the combination of yeast two-hybrid, co-immunoprecipitations, and BiFC assays has created a unique NRIP1 interaction dataset that no single technique could produce.
Our interaction dataset suggests that NRIP1 is a host factor that facilitates or supports the association of N and p50 within an immune receptor complex. Multiple lines of evidence support this hypothesis. First, N and p50 do not directly associate in vivo, and therefore, the association is likely mediated by other host factors (Burch-Smith et al., 2007). NRIP1 interacts with the TIR domain of N, which mediates the indirect association of N and p50. In coimmunoprecipitation assays, p50 complexes contain both N (Burch-Smith et al., 2007) and NRIP1 (Figure 3); and, N complexes contain both NRIP1 (Figure 1) and p50 (data not shown). Finally, NRIP1 only associates with N in the presence of p50, suggesting that a NRIP1-N complex never forms, and only a p50-NRIP1-N complex activates defense. The existence of such complexes containing plant immune receptors, host proteins, and their pathogen effectors is in agreement with the guard hypothesis (Van der Biezen and Jones, 1998), which proposes immune receptors monitor key host factors to detect pathogens and subsequently activate defense responses. However, in a significant departure from the original guard hypothesis, N does not constitutively associate with the host factor NRIP1, but instead, is part of a pre-recognition complex containing p50 and NRIP1. As a result, NRIP1 is not a traditional guardee of N and reveals a putative novel mode of pathogen detection by immune receptors.
Activation of an N immune receptor complex
In all other known cases that adhere to the postulates of the guard hypothesis, activation of the immune receptor occurs when a constitutively associated host factor is modified by the pathogen effector (Jones and Dangl, 2006 and refs therein). We had therefore speculated that the modification recognized by N immune receptor was the mislocalization of NRIP1 to the cytoplasm and nuclei. To our surprise, the colocalization of (-SP)NRIP1 and N was not sufficient for the two proteins to interact, or to activate N-mediated defense in the absence of p50 effector (Figure 5). Furthermore, both the eliciting p50 and N-evading p50-U1-Ob altered the localization of NRIP1. This implies that an additional modification besides NRIP1's change in localization must be recognized for N to activate defense.
Intriguingly, both the eliciting p50 from TMV-U1 and the N-evading chimera p50-U1-Ob, can interact with NRIP1 in vivo, but only p50 from TMV-U1 is recognized by N. Also, p50-U1-Ob can direct the change in localization of NRIP1 and allow NRIP1, (-SP)NRIP1, and NRIP1(C145S) to associate with N in a BiFC assay (data not shown); in all these functions p50-U1-Ob is indistinguishable from the eliciting p50-U1. Hence, the association of N and NRIP1 alone cannot account for N's ability to discriminate between p50 and p50-U1-Ob. How does N discriminate between p50 and p50-U1-Ob in this complex? The eliciting p50 may make an unknown modification of NRIP1 that is not induced by p50-U1-Ob.
One possibility is N recognizes a modification of NRIP1's sulfurtransferase activity. NRIP1 has in vitro thiosulfate sulfurtransferase activity and its activity is not required for its change in localization or the association with N or p50 in vivo (Figure 6). However, it is important to distinguish between associations and activation. NRIP1's sulfurtransferase activity is not required for a NRIP1-N association, but it may be required for activation of N. Unfortunately, a lack of true knockouts in N. benthamiana prevented us from doing complementation experiments with the mutant NRIP1(C145S) to unambiguously answer this question. We have not identified a possible in vivo substrate for NRIP1 during an N-mediated defense response; indeed, the identification of the biological substrates of rhodaneses over the past 50 years has been largely unsuccessful (Bordo and Bork, 2002). Given that NRIP1 changes localization in the presence of p50, NRIP1's substrate may also change upon virus infection as it moves from the chloroplasts to the cytoplasm and nucleus. NRIP1 may modify p50, N, or an unknown component only when p50 is present, and N may recognize this modification.
Alternatively, the p50-NRIP1 complex may bind in a different conformation that is recognized by N, whereas an inactive p50-U1-Ob-NRIP1 complex goes undetected. Another possibility is that after the association with N's TIR domain, the specific activation of N occurs only when p50-U1 interacts with N's LRR domain. Lastly, we must entertain the possibility that an unknown host factor is brought to this complex only by the eliciting p50. NRIP1 may simply bring p50 and N together, and an unknown factor that binds only to the eliciting p50, but not p50-U1, may be required for that association to induce the activation of N.
Model for N perception of TMV's p50 effector
We propose that N perception of p50 occurs in three distinct phases: association, specific recognition, and immune receptor activation. This investigation focuses on the first association phase, and will provide the necessary framework for future studies. In our model, NRIP1 normally localizes to the chloroplasts and N to the cytoplasm (Figure 7A) and nucleus (not shown). In the presence of p50, NRIP1 is recruited to the same subcellular compartments that contain the N immune receptor (Figure 7B). There it forms a pre-recognition complex with p50 (Figure 7B). The p50-NRIP1 complex associates with N, possibly along with other host factors (X) and cytoplasmic N is activated by an unknown mechanism (Figure 7C). Once cytoplasmic N is activated, it either enters the nucleus to become nuclear activated N or sends a signal to activate the nuclear pool of N. Nuclear N then signals a defense response. To carefully study NRIP1's role during the activation of N, a detailed analysis that is beyond the scope of this paper will be required. Ideally, the components found in NRIP1-containing and N complexes will be isolated. Preliminary results show that NRIP1 belongs to a high molecular weight complex in the presence of p50 (data not shown). Second, we need to conduct a detailed analysis of NRIP1 and N modifications in the presence and absence of p50 and p50-U1-Ob.
We have identified NRIP1 as a host protein that interacts with both N and p50. NRIP1 mediates the association of the N immune receptor and the pathogen-encoded p50 effector. Together, N, NRIP1 and p50 constitute a core recognition complex that allows the plant to detect the presence of TMV in infected cells. Our data implicates NRIP1 as a chloroplastic protein intimately involved in pathogen recognition by a plant immune receptor; and as a result, it emphasizes the growing importance of studying plant innate immunity in the context of the subcellular localization of immune receptors, pathogen effectors, and associated host proteins.
Experimental Procedures
Transient expression by Agroinfiltration
GV2260 Agrobacterium containing expression vectors were grown overnight, pelleted, resuspended in infiltration medium containing 10 mM MgCl2, 10 mM 2-Morpholinoethanesulfonic acid and 200 μM acetosyringone, and induced at room temperature for 2 hours. Strains containing N-derived constructs were infiltrated into N. benthamiana leaves at OD600=1.8 and those containing p50-, NRIP1-, or AtSEN1-derived constructs were infiltrated at OD600=1.0-1.2. For co-infiltration, equal volumes of Agrobacterium were mixed. 4-5 week-old N. benthamiana plants grown on 24 h light carts were used for all assays.
Monitoring protein expression levels and coimmunoprecipitation assays
Plant tissue expressing proteins of interest was collected and ground in liquid nitrogen. Protein was extracted with buffer containing 50 mM NaCl, 20 mM Tris/HCl pH 7.5, 1 mM EDTA, 0.1 % Triton X-100, 10 % glycerol, 3 mM DTT, 2 mM NaF, 1mM PMSF and Complete protease inhibitors (Roche). Equal amounts of protein were loaded onto polyacrylamide gels and transferred to PVDF membrane (Millipore) for Western blot analysis. Antibodies used were as follows: mouse anti-MYC (Santa Cruz), mouse anti-GFP (Covance or AbCam), rabbit anti-PEPC (Rockland) and anti-mouse or anti-rabbit horseradish peroxidase conjugate (Sigma). For coimmunoprecipitation assays, protein extracts were incubated with 100μl of IgG-Sepharose bead slurry (GE Healthcare) equilibrated with extraction buffer and tumbled for 2-3 hours at 4°C. The beads were washed three times with extraction buffer containing 150 mM NaCl. 60 μl of 2xSDS loading buffer was added to each sample and boiled for 4 minutes. 30μl of IP was used for immunoblotting as described in (Burch-Smith et al., 2007).
Fluorescence microscopy
Live plant imaging was performed on a Zeiss LSM510 META confocal microscope (Carl Zeiss) using a 40x C-Apochromat water immersion objective lens (NA 1.2). Tissue samples were cut from N. benthamiana leaves at approximately 46 hpi. The 458 nm and 514 nm laser lines of a 25mW argon laser (Coherent) with appropriate emission filters were used to image Cerulean and Citrine with chloroplasts respectively. Alternatively, a 458 nm laser line of a 25 mW argon laser and a META detector was used for chloroplast autofluorescence. Images within a panel were taken using the same confocal settings and adjusted together in Photoshop CS.
VIGS assay and GFP imaging
All Agrobacterium cultures were adjusted to an OD600=1.0 and pTRV1 was mixed 1:1 with pTRV2 control, pTRV-N, or pTRV2-NRIP1. These cultures were infiltrated onto N-containing N. benthamiana plants grown at 24-26°C under continuous light. Eight days post-infiltration, plants were rub-inoculated with TMV-GFP. GFP imaging was conducted under a 100W UV spotlight (Sylvania) using a CAMEDIA E20 digital camera (Olympus) fitted with a MCON-35 macro lens (Olpymus) and a yellow filter (HOYA, K2).
RNA isolation and RT-PCR analysis
Total RNA was extracted from NRIP1(3')-, NRIP1(FL)-, N-, STR14-, and vector control-silenced N-containing plants using RNeasy Mini Kit (Qiagen). First strand cDNA was synthesized using 2 μg of total RNA and SuperScript II reverse transcriptase (Invitrogen). Semi-quantitative RT-PCR was performed as described in (Liu et al., 2002).
Rhodanese sulfurtransferase assay
Proteins were expressed and purified from E. coli cells (see Supplementary Experimental Procedures). Thiosulfate and 3-mercaptopyruvate rhodanese sulfurtransferases activities were assayed by quantifying the formation of thiocyanate (SCN−) as the formation of the red Fe(SCN)3 complex as described in (Papenbrock and Schmidt, 2000).
Supplementary Material
Acknowledgements
We thank Montrell Seay, Patrick Cournoyer and Meenu Padmanabhan for helpful comments on the manuscript. Supported by NIH grant GM62625 to S.P.D-K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbink TE, de Vogel J, Bol JF, Linthorst HJ. Induction of a hypersensitive response by chimeric helicase sequences of tobamoviruses U1 and Ob in N-carrying tobacco. Mol Plant Microbe Interact. 2001;14:1086–1095. doi: 10.1094/MPMI.2001.14.9.1086. [DOI] [PubMed] [Google Scholar]
- Abbink TE, Peart JR, Mos TN, Baulcombe DC, Bol JF, Linthorst HJ. Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology. 2002;295:307–319. doi: 10.1006/viro.2002.1332. [DOI] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bauer M, Dietrich C, Nowak K, Sierralta WD, Papenbrock J. Intracellular localization of Arabidopsis sulfurtransferases. Plant Physiol. 2004;135:916–926. doi: 10.1104/pp.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 2002;3:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A Novel Role for the TIR Domain in Association with Pathogen-Derived Elicitors. PLoS Biol. 2007;5:501–514. doi: 10.1371/journal.pbio.0050068. e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Gogvadze V, Orrenius S, Zhivotovsky B. Multiple pathways of cytochrome c release from mitochondria in apoptosis. Biochem Biophys Acta. 2006;1757:639–647. doi: 10.1016/j.bbabio.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Holt BF, 3rd, Boyes DC, Ellerstrom M, Siefers N, Wiig A, Kauffman S, Grant MR, Dangl JL. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell. 2002;2:807–817. doi: 10.1016/s1534-5807(02)00174-0. [DOI] [PubMed] [Google Scholar]
- Holzinger A, Buchner O, Lutz C, Hanson MR. Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma. 2007;230:23–30. doi: 10.1007/s00709-006-0222-y. [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Masuda K, Naito S, Meshi T, Ishikawa M. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc Natl Acad Sci. 2007;104:13833–13838. doi: 10.1073/pnas.0703203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kwok EY, Hanson MR. Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 2004;23:188–195. doi: 10.1007/s00299-004-0824-9. [DOI] [PubMed] [Google Scholar]
- Lehto K, Tikkanen M, Hiriart JB, Paakkarinen V, Aro EM. Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Mol Plant Microbe Interact. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Martini DM, Hua S, Horowitz PM. Cysteine 254 can cooperate with active site cysteine 247 in reactivation of 5,5′-dithiobis(2-nitrobenzoic acid)-inactivated rhodanese as determined by site-directed mutagenesis. J Biol Chem. 1994;269:12414–12418. [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. Stromules: a characteristic cell-specific feature of plastid morphology. J Exp Bot. 2005;56:787–797. doi: 10.1093/jxb/eri088. [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Schmidt A. Characterization of a sulfurtransferase from Arabidopsis thaliana. Eur J Biochem. 2000;267:145–154. doi: 10.1046/j.1432-1327.2000.00980.x. [DOI] [PubMed] [Google Scholar]
- Rooney HC, Van't Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ. The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol Biochem. 2005;43:997–1005. doi: 10.1016/j.plaphy.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Iwai T, Iwano M, Fukui K, Isogai A, Nakajima N, Ohashi Y. Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. Plant Cell. 2000;12:917–932. doi: 10.1105/tpc.12.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Shapiro AD. Nitric oxide signaling in plants. Vitam Horm. 2005;72:339–398. doi: 10.1016/S0083-6729(05)72010-0. [DOI] [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci. 2002;99:11640–11645. doi: 10.1073/pnas.182427699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP. Mechanisms of plant resistance to viruses. Nat Rev Microbiol. 2005;3:789–798. doi: 10.1038/nrmicro1239. [DOI] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H. Direct Interaction between the Tobacco Mosaic Virus Helicase Domain and the ATP-bound Resistance Protein, N Factor during the Hypersensitive Response in Tobacco Plants. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yang SH, Berberich T, Miyazaki A, Sano H, Kusano T. Ntdin, a tobacco senescence-associated gene, is involved in molybdenum cofactor biosynthesis. Plant Cell Physiol. 2003;44:1037–1044. doi: 10.1093/pcp/pcg122. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



