What is a module? This must be one of the most commonly asked questions in systems biology and possibly the question with the most variety of answers. A bioinformatician with an eye on graph theory will view modules as loosely linked islands of densely connected nodes, whereas a geneticist might see modules as groups of coexpressed genes. The paper by Del Vecchio et al (2008) in this issue takes an engineer's approach to understanding modularity and develops a concept called retroactivity. This term was originally introduced by Saez-Rodriguez et al (2005) but Del Vecchio et al have expanded the idea into a more quantitative theory of modularity.
To explain retroactivity, imagine that a cell contains a network that behaves as an oscillator and the oscillator in turn is used to signal another process. Such an example might include the P35/Mdm2 oscillator that signals DNA repair (Tyson, 2006). Now consider the process by which the oscillator transmits its signal; it must and can only transmit this signal through a physical connection. Therefore, the downstream process must bind to proteins generated within the oscillator in order to ‘perceive' the signal. However, in the process of binding proteins from the oscillator, there is the potential to disrupt the functioning of the oscillator since part of the oscillator is effectively being sequestered. The effect a downstream process has on an upstream process is called retroactivity and is at the heart of defining what is and what isn't a module.
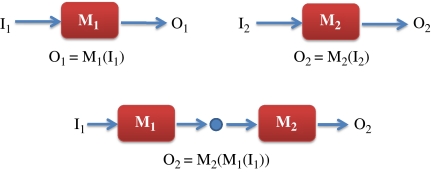
In engineering, a module is defined as a functional unit that is capable of maintaining its intrinsic properties irrespective of what it is connected to. This is an important concept because it allows engineers to connect diverse elements together while achieving predicable outcomes (Figure 1). Moreover, the use of modular components reduces costs and makes the design process much easier than it otherwise would be. This methodology is most commonly employed in electronics and anyone who has dabbled with digital electronics will be familiar with how easy it is to connect TTL chips together. Engineers achieve modularity by minimizing the effect a downstream process has on an upstream process, usually by actively altering the input and output resistances (impedances) of the module. Del Vecchio et al, in their paper, quantify this effect and call it the retroactivity R. The less the retroactivity (a smaller R), the more the confidence we have in making the assertion that the upstream and downstream systems can be classed as modules with respect to each other. Note however that if two systems A and B have a low retroactivity with respect to each other, then it is possible that an additional connection, say between A and a system C, will have a high retroactivity. Retroactivity is in general a shared property although engineers frequently devise mechanisms to make low retroactivities that are robust to downstream variability (Figure 2).
Figure 1.
Definition of a module: two biochemical systems, M1 and M2, have defined input and output characteristics. M1 and M2 can be considered modules when we are able to predict the behavior of the composite network M2(M1) from the input/output characteristics of the individual systems. Interestingly, random biochemical networks connected together do not behave this way and therefore cannot be considered modules. The ability to predict the behavior of M2(M1) requires minimal retroactivity between M1 and M2 and the networks have to be either specifically engineered or evolved through a genetic algorithm for this to be the case.
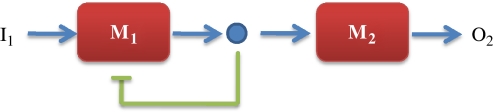
Figure 2.
Reducing retroactivity: one of the commonest methods to reduce retroactivity between two systems is to employ negative feedback. Here M1 and M2 are two biochemical systems joined at a common node to form a composite system, M2(M1).
In their paper, the authors cover some common biological examples including genetic networks and protein signaling networks illustrating how retroactivity manifests in these systems and what conditions and mechanisms reduce the level of retroactivity. More interesting is that retroactivity can in principle be measured experimentally using the operational definition provided by the authors. Retroactivity is always measured with respect to a particular node in the network such as a protein or a metabolite. Operationally, retroactivity measures the relative difference between the dynamics of two systems, one intact and another disconnected at the designated node. The modular structure of the entire network can be determined from the pattern of retroactivity. Moreover, modules defined in this manner have a very useful operational meaning; they can in principle be excised from the network and reinserted elsewhere with the prediction that they will operate as expected.
One might ask how important is this kind of modularity to biology? From an evolutionary perspective there is a growing awareness that modularity may facilitate evolutionary change by encouraging the ability to rewire modules while maintaining modular function (McAdams et al, 2004; Kirschner and Gerhart, 2005). This concept has been termed facilitated variation. Another question is how retroactivity is reduced in a biological pathway. As the paper by Del Vecchio et al suggests, retroactivity can be influenced by a number of factors but possibly one of the most important is through negative feedback. This should come as no surprise since negative feedback is an important concept that engineers use to modularize man-made devices. A classic example from electronics is the operational amplifier, the workhorse in analog circuits. High-gain amplifiers coupled with feedback ensure modularization of function and allow engineers to swap and connect devices without the fear of disrupting modular behavior.
There are numerous examples of this kind of modularity in biology. For example, bacterial amino-acid biosynthesis is riddled with negative feedback loops, presumably to stabilize amino-acid concentrations in the face of varying protein synthesis demand. Negative feedback reduces the retroactivity at the amino-acid nodes, thus ensuring a clean functional separation of amino-acid synthesis and protein synthesis. The MAPK pathway might also serve as another example of negative feedback employed to reduce the retroactivity between doubly phosphorylated ERK and its nuclear target.
What of the future? I believe two areas stand out—one is to generalize the idea of retroactivity further and understand its consequences in more detail. This will involve linking retroactivity to other quantitative theories in systems biology such as structural network analysis, cellular control analysis and stochastic biochemical dynamics; the other is to apply modularity principles to design new cellular circuits in the field of synthetic biology. Synthetic biology will depend on being able to define reusable circuits such that they can be connected together without the individual units loosing functional cohesion. The paper by Del Vecchio et al highlights an important problem and suggests a way forward.
References
- Del Vecchio D, Ninfa AJ, Sontag ED (2008) Modular cell biology: retroactivity and insulation. Mol Syst Biol 4: 161 10.1038/msb4100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J (2005) The Plausibility of Life, Resolving Darwin's Dilemma. Yale University Press: New Haven [Google Scholar]
- McAdams HH, Srinivasan B, Arkin AP (2004) The evolution of genetic regulatory systems in bacteria. Nat Rev Genet 5: 169–178 [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Kremling A, Gilles ED (2005) Dissecting the puzzle of life: modularization of signal transduction networks. Comput Chem Eng 29: 619–629 [Google Scholar]
- Tyson JJ (2006) Another turn for p53. Mol Syst Biol 2: 2006.0032 10.1038/msb4100060 [DOI] [PMC free article] [PubMed] [Google Scholar]