Abstract
Biological networks are inherently modular, yet little is known about how modules are assembled to enable coordinated and complex functions. We used RNAi and time series, whole-genome microarray analyses to systematically perturb and characterize components of a Caenorhabditis elegans lineage-specific transcriptional regulatory network. These data are supported by selected reporter gene analyses and comprehensive yeast one-hybrid and promoter sequence analyses. Based on these results, we define and characterize two modules composed of muscle- and epidermal-specifying transcription factors that function together within a single cell lineage to robustly specify multiple cell types. The expression of these two modules, although positively regulated by a common factor, is reliably segregated among daughter cells. Our analyses indicate that these modules repress each other, and we propose that this cross-inhibition coupled with their relative time of induction function to enhance the initial asymmetry in their expression patterns, thus leading to the observed invariant gene expression patterns and cell lineage. The coupling of asynchronous and topologically distinct modules may be a general principle of module assembly that functions to potentiate genetic switches.
Keywords: C. elegans, gene regulatory network, module, transcription factor
Introduction
Gene regulatory networks control spatial and temporal patterns of gene expression during embryonic development to produce a wide array of cell types (Davidson, 2006). Our knowledge of the structure and function of these gene regulatory networks has been largely gained through focused efforts to understand one or two components at a time. These studies gradually coalesce to reveal interactions among components, thus providing mechanistic insight into development, evolution, and disease (von Dassow et al, 2000; Davidson et al, 2002a; Maduro and Rothman, 2002). The development of quantitative genome-scale methods is providing new opportunities for discovery and analysis that are not limited to abundantly expressed genes or mutants that produce highly penetrant phenotypes. For example, the structure of transcriptional regulatory networks is being inferred by physical methods that identify, on a genome-wide scale, transcription factor (TF)-binding sites (Boyer et al, 2005; Zeitlinger et al, 2007). Although these methods offer the possibility of detecting direct protein–DNA interactions, binding is necessarily assumed to correspond to functional gene regulation, which could be positive or negative and weak or strong. Therefore, others are exploiting the specific experimental advantages of selected model systems to add traditional functional approaches with genome-scale experimental and computational methods to discover and validate the gene regulatory circuitries that specify cell types (Davidson et al, 2002a, 2002b; Stathopoulos et al, 2002; Baugh et al, 2005a). As more precise models of developmental regulatory networks become available, we expect to discover general principles regarding their organization and function and better understand the evolution and robustness of development mechanisms.
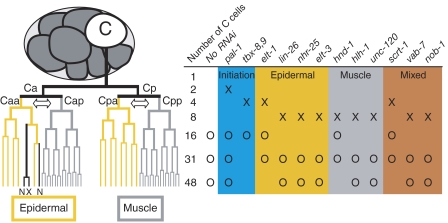
Caenorhabditis elegans embryonic development is characterized by an invariant cell lineage (Sulston et al, 1983) making it possible to define discrete gene expression states for each cell in the developing embryo, and thus provides a powerful experimental platform for investigating the structure and function of gene regulatory networks. The ParaHox gene pal-1 is the C. elegans caudal ortholog and is necessary and sufficient to specify the identity of the C blastomere (Hunter and Kenyon, 1996), which is born at the eight-cell stage of embryogenesis and produces primarily posterior body-wall muscle and epidermis (Sulston et al, 1983; Figure 1). We previously identified a set of 13 putative TFs whose expression in the C lineage is dependent on pal-1 (Baugh et al, 2005a). Transcription of these TFs is initiated in distinct temporal phases, which together with their tissue-restricted expression patterns led us to propose a provisional model of the regulatory network specified by pal-1 (Baugh et al, 2005a).
Figure 1.
Development of the C lineage and experimental design. The C blastomere is born at the eight-cell embryonic stage and follows an invariant cell lineage to produce 32 muscle cells, 13 epidermal cells, 2 neurons (N), and 1 cell death (X). The two arrows indicate the asymmetric cell divisions where muscle and epidermal cell fates are segregated in the lineage, as depicted by gray and yellow lines. Temporal and spatial expression patterns for 13 TFs expressed in the C lineage under the control of pal-1 are summarized to the right of the lineage. The background color indicates the temporal and spatial expression pattern of each TF. ‘X' indicates time of initial detection of zygotic gene product and ‘O' indicates a time point collected for mRNA expression analysis following RNAi of each TF. ELT-1 is initially detected in Cap and Cpp, but diminishes quickly while persisting in the epidermal precursors and their descendants as shown (Page et al, 1997). hnd-1 expression is first detected at the 4C stage (Baugh et al, 2005a), but expression in the C lineage is first detected at the 8C stage (Mathies et al, 2003).
We used a genetic approach coupled to microarray analysis of precisely staged embryos to comprehensively identify interactions between these regulators and their targets. We reasoned that analysis of a set of systematic perturbations of TF function and the resulting gene expression patterns should reveal the set of transcriptional regulatory relationships comprising the C-lineage regulatory network. The resulting network architecture, or topology, would consequently provide insight into the mechanisms that ensure fidelity of cell fate determination and patterning. We used whole-genome microarrays to collect transcript abundance data from precisely staged embryos depleted for each of the 13 TFs by RNAi. TF-DNA-binding studies and computational analysis were also integrated with the expression data. Our results indicate that the epidermal and muscle TFs are arranged in topologically distinct regulatory subnetworks, or modules, and that these modules repress one another and compete to specify cell fate. Specification of other cell fates in C. elegans as well as in other organisms is likely to rely on similar fate-specific regulatory modules, and we speculate that these modules may also compete via direct, mutual repression, thus accounting for the mutually exclusive nature of cell fate decisions.
Results
Inference of network topology
To study the function of the C-lineage gene regulatory network, we used RNAi to knock down expression of the 13 TFs expressed early in the C lineage, and then used whole-genome microarrays to assess each perturbation for changes in mRNA abundance. RNAi has several advantages as a method to perturb gene function: it reduces both maternal and zygotic transcripts, thus avoiding maternal rescue effects; for essential genes, it allows collection of fully affected progeny; and it enables comparison between perturbations. Measuring transcript levels on microarrays is also advantageous, because it is highly parallel, allowing direct comparisons between genes within each perturbation, and it is comprehensive, thus enabling discovery of regulated genes. In addition, it allows direct comparison to previous efforts to characterize the pal-1 regulatory network (Baugh et al, 2005a, 2005b). To increase sensitivity and specificity, we used mex-3 mutant mothers whose progeny are primarily composed of C-like lineages (Draper et al, 1996). In each case, RNA was collected from embryos two and three cell cycles after the initial zygotic expression of the targeted TF mRNA (Figure 1). We note that our sampling of only two time points might underestimate the magnitude of the observed effect of RNAi on mRNA levels, particularly if the expression level of the assayed mRNA peaks before or after the selected time points. The paralogous tbx-8 and tbx-9 genes, which were previously demonstrated to be functionally redundant (Pocock et al, 2004b; Baugh et al, 2005b), were simultaneously targeted by RNAi to assay a more penetrant effect on gene expression. A principal components analysis of the microarray data from the 12 perturbations showed that the effects of pal-1 RNAi and the double RNAi of tbx-8 and tbx-9 (tbx-8,9) are distinct from untreated and all other RNAi perturbations (Supplementary Figure 1). A closer inspection of the microarray data revealed that those were the only TFs whose perturbations had a large effect on the expression of transcripts of other C lineage enriched genes (Baugh et al, 2005a). This suggests that, despite RNAi being effective in each case (see below), depletion of most of the TFs involved in the specification of the C lineage did not grossly alter progression of C lineage development.
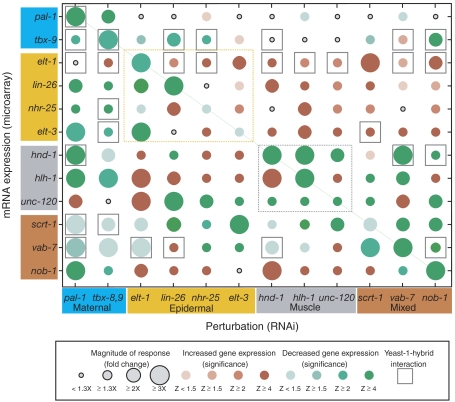
Analysis of the Drosophila segment polarity and dorsoventral gene regulatory networks has revealed substantial regulatory interactions among and between TFs and signaling molecules (Lawrence and Struhl, 1996; Stathopoulos et al, 2002). To identify potential regulatory effects among the set of all pal-1-regulated TFs, we examined the expression of each TF mRNA following RNAi of each other TF. The results are presented graphically in Figure 2. We found that the RNAi treatment was effective at decreasing transcript abundance of the target mRNA as indicated by the green circles along the diagonal of Figure 2. The left-most column of Figure 2 shows the effect of pal-1 RNAi on expression of each of the TFs, demonstrating that the TFs in the experiment do in fact mostly behave like PAL-1 targets, as their expression decreased following pal-1 RNAi. However, unc-120 expression appears to increase following pal-1 RNAi. This result is at odds with multiple microarray and reporter gene experiments and may indicate either a transient response to pal-1 inhibition that is subsequently resolved (Baugh et al, 2005a; Fukushige et al, 2006), or could be due to the mex-3 mutant background used for these studies. The other exception is elt-1, which despite the microarray data, we have validated as a pal-1 target by analysis of reporter constructs (see below). In addition, the known dependence of elt-3 and lin-26 expression upon elt-1 function (Gilleard et al, 1999b; Landmann et al, 2004) and the positive regulation of vab-7 by tbx-8,9 (Pocock et al, 2004b) are readily apparent in the data (Figure 2).
Figure 2.
A perturbation–expression matrix for a set of lineage-specific TFs. The graph indicates the fold increase or decrease (RNAi relative to untreated) and statistical significance (Z-score) for the effect of each TF RNAi (columns) on each TF expression level (rows). The time point with the largest fold-change was selected. Where the fold change is less than 1.3 fold, a small gray circle is shown. The rows and columns are organized by spatial expression pattern of each TF with the same color scheme as in Figure 1. Muscle and epidermal submatrices are outlined by gray and yellow dashed lines, respectively. Positive yeast one-hybrid results are indicated by gray squares and represent the interaction between the TF protein (column) and 1.5 kb of noncoding DNA sequence 5′ of a particular TF gene (row). All yeast one-hybrid combinations were tested with the exception of those involving the promoter of lin-26, which is a member of an operon. tbx-8 and tbx-9 are chromosomal neighbors transcribed in opposing directions, and consequently share a putative promoter region. Positive results for TBX-8 and TBX-9 have been combined (See Supplementary Figure 2 for complete data set).
The principal components analysis showed that tbx-8,9 RNAi and pal-1 RNAi uniquely had large effects on embryonic gene expression, and this analysis suggests that they have strikingly similar effects on C-lineage gene expression (Figure 2; Figure 5; Supplementary Figure S1). It is interesting to note that at this level of analysis the role of tbx-8,9 and pal-1 appear to be equivalent. This observation, combined with the similarity between the terminal embryonic loss-of-function phenotypes of tbx-8,9 and pal-1 (Hunter and Kenyon, 1996; Edgar et al, 2001; Pocock et al, 2004a; Baugh et al, 2005b), leads us to propose that the tbx-8,9 genes function at a high level in the network, perhaps supporting pal-1 to specify the C lineage. Indeed, induced expression of either pal-1 or tbx-8 or tbx-9 can induce expression of the other two (JJS and CPH, in preparation).
A simplifying assumption is that the initial regulatory interactions will be among the coexpressed genes. Thus our initial analysis focuses on TFs expressed exclusively in either epidermal or muscle precursors (yellow and gray boxes in Figure 2). Our goal is to use the microarray and reporter gene analyses to infer probable regulatory relationships among these subnetworks of epidermal- and muscle-specific TFs. We next investigate the interactions between these subnetworks, or modules, to gain insight into the regulatory mechanisms that lead to the patterned specification of multiple cell fates among sister cells.
A regulatory module controlling epidermal specification
ELT-1 is necessary and sufficient to induce epidermal development (Gilleard et al, 1999a), and analysis of an elt-1p∷GFP reporter gene (Figure 3A and B) confirmed previous expectations that elt-1 is a PAL-1 target in the C lineage epidermal cells (Baugh et al, 2005a). To generate a model of the regulatory relationships among the TFs expressed exclusively in epidermal cells, we inferred putative regulatory relationships from a combination of microarray and reporter gene analysis. Effects on epidermal TF expression following epidermal TF RNAi were graphed as either positive or negative regulatory interactions based on whether expression was decreased or increased following RNAi (Figure 4A and C). Previous work has demonstrated that elt-1 is required for expression of elt-3 and lin-26 (Gilleard et al, 1999b; Landmann et al, 2004), and during wild-type development, elt-1 mRNA, and protein are detected a cell cycle before elt-3, lin-26, and nhr-25 (Labouesse et al, 1996; Page et al, 1997; Gilleard et al, 1999b; Baugh et al, 2005a). Following pal-1 RNAi we failed to detect a significant decrease in elt-1 mRNA by microarray, although we did detect the expected decrease in mRNA levels of the epidermal PAL-1 targets elt-3, lin-26, and nhr-25 (Figures 2 and 4A). Thus, our microarray data are consistent with ELT-1 acting as a positive regulator for elt-3 and lin-26 and suggests that nhr-25 is likewise regulated by ELT-1 (Figures 2 and 4A). The inferred positive regulation of nhr-25 by elt-1 was validated using an nhr-25p∷YFP reporter (Figure 3E and F). Surprisingly, RNAi of each of the ELT-1 targets generally increased transcript abundance of the other targets as well as of elt-1 (Figure 2). Such an increase suggests that LIN-26, NHR-25, and ELT-3 feed back to negatively regulate elt-1 expression and directly or indirectly repress each other's expression. Negative feedback regulation of elt-1 expression by ELT-1 targets is consistent with the temporal expression pattern of elt-1 mRNA, which peaks early and then decreases (Baugh et al, 2003). The apparent inhibition among the ELT-1 target genes likely reflects this negative feedback on their common inducer. This inference is supported by TF-DNA interaction studies described below. In summary, we propose that elt-1, elt-3, lin-26, and nhr-25 function as a regulatory module to control early epidermal development, and we refer to this subnetwork as the epidermal module.
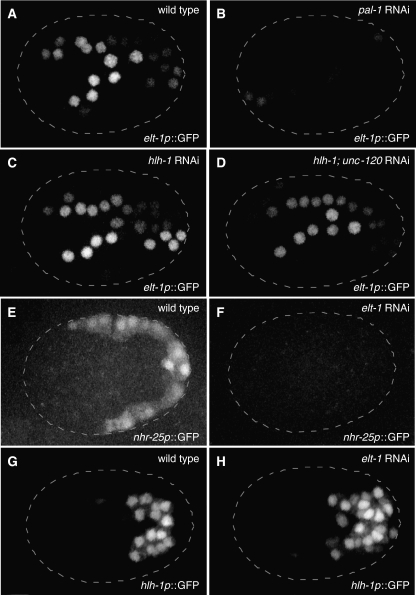
Figure 3.
Reporter gene analysis validates key inferred interactions. Two-dimensional projections of three-dimensional confocal microscopy images are shown for elt-1p∷GFP reporter gene 150 min after the 4-cell stage (22°C; ∼16C stage) for wild-type (A), pal-1 RNAi (B), hlh-1 RNAi (C), and hlh-1; unc-120 double RNAi (D). Similar projections are shown for an nhr-25p∷YFP reporter gene 200 min after the 4-cell stage (∼31C stage) for wild-type (E) and elt-1 RNAi (F), and for an hlh-1p∷YFP reporter gene at the same time for wild-type (G) and elt-1 RNAi (H). The number of cells expressing elt-1p∷GFP was not found to differ from wild-type (A) following hlh-1 RNAi (C) or hlh-1; unc120 RNAi (D) (data not shown). The number of cells expressing hlh-1p∷YFP was found to increase following elt-1 RNAi, from a wild-type average of 30 (n=5) to an elt-1 RNAi average of 42 (n=8), while 100% transformation from epidermal-to-muscle cell fate (between sister cells) should result in an increase of 16 hlh-1p∷YFP-positive cells.
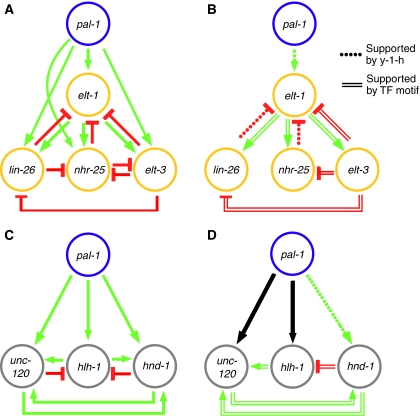
Figure 4.
Inference of cell type-specific regulatory interactions. A network representation of the perturbation-expression data is presented for the epidermal (A) and muscle (C) TFs. Each edge or arrow represents an inferred regulatory interaction (Figure 2, fold-difference >1.5, though most are >2), where decrease and increase in mRNA expression following RNAi is interpreted as activation (green) and repression (red), respectively. In the graphs in panels B and D, the red and green edges are those supported by either a yeast one-hybrid interaction (Figure 2, Supplementary Figure 2) or the presence of a significant number of putative TF-binding sites in the inferred target promoter region (Table I); the black arrows correspond to edges supported by microarray and reported genetic data, but not supported by either yeast one-hybrid or presence of putative TF-binding sites. Although none of these motifs has been validated as a functional cis-regulatory binding site by mutational analysis or otherwise, nor have the yeast one-hybrid interactions been confirmed in vivo, all are consistent with the topologies inferred for the epidermal and muscle modules, and they provide hypotheses regarding which regulatory relationships are likely to be direct.
A regulatory module controlling muscle specification
Previous phenotypic and molecular analyses indicate that muscle specification activity is distributed among the three muscle-expressed TFs, hlh-1, hnd-1, and unc-120, as the function of at least two of these TFs must be simultaneously perturbed to disrupt muscle development, whereas only the depletion of all three can eliminate muscle (Baugh et al, 2005b; Fukushige et al, 2006). Consistent with previous work demonstrating that hlh-1 and hnd-1 are PAL-1 targets (Baugh et al, 2005a; Fukushige and Krause, 2005b; Fukushige et al, 2006), our microarray results show that mRNA levels for hlh-1 and hnd-1 decreased following RNAi of pal-1 (Figure 2). Inexplicably, in this experiment with mex-3 mutant embryos, unc-120 mRNA levels were not reduced by pal-1 RNAi (Figure 2); however, given the consistency of the published results (Baugh et al, 2005a; Fukushige et al, 2006), we henceforth include unc-120 as a PAL-1 target gene.
A hypothesis to explain the observed genetic redundancy is that each of the three muscle TFs coactivate each other's expression as seen with their vertebrate homologs (Weintraub, 1993; Yun and Wold, 1996; Baugh and Hunter, 2006). Consistent with this expectation, expression of hnd-1 and unc-120 decreased following RNAi of each other or hlh-1. In contrast, hlh-1 expression appears to increase following RNAi of hnd-1 or unc-120, suggesting that they each repress hlh-1 transcription (Figures 2 and 4C). Consistent with negative feedback regulation of hlh-1, hlh-1 mRNA levels decline following a peak of two cell cycles after induction (supplementary Table 1). Finally, in contrast to the epidermal TFs, expression of all three muscle TFs is detected at the same time in the C-muscle progenitors (Figure 1; JMC and CPH, unpublished results), thus we infer a single, pal-1-dependent, regulatory step that initiates a self-sustaining subnetwork, or muscle module that controls muscle specification.
Limitation of the approach in addressing ‘patterning' TFs
RNAi of the three genes homologous to known patterning genes—vab-7, scrt-1, and nob-1—did not produce an effect on gene expression that aligned the activity of these genes with the high-level regulators (pal-1 and tbx-8,9), either cell-type module or between each other. We thus elected to not attempt to include them in the topology inference. In part, this is to be expected as these genes likely function to control the spatial organization of a subset of the muscle and epidermal cells. For example, vab-7 is expressed in only a subset of C-derived cells and controls the spatial organization of muscle and epidermal cells (Ahringer, 1996).
Potential transcription factor–DNA interactions
The microarray data provide genetic information regarding the transcriptional regulation of each of the TFs. In particular, the data suggest how the expression of each TF depends on the function of each other TF, but they do not indicate whether regulation is direct or indirect. Consequently, the number of regulatory interactions to be considered is numerous and can obscure the logic of a module topology. Relying on simple parsimony, attempting to explain all the data with a minimal number of regulatory steps, is a common means of inferring regulatory topologies but can be misleading, as networks are not necessarily designed for such efficiency. An alternative approach that complements functional data quite well is to identify direct TF–promoter interactions. To identify potential direct TF–promoter interactions, we used the yeast one-hybrid assay to systematically ask whether each TF could bind 1.5 kb of noncoding DNA sequence 5′ of the translation start site of each gene encoding every other TF. Detected interactions are shown as gray squares in Figure 2 (full data set is presented in Supplementary Figure 2). Two general patterns are apparent: PAL-1 and the TBX-8,9 proteins account for over one-third of the detected interactions, consistent with their functioning at a high level in the network, and 10 of the 13 tested TFs bound to the elt-1 promoter region, suggesting that elt-1 is a highly connected node in the network with complex regulation.
To complement the yeast one-hybrid analysis, we searched for putative TF-binding motifs among short, conserved noncoding sequences. We reasoned that functional binding sites were likely to be ancient, and therefore embedded within conserved sequences, whereas nonfunctional sequences are likely to arise randomly and are more likely to be found in nonconserved regions. For this analysis, we relied on generalized domain-binding sites: the GATA factors ELT-1 and ELT-3 were assumed to bind to the relatively ubiquitous GATA site, HLH-1 and HND-1 to the E-box site, and UNC-120 to the CaRG site (Vlieghe et al, 2006). The number of evolutionarily conserved occurrences of each binding site and the associated Z-score are presented in Table I. As both the detection of TF-promoter-binding and putative TF-binding sites operate on a relatively small subsequence of the entire promoter, they are more likely to suffer from higher false-negative than false-positive rates.
Table 1.
Putative TF-binding sites
Each cell indicates the number of occurrences (bold) of the particular site in conserved, noncoding sequences 5′ of the specified gene and the corresponding Z-score (see Materials and methods). Occurrences with ⩾2 sites and with Z-score ⩾1.4 are highlighted and indicated in Figure 4B and D. TFs are colored as in Figure 1.
aFor the putative hnd-1 promoter, noncoding sequence was not filtered by evolutionarily conservation, as the C. briggsae sequence is incomplete and synteny with respect to C. remanei is disrupted.
Where multiple regulatory interactions or edges are consistent with the microarray data (Figure 4 A and C), we used the predictions inferred by the yeast one-hybrid results and presence of conserved TF-binding motifs to describe minimal module topologies (Figure 4B and D) and infer functional properties. Thus the proposed epidermal module is composed of two sequential positive regulatory steps with negative feedback on elt-1. This negative feedback may delay commitment to the epidermal fate. In contrast, the proposed muscle module is a self-reinforcing feed-forward loop that would be expected to commit cells irreversibly to the muscle fate. These ideas are more fully explored in the Discussion.
Mutual repression between epidermal and muscle modules
The C lineage produces muscle and epidermal cells in a reproducible pattern, yet C-lineage expression of both epidermal and muscle TFs depends on pal-1 function, raising the question of how one factor can reliably specify two opposing fates. It is logical to hypothesize that the epidermal and muscle modules promote mutually exclusive gene expression states by at least one module inhibiting the expression of the other. Examination of Figure 2 shows strong asymmetric inhibition between the muscle and epidermal modules or their component parts. For example, with one exception, RNAi of any of the three muscle TFs resulted in increased expression of all four epidermal TFs, suggesting that the muscle module represses the epidermal module. Given the topology of the epidermal module (Figure 4A) combined with the yeast one-hybrid data (Figure 2, Supplementary Figure 2), the simplest hypothesis is that the muscle TFs repress elt-1 transcription and thus, indirectly, each ELT-1 target TF. Similarly, the RNAi data suggest inhibition of the muscle module by elt-1 and some of its targets (Figure 2).
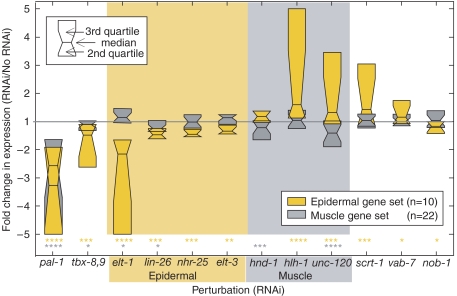
Given the complexities of the interactions among module components and our limited temporal data, we reasoned that an independent set of ‘downstream' muscle and epidermal genes would prove to be a reliable indicator of muscle and epidermal module activity. Therefore, by referring to independent published studies we identified a set of 10 epidermal and 22 muscle genes (see Materials and methods). As expected (Hunter and Kenyon, 1996), pal-1 RNAi depressed median expression of both gene sets (Figure 5). Furthermore, median expression of the epidermal gene set decreased significantly following elt-1 RNAi and to a lesser, but significant, extent following RNAi of lin-26, nhr-25, and elt-3—consistent with ELT-1 initiating the module and the ELT-1 target genes regulating distinct subsets of epidermal genes. Likewise, the muscle module is required for full expression of the muscle gene set as RNAi of hnd-1 or unc-120 causes a modest but significant decrease in median expression of the muscle gene set (Figure 5). However, hlh-1 RNAi does not appear to affect expression of this specific set of downstream muscle genes. The lack of a detectable effect of hlh-1 RNAi on muscle gene expression is not due to incomplete RNAi, because hlh-1 transcript levels were reduced over 10-fold and other genes were clearly affected, including the epidermal gene set (Figures 2 and 5). It is possible that hlh-1 is not involved in the regulation of this specific set of muscle genes or that the topology of the muscle module (Figure 4) enables robust muscle specification in the absence of hlh-1; however, it is also likely that these transcript levels decrease after our sampled time points.
Figure 5.
The muscle and epidermal modules are differentially sensitive to perturbation and mutually repress each other. The effects of RNAi on epidermal- and muscle-specific gene sets are presented as quartile boxplots. For clarity, only median (horizontal line) and two quartiles of each distribution are shown. Effects greater than fivefold are trimmed to fivefold. P-values (see Experimental Procedures) of the deviation of the mean from the expected are indicated below the plots: *P⩽0.05; **P⩽10−2; ***P⩽10−3; ****P⩽10−4. As in Figure 2, the time point with the largest fold effect was selected.
Revisiting cross-inhibitory interactions using expression of the muscle and epidermal gene sets as a proxy for muscle and epidermal module activity, we find that hlh-1 RNAi and unc-120 RNAi resulted in a significant increase in epidermal gene set expression. These observations support the conclusion that the muscle module represses epidermal module activity. However, RNAi of hlh-1 or the double RNAi of hlh-1 and unc-120 did not affect the number of cells expressing an elt-1p∷GFP reporter gene (Figure 3A, C and D). This is consistent with a lack of muscle-to-epidermal cell fate transformation, even in the absence of all three muscle module TFs (Fukushige et al, 2006). This indicates that the significant increase in epidermal gene expression detected by our quantitative microarray data (Figures 2 and 5) is likely restricted to the epidermal precursors. These results indicate that, although expression of hlh-1, hnd-1, and unc-120 is not detectable in epidermal cells, the muscle module partially inhibits epidermal module function in epidermal cells.
A reciprocal inhibition of muscle module activity by the epidermal module is detected, but the increase in muscle gene expression is limited to perturbation of elt-1 and is modest (Figure 5). Consistent with the epidermal-to-muscle transformation that occurs in the C lineage of an elt-1 mutant (Page et al, 1997), elt-1 RNAi results in an increase in the number of cells expressing an hlh-1p∷YFP reporter gene (Figure 3G and H). This validates the microarray data by clearly demonstrating that elt-1 is required for repression of hlh-1 transcription. RNAi of elt-3, nhr-25, and lin-26, alone or in combination, did not affect the number of cells expressing the hlh-1p∷YFP reporter gene (data not shown). These results suggest that the reliable restriction of muscle specification to two of the four C granddaughters requires repression of the muscle module in the epidermal precursors. We discuss below the implications of the differing module topologies and their asymmetric patterns of cross-inhibition. We note that ELT-1, the epidermal module master regulator, is initially expressed broadly in all C descendants, whereas the muscle module is induced subsequently in cells that produce exclusively muscle descendants.
Discussion
We investigated a transcriptional regulatory network that controls the development of an embryonic cell lineage to produce muscle and epidermal cell fates. The goal was to infer the topology of the regulatory network by determining the near immediate transcriptional consequences of knocking down each constituent TF. The microarray analysis afforded a direct, unbiased, and highly parallel approach to determine the phenotype of the network at discrete time points; however, it was also limiting as the sparse temporal sampling likely missed some significant changes in gene expression. The expression data were complemented by a systematic yeast one-hybrid binding analysis of all C lineage TFs as well as computational analysis to identify likely functional cis-binding sites for these TFs. From these data, we are able to infer whether and how the expression of any gene depends on the function of any of the 13 TFs that comprise the C lineage transcriptional regulatory network. We found that two highly connected subnetworks, or modules, control specification of muscle and epidermal cell fates and that these modules repress each other, effectively competing to specify alternative cell fates (Figure 6). Here, we discuss these results and their implications in our efforts to understand how a single TF (pal-1) can specify and pattern multiple, mutually exclusive cell fates in a single-cell lineage. Future experiments will undoubtedly further refine the model and promise to yield new insight into cell fate specification.
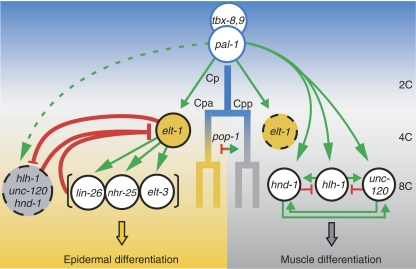
Figure 6.
A model for the C lineage developmental regulatory network. A model for how pal-1 robustly specifies and patterns multiple cell fates. The expression and inferred regulatory interactions among C lineage TFs in only the posterior C daughter and its immediate descendants are shown, but the model is the same for the anterior C daughter. PAL-1 and TBX-8,9 initiate C blastomere development, inducing first elt-1 such that ELT-1 is present in all cells at the 4C stage. In the daughters of the anterior cell (8C stage), ELT-1 continues to be expressed and is active, inducing the second stage epidermal TFs and repressing the muscle module. In the posterior daughter cell, ELT-1 expression is not maintained and all three muscle module TFs are strongly activated, rapidly leading to a self-sustaining (inducer independent) expression state—the presented muscle module topology is one of the several self-sustaining topologies consistent with the data (see text). The TF POP-1 is asymmetrically deployed in these anterior-posterior daughters at the 4C stage, but it is unknown whether POP-1 represses muscle development in the anterior daughter cells and/or activates muscle module expression in posterior daughter cells. POP-1 and/or its regulators may also influence ELT-1 activity.
Cell fate-specific regulatory modules
The GATA factor ELT-1 specifies epidermis in the C. elegans embryo: although loss of elt-1 function results in a failure to specify all major epidermal cells (Page et al, 1997), ectopic elt-1 expression in all early blastomeres is sufficient to convert the entire embryo into epidermis (Gilleard and McGhee, 2001). We show that PAL-1 binds to the elt-1 promoter and that depletion of pal-1 eliminates elt-1 expression in the C lineage. These results indicate that PAL-1 activates transcription of elt-1. Depletion of elt-1 reduces the expression of the other epidermal TFs (elt-3, lin-26, and nhr-25), and the promoters for each of these genes are enriched for putative functional GATA-binding sites, suggesting that ELT-1 directly activates these genes. Depletion of elt-1 and each of these ELT-1 target genes also reduces the expression of a cohort of epidermal differentiation genes. These results suggest a multistep cascade of gene activation. However, depletion of each ELT-1 target gene results in an increase in elt-1 levels and the other, nontargeted, epidermal TFs, suggesting negative feedback on elt-1 expression (see below). The topology of the epidermal module is thus composed of the inferred activation and feedback regulatory steps as substantiated by positive yeast one-hybrid results and/or predicted functional binding motifs in the presumed promoter regions. This topology highlights the critical role of elt-1 as master regulator of epidermal specification and suggests that epidermal specification occurs via a two-step process.
The sensitivity of the epidermal module to elt-1 function is demonstrated by a significant decrease in downstream epidermal gene expression following elt-1 RNAi (Figure 5). By the same measure, epidermal module function is less sensitive to the functions of lin-26, nhr-25, or elt-3. Although disruption of each of these genes affects epidermal development, it is not to the same phenotypic extent as disruption of elt-1 (Labouesse et al, 1996; Page et al, 1997; Gilleard, 2001; Chen et al, 2004). This is likely due to distributed activation of the differentiation genes by each of the ELT-1 target genes and to the topology of the epidermal module. Our data show that depletion of each ELT-1 target gene results in increased abundance of the other two, suggesting that these genes negatively regulate one another. We propose that this feedback is mediated indirectly through effects on elt-1 expression (Figure 4). Because all the regulatory interactions are mediated by elt-1, this topology has the effect of stabilizing expression among the ELT-1 target genes. The other possibility is that each gene directly inhibits expression of the other two ELT-1 target genes. This topology would have the effect of amplifying initial heterogeneities among ELT-1 target gene expression levels. As these genes are coexpressed in at least some cells, we favor the model whereby elt-1 integrates activation and feedback. Thus characterization of the elt-1 promoter, which was bound by 10 of 13 tested TFs (Figure 2), should reveal much about how this module is integrated with other cell fate and patterning modules.
The specification of muscle in animals relies on functionally redundant networks of TFs (Molkentin and Olson, 1996; Yun and Wold, 1996). In C. elegans, hlh-1, unc-120, and hnd-1 comprise the redundant gene set that specifies the bodywall muscle fate: embryos that lack all three fail to produce bodywall muscle cells, whereas embryos in which any one of the three remains functional produce an apparently normal number of muscle cells, although the function of those cells is compromised (Baugh et al, 2005b; Fukushige et al, 2006). The robustness to perturbation of these genes, and by extension the muscle module, presents an obstacle to phenotypic analysis. Our objective was to use microarray analysis to reveal transcriptional responses that would enable us to propose a topology for the regulatory interactions among these three genes. However, because microarray data reflect both direct and indirect effects, our results are consistent with a number of network topologies. The yeast one-hybrid interactions (Figure 2) and the presence of conserved TF-binding sites (Table I) provide positive evidence for some of these edges, but because of the high probability of false negative results, these data cannot be used to exclude other edges. Thus, in Figure 4D, we show a minimal topology for the muscle module that is consistent with all available data, while in Figure 6, we attempt to account for the phenotypic robustness of muscle specification by including additional edges supported by expression data alone (Figure 4C). However, given the experimental challenges introduced by the robustness of this module, the proposed topology should be considered provisional. An important aspect of the proposed muscle module topology is the self-sustaining interaction between unc-120 and hnd-1, which is also supported by the presence of predicted functional binding motifs in the presumed promoter regions of both genes. The significance of this proposed regulatory relationship was demonstrated when knockdown of either gene produced a significant, but minimal, effect on the median expression level of the cohort of 22 muscle differentiation genes, whereas knockdown of hlh-1 had no effect (Figure 5). As deletion alleles of these genes do not affect muscle specification (Fukushige et al, 2006), it is likely that the immediate targets of these genes are regulated by at least two of the three muscle-module TFs or that the regulatory topology of the downstream genes is also self-sustaining.
Our analysis suggests that the self-sustaining regulatory interaction between unc-120 and hnd-1 (Figure 4D) likely underlies the muscle module's robustness to perturbation. However, the fact that all three muscle genes are simultaneously induced bypasses the requirement for these interactions for initiation of module expression. Furthermore, double mutants between any two of these genes still produce bodywall muscle; therefore, the expression of any single gene must be sufficient to specify muscle (Baugh et al, 2005a). Thus the topology of the muscle module is not necessary for muscle specification. We propose that the irreversible gene expression state produced by this self-sustaining arrangement functions to commit cells to the muscle fate. Given such irreversibility, other factors or pathways are likely necessary to restrict induction of the muscle module to the muscle precursors. For example, inhibition of the epidermal module expands muscle module expression (Figures 3 and 5), suggesting that the epidermal module acts to restrict muscle module activation. However, the reverse is not true: depletion of muscle factors does not result in an expansion of epidermal gene expression (Figures 3 and 5). Thus, within the C lineage, the muscle module is not involved in patterning, only in specifying cell fate. The critical issue is when and where to deploy the muscle module and how to contain its activity.
Competing modules and developmental patterning
PAL-1 specifies muscle and epidermal cell types in the C lineage by activating epidermal- and muscle-specifying modules that cannot be coexpressed at high levels in the same cell, because they repress each other. Here, we discuss how these mutually antagonistic modules are deployed so as to avoid stochastic outcomes and robustly pattern muscle and epidermal development in the C lineage. Symmetry breaking within the C lineage is essential to selectively activate the muscle module in the two posterior daughter cells (myoblasts) at the four-C-cell stage. Once activated, the double feed-forward topology of this module rapidly renders its activation nearly irreversible, committing cells to the muscle fate. The initial symmetry breaking among the anterior–posterior daughter cells likely involves the TCF/LEF homolog POP-1, which accumulates to high levels in the nucleus of anterior daughter cells and high cytoplasmic levels in posterior daughter cells (Kaletta et al, 1997; Lin et al, 1998). Previous work has shown that pop-1 and/or its regulators and effectors are critical regulators of muscle specification (Fukushige and Krause, 2005a). Whether POP-1 inhibits myogenesis in anterior cells and/or activates myogenesis in posterior cells is unknown. However, given the relative irrepressibility of the muscle module, a requirement for combinatorial activation with PAL-1 would provide a check on inappropriate expression of this module. Based on the presence of conserved TCF/LEF-binding sites in the promoters of hlh-1, unc-120, and hnd-1 (Table I), we predict that POP-1 directly regulates their transcription. Our model for how the epidermal and muscle modules are temporally and spatially activated in the C lineage is summarized in Figure 6 and discussed below.
To specify multiple cell fates, PAL-1 first activates elt-1 in the C lineage (Figure 3), and ELT-1 is readily detected at equivalent levels in all four C granddaughters (4C stage) (Page et al, 1997). In the anterior C granddaughters, ELT-1 then induces expression of the second stage epidermal TFs. At this time, PAL-1 activates the muscle module, but it becomes irreversibly self-sustaining in only the posterior cells. However, in elt-1 mutants, muscle module expression is also established in the anterior cells and they are transformed to muscle. This suggests that the muscle module is at least minimally coexpressed with the epidermal module and that elt-1 is required to restrict muscle module activity to the posterior daughters. In contrast, even in the absence of all three muscle-module TFs, epidermal cell types are not produced by the posterior daughter cells (Fukushige et al, 2006), and we failed to detect any change in the spatial pattern of elt-1p∷YFP expression following hlh-1 RNAi or hlh-1;unc-120 double RNAi (Figure 3). Therefore, the measurable reduction in elt-1 mRNA levels mediated by the muscle module is occurring primarily in the anterior daughters (epiblasts). These observations suggest that PAL-1 initiates muscle module expression in all C granddaughters, but that in the anterior cells, ELT-1 prevents its transition to a self-sustaining, irreversible myogenic state.
C. elegans is famous for developing from an invariant cell lineage (Sulston et al, 1983); however, the coexpression of these opposing regulatory modules in the anterior granddaughter cells presumably leads to uncertainty as to which module will dominate. We argue here that the relative timing of epidermal and muscle module activation coupled with their mutual repression and respective topology-dependent repressibility and irrepressibility form the mechanistic basis for the robustness of cell fate choice in the C lineage. The epidermal module, because it employs two successive rounds of gene expression coupled with negative feedback on elt-1, is expected to act relatively slowly (Figure 4). In contrast, the single-step, self-sustaining muscle module is expected to rapidly become irrepressible, thus irreversibly committing cells to the muscle fate. Although the muscle module is primarily activated in the posterior daughter cells, it is sufficiently active in the anterior cells such that in the absence of the epidermal module, it commits these cells to the muscle fate. Because the muscle module can rapidly become irrepressible, it is important that the slow-acting epidermal module be well established in the anterior cell. Consequently, the slow acting two-step module is induced a cell cycle before the muscle module. Alternatively, if both modules were activated simultaneously by this asymmetric signal, then to compete effectively with the muscle module, the epidermal module would need to rapidly progress to a determined state. This would require a more precise regulator of anterior-posterior expression to reduce the variable outcomes. It thus appears that the pairing of a strong, rapidly acting module (muscle) with a weak, slow-acting module (epidermal) functions to enhance an initial asymmetric signal and reliably pattern embryonic development.
Materials and methods
Microarray analysis
mex-3 (zu155) mutant worms (JJ518) were used in this experiment. RNAi was administered by soaking and RNA was collected from embryos, both as described previously (Baugh et al, 2005a, 2005b). Cohorts of 10 embryos were used for each RNA preparation, and half of the RNA was used for linear amplification. The Artus ‘ExpressArt mRNA Amplification Kit' was used for amplification. The manufacturer's protocol was modified in round one with a 10-fold increase in concentration of primers A, B, and C and a 4-fold decrease in cDNA synthesis reaction volumes. Microarray hybridization onto Affymetrix C. elegans GeneChip, scanning, data reduction, and analysis were done as described previously (Baugh et al, 2003). Expression values were normalized using RMA (Bolstad et al, 2003). The epidermal gene set of ‘down-stream' genes is composed of 10 cuticular collagens: col-76, col-93, col-94, col-117, col-125, col-154, dpy-4, dpy-14, dpy-17, and sqt-3. The muscle gene set is composed of those genes previously identified to be enriched in muscle (Roy et al, 2002; Schwarz et al, 2006) that are also C-lineage enriched (decreased by pal-1 RNAi at 186′ or 230′, P-value 0.05) and contain at least two E-box or two CArG motifs in their filtered promoters (see below): dgk-2, rpl-1, T28B4.3, grl-6, K11D12.11, unc-27, alh-8, rpl-31, and rps-5. To this list, we also added C-lineage genes (Baugh et al, 2005a) whose Wormbase (Schwarz et al, 2006) gene annotations provided support for expression in body-wall muscle: pat-4, pat-10, dhp-2, gas-1, gpd-2, gta-1, let-268, sup-12, T09B4.8, F10E7.4, F56C11.3, F53F10.1, and zig-7. The complete data set has been deposited in the Gene Expression Omnibus with accession code GSE9665.
Yeast one-hybrid
TF cDNAs were cloned into the yeast expression vector pPC86 (Chevray and Nathans, 1992). Promoter fragments including 1.3–1.5 kb upstream of the translation start site were cloned into one of three yeast reporter vectors; pHisi-1 (Clontech), pLacZi (Clontech), or pJJS50 (a yCp variant of pHisi-1 with a LEU2 selectable marker). Reporter and TF plasmids were cotransformed into YM4271 yeast (BD Biosciences) and tested for interaction by growth on selective medium or by β-galactosidase activity. Promoter fragments driving HIS3 were tested for growth on selective medium with varying concentrations of 3-AT. Pscrt-1 and Pelt-3 were cloned into pHisi-1. Pelt-1, Phnd-1, Pnhr-25, Pnob-1, Ptbx-8, Ptbx-8,9, and Pvab-7 were cloned into pJJS50. Phlh-1, Ppal-1, Ptbx-9, and Punc-120 were cloned into pLacZi and tested for interaction by β-galactosidase activity. In all cases, interactions were deemed positive if they showed reporter activity above background levels seen in corresponding yeast transformed with empty pPC86. The lin-26 promoter was not cloned due to its location in an operon.
Reporter analysis
Fluorescent reporters for hlh-1 (PD7963), elt-1 (JG33), and nhr-25 (HC395) expression were obtained from previous reports (Chen et al, 1994; Baugh et al, 2005a; Smith et al, 2005). dsRNA was made by in vitro transcription. DNA templates were amplified by PCR from cDNAs using primers containing minimal T7 or T3 promoters. Following purification, the dsRNA was reannealed by heating to 90°C and then cooling 6°C/min to 25°C and diluted to approximately 0.3 μg/μl. Primer sequences used are available in Baugh et al, 2005b. Young adult animals were injected with dsRNA and allowed to recover overnight. Four-cell stage embryos were collected from injected mothers and mounted on 2.0% agarose pads. After incubation of 60–90 min at 22°C, embryos were imaged using a Zeiss Axiovert 200 m spinning disc confocal microscope equipped with a PerkinElmer UltraView confocal scanner unit, Melles Griot ion laser, and Hamamatsu Orca-ER digital camera. A total of 20 optical sections of 1.5 μm were taken along the z-axis every 10 min with exposure times of 350 or 450 ms. Two-dimensional projections of three-dimensional images were generated using Axiovision software release 4.4.
Statistical analysis
To assess the statistical significance of an TF RNAi on a gene's expression (Figure 2), we calculated Z-scores: Z=∣e−m∣/s, where e is the mean perturbed expression, m is mean unperturbed expression, and s is the standard deviation of the unperturbed expression. To assess the significance of effect of RNAi on epidermal and muscle gene sets (Figure 5), we compared the observed effects to the effects on 10,000 equally sized sets of randomly chosen embryonically expressed genes, as previously identified (Baugh et al, 2003). We computed the corresponding P-value as the fraction of randomly selected sets whose mean fold-change is larger than that of the observed.
Computational identification of cis-binding sites
For each TF, C. briggsae (Stein et al, 2003) and C. remanei (Schwarz et al, 2006) orthologs were identified using Wormbase (Schwarz et al, 2006). Promoter sequences were defined as the 5′ intergenic sequence (up to 1.5 kb upstream from the start of transcription) concatenated with the first intron (up to 1 kb). wuBLATN was used to identify regions of similarity between the C. elegans TF promoter and either of the C. briggsae and C. remanei sequences (mismatch penalty, 2; Word-size, 6; E-value, 3). These filtered sequences were produced by converting each base to ‘N' if it did not appear in an aligned region in either search. To determine the significance of the appearances of GATA, E-box, TCF, and CArG (see Table I for motifs), we permuted the filtered sequence 1,000 times by randomly re-ordering the nucleotides without accounting for di- and trinucleotide frequencies—while constraining the locations of the non-aligned segments—and computed the number of sites corresponding to each motif. The mean and standard deviations of 1,000 permutations were used to determine the Z-score of the actual number of observed sites.
Supplementary Material
Supplementary Information
Acknowledgments
We thank M Lercher, C Camacho, A Derti, R Milo, R Kafri, Y Raz, A Jose, K Ragkousi, and D Segre for a critical reading of this manuscript. We also thank all the members of the Hunter lab for discussion. LRB is supported by the American Cancer Society; CR by the NSF; IY by an NIH NRSA fellowship. This work was funded by NIH grant GM64429 to CPH.
References
- Ahringer J (1996) Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev 10: 1120–1130 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP (2005a) The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development 132: 1843–1854 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP (2003) Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hunter CP (2006) MyoD, modularity, and myogenesis: conservation of regulators and redundancy in C. elegans. Genes Dev 20: 3342–3346 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Wen JC, Hill AA, Slonim DK, Brown EL, Hunter CP (2005b) Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biol 6: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A (1994) The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120: 1631–1641 [DOI] [PubMed] [Google Scholar]
- Chen Z, Eastburn DJ, Han M (2004) The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol 24: 7345–7358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevray PM, Nathans D (1992) Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA 89: 5789–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH (2006) The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press/Elsevier [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI et al. (2002a) A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Schilstra MJ, Clarke PJ, Rust AG, Pan Z, Arnone MI et al. (2002b) A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol 246: 162–190 [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR (1996) MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216 [DOI] [PubMed] [Google Scholar]
- Edgar LG, Carr S, Wang H, Wood WB (2001) Zygotic expression of the caudal homolog pal-1 is required for posterior patterning in Caenorhabditis elegans embryogenesis. Dev Biol 229: 71–88 [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M (2006) Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev 20: 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M (2005a) The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 132: 1795–1805 [DOI] [PubMed] [Google Scholar]
- Fukushige T, Krause M (2005b) The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 132: 1795–1805 [DOI] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD (2001) Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol 21: 2533–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD (1999a) ELT-3: a Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Devl Biol 208: 265–280 [DOI] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD (1999b) ELT-3: a Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev Biol 208: 265–280 [DOI] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C (1996) Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87: 217–226 [DOI] [PubMed] [Google Scholar]
- Kaletta T, Schnabel H, Schnabel R (1997) Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature 390: 294–298 [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR (1996) The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development 122: 2579–2588 [DOI] [PubMed] [Google Scholar]
- Landmann F, Quintin S, Labouesse M (2004) Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev Biol 265: 478–490 [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G (1996) Morphogens, compartments, and pattern: lessons from Drosophila? Cell 85: 951–961 [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR (1998) POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92: 229–239 [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH (2002) Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev Biol 246: 68–85 [DOI] [PubMed] [Google Scholar]
- Mathies LD, Henderson ST, Kimble J (2003) The C. elegans hand gene controls embryogenesis and early gonadogenesis. Development 130: 2881–2892 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN (1996) Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA 93: 9366–9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR (1997) ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev 11: 1651–1661 [DOI] [PubMed] [Google Scholar]
- Pocock R, Ahringer J, Mitsch M, Maxwell S, Woollard A (2004a) A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development 131: 2373–2385 [DOI] [PubMed] [Google Scholar]
- Pocock R, Ahringer J, Mitsch M, Maxwell S, Woollard A (2004b) A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development 131: 2373–2385 [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK (2002) Chromosomal clustering of muscle-expressed genes in. Nature 418: 975–979 [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, Chan J, Chen N, Chen WJ, Davis P, Fiedler TJ, Girard L, Harris TW, Kenny EE, Kishore R, Lawson D, Lee R, Muller HM, Nakamura C, Ozersky P et al. (2006) WormBase: better software, richer content. Nucleic Acids Res 34: D475–D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, McGarr P, Gilleard JS (2005) The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. J Cell Sci 118: 5709–5719 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M (2002) Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111: 687–701 [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, Coulson A, D'Eustachio P, Fitch DH, Fulton LA, Fulton RE, Griffiths-Jones S, Harris TW, Hillier LW, Kamath R, Kuwabara PE et al. (2003) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res 34: D95–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM (2000) The segment polarity network is a robust developmental module. Nature 406: 188–192 [DOI] [PubMed] [Google Scholar]
- Weintraub H (1993) The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Yun K, Wold B (1996) Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol 8: 877–889 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M (2007) Whole-genome ChIP-chip analysis of dorsal, twist, and snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information