Abstract
Modularity plays a fundamental role in the prediction of the behavior of a system from the behavior of its components, guaranteeing that the properties of individual components do not change upon interconnection. Just as electrical, hydraulic, and other physical systems often do not display modularity, nor do many biochemical systems, and specifically, genetic networks. Here, we study the effect of interconnections on the input–output dynamic characteristics of transcriptional components, focusing on a property, which we call ‘retroactivity', that plays a role analogous to non-zero output impedance in electrical systems. In transcriptional networks, retroactivity is large when the amount of transcription factor is comparable to, or smaller than, the amount of promoter-binding sites, or when the affinity of such binding sites is high. To attenuate the effect of retroactivity, we propose a feedback mechanism inspired by the design of amplifiers in electronics. We introduce, in particular, a mechanism based on a phosphorylation–dephosphorylation cycle. This mechanism enjoys a remarkable insulation property, due to the fast timescales of the phosphorylation and dephosphorylation reactions.
Keywords: feedback, insulation, modularity, singular perturbation
Introduction
In their influential paper, Hartwell et al (1999) argued for the recognition of functional ‘modules' as a critical level of biological organization, contending that modularity is a key feature that makes biology close to synthetic disciplines such as computer science and engineering. Hartwell et al defined modules as discrete entities, whose relatively autonomous functions are separable (through spatial localization or chemical specificity) from those of other modules, and designed (or evolved) so that interconnecting modules allow higher level functions to be built. Examples of modules are signaling subsystems such as MAPK cascades, or machinery for protein synthesis or DNA replication. Lauffenburger (2000) (see also Asthagiri and Lauffenburger, 2000) further elaborates that biology could be understood in a hierarchical or nested manner, analogous to engineering design, where components are studied first in isolation, tested and individually characterized, prior to their incorporation into larger systems. From an evolutionary perspective, ‘modular structures may facilitate evolutionary change [because] embedding particular functions in discrete modules allows the core function of a module to be robust to change, but allows for changes in the properties and functions of a cell (its phenotype) by altering the connections between different modules' (Hartwell et al, 1999). This argument has much in common with facilitated variation (Kirschner and Gerhart, 2005), in which highly conserved ‘core processes' are tuned and rearranged via ‘weak regulatory linkages', thus allowing an acceleration of evolutionary change. It is also consistent with François Jacob's view of ‘evolution as a tinkerer' (Jacob, 1977): ‘organisms are historical structures… They represent, not a perfect product of engineering, but a patchwork of odd sets pieced together when and where opportunities arose.' In recent work, Alon (2007) revisited Jacob's idea (‘evolution as a tinkerer works with odds and ends, assembling interactions until they are good enough to work') and framed it from the point of view of a quantitative systems biologist, identifying modularity as a central principle in biological networks.
A fundamental systems engineering issue that arises when interconnecting subsystems is how the process of transmitting a signal to a ‘downstream' component affects the dynamic state of the sending component. Indeed, after designing, testing, and characterizing the input–output behavior of an individual component in isolation, it is certainly desirable if its characteristics do not change substantially when another component is connected to its output channel. This issue, the effect of ‘loads' on the output of a system, is well understood in many fields of engineering, for example in electrical circuit design. It has often been pointed out that similar issues arise for biological systems. Alon states that ‘modules in engineering, and presumably also in biology, have special features that make them easily embedded in almost any system. For example, output nodes should have ‘low impedance', so that adding on additional downstream clients should not drain the output to existing clients (up to some limit).' An extensive review on problems of loads and modularity in signaling networks can be found in Sauro (2004), Sauro and Kholodenko (2004) and Sauro and Ingalls (2007), where the authors propose concrete analogies with similar problems arising in electrical circuits.
These questions are even more delicate in synthetic biology. For example, suppose that we have built a timing device, a clock made up of a network of activation and/or repression interactions among certain genes and proteins, such as the one of Atkinson et al (2003). Next, we want to employ this clock (upstream system) to drive one or more components (downstream systems), by using as its output signal the oscillating concentration A(t) of one of the proteins A involved in its implementation. Typically, A will be an activator or a repressor of a gene involved in the network constituting a downstream system. From a systems/signals point of view, A(t) becomes an input to the second system. The terms ‘upstream' and ‘downstream' reflect the direction in which we think of signals as traveling, from the clock to the systems being synchronized. However, this is only an idealization, because the binding and unbinding of A to promoter sites in a downstream system competes with the biochemical interactions that constitute the upstream block (retroactivity) and may therefore disrupt the operation of the clock itself. One possible approach to avoid disrupting the behavior of the clock, motivated by the approach used with reporters such as GFP, is to introduce a gene coding for a new protein X, placed under the control of the same promoter as the gene for A, and using the concentration of X, which presumably mirrors that of A, to drive the downstream system. This approach, however, has still the problem that the behavior of the X concentration in time may be altered and even disrupted by the addition of downstream systems that drain X. The net result is still that the downstream systems are not properly timed.
The above considerations strongly motivate the need for a novel theoretical framework to formally define and quantify retroactivity effects in biological systems, such as cell signaling or gene transcriptional networks. In this paper, we first present such a formalism, and then study a general approach to the reduction of retroactivity by means of feedback. Our work complements, but is different from, questions of optimally partitioning large networks into ‘modules' for which retroactivity-like effects are minimized and the identification of possible functional modules from co-expression and other data, which typically employ graph, information theoretic, and statistical approaches (Snel et al, 2002; Papin et al, 2004; Saez-Rodriguez et al, 2005; Andrianantoandro et al, 2006; Mason and Verwoerd, 2006; Kremling and Saez-Rodriguez, 2007). In contrast, and closer to the work in Sauro (2004), we are less concerned with network topology and more with the understanding of dynamical behavior. Our ultimate goal is not top–down partitioning, nor necessarily to ignore, or even to necessarily minimize, retroactivity, but to formally define and characterize these effects, thus making the problem amenable to theoretical analysis and concrete in vivo solutions, and in particular, in the context of gene transcriptional networks. One of our theoretical contributions is a new paradigm for input–output systems analysis that allows us to characterize the equivalents of ‘impedance', which we call retroactivity, for biochemical networks. The standard model, used in virtually every control and systems theory mathematical and engineering textbooks since the 1950s, e.g. Sontag (1998), is based on the view of devices described solely in terms of input channels, output channels, and state (internal, non-shared) variables. This view is also prevalent in biology; for example, Alon (2003, 2007) defines modules as sets of nodes, each representing a protein or some other type of chemical species, that have strong interactions and a common function, specifying that a module should have ‘defined input nodes and output nodes that control the interactions with the rest of the network' as well as ‘internal nodes that do not significantly interact with nodes outside the module.' A notable exception to this standard model is found in the work of Polderman and Willems (2007), which blurs the distinction between inputs, states, and outputs; in our work, in contrast, we keep these three distinct entities, and augment the model with two additional signals, namely the retroactivities to inputs and outputs, respectively. In our formalism, achieving low output impedance becomes the problem of attenuating retroactivity to the output. Accordingly, insulators can be designed and inserted between two systems that one wishes to interconnect. Insulators are devices that have low retroactivity to the input (thus they do not affect the upstream system from which they receive the signal) and are capable of attenuating the retroactivity to the output (thus they can keep the same output independently of the downstream systems connected to such an output).
In this paper, we analyze and quantify retroactivity in transcriptional components using tools from singular perturbation theory. This leads to the identification of a key retroactivity measure, which can be interpreted as the sensitivity of the quasi-steady-state dynamics of the concentration of a protein, with respect to its dynamics if the downstream system were not present. Retroactivity is large when the amount of transcription factor is comparable to or smaller than the amount of promoter-binding sites or when the affinity of such binding sites is high. To attain modularity in the general case in which one cannot alter the hardware features of the downstream systems, the upstream system must be designed so as to attenuate the retroactivity to its output. We thus suggest a mechanism similar to that used to design non-inverting amplifiers employing operational amplifiers (OPAMPs; Schilling and Belove, 1968) to attenuate retroactivity. This simple mechanism employs a large input gain and a similarly large negative feedback. We then propose and analyze two biological instances of this mechanism, for gene and protein networks. The first one involves a strong, non-leaky promoter to implement a large input gain, combined with an abundant protease that degrades the protein product and hence implements a high-gain negative feedback. The second one involves a post-translational modification mechanism through a phosphorylation–dephosphorylation cycle, such as found in MAPK cascades. Our dynamic analysis reveals that a simple phosphorylation–dephosphorylation cycle enjoys a remarkable insulation property. This property is in part due to the fast timescales of phosphorylation–dephosphorylation reactions. Such a mechanism, as a signal transduction system, has thus an inherent capacity to provide insulation and hence to increase the modularity of the system in which it is placed.
Related work
Our work makes contact with several other areas of biochemical systems analysis, including metabolic supply and demand analysis (Hofmeyr, 1997; Hofmeyr and Cornish-Bowden, 2000), metabolic control analysis (MCA) (Fell, 1992; Heinrich and Schuster, 1996), and the closely related Biochemical Systems Theory (Savageau, 1976).
The field of metabolic supply and demand analysis (Hofmeyr, 1997; Hofmeyr and Cornish-Bowden, 2000) is concerned with understanding how the equilibrium value of the product of a supply process can be controlled by the processes that consume the product. Therefore, it is related to our retroactivity concept and analysis. Our definition and analysis of the retroactivity, however, is mainly concerned with the quantification of the difference between the dynamics of an isolated component and the dynamics of the same component when it is connected. We are not only concerned with steady-state behavior, which in transcriptional networks is often preserved upon interconnection. We are especially concerned with transient and non-equilibrium situations, such as those found in oscillatory regimes. Our definition of retroactivity incorporates transient, steady state, and permanent non-constant effects of an interconnection.
Similarly MCA, which focuses on the computation of sensitivities to parameters, may be used to quantify the retroactivity at steady state due to parameter variations in downstream modules. A generalization of MCA, to deal with non-steady-state behavior, was proposed for specific types of time-varying behaviors in various papers (e.g. Acerenza et al, 1989; Demin et al, 1999) and for general reference solutions in Ingalls and Sauro (2003). Moreover, the paper by Bruggeman et al (2002) proposed a method for analyzing MCA sensitivities based exclusively upon ‘communicating intermediates' among submodules of a large network, hence adapting MCA techniques to the analysis of modularly decomposed systems. Nonetheless, by its nature, MCA is a ‘local' method, analyzing only small perturbations around a steady state or around a specific reference trajectory. In contrast, in our paper we emphasize dynamical, non-stationary, global aspects, including transients and non-constant permanent responses, in which signals may substantially change with time, and we study these time-varying signals as key ingredients of the formalism, as opposed to only looking for sensitivities.
Results
Modeling retroactivity
In traditional systems theory, a system is usually modeled as an input–output device with internal dynamics. Such an input–output abstraction has been very useful for composing systems and for deriving properties of an interconnection by the properties of the composing systems. Such an abstraction, however, tacitly assumes that the input–output response and internal dynamics of a system does not change upon interconnection. As it has been noticed by Polderman and Willems (2007) and Willems (1999), viewing interconnections as input-to-output assignments and viewing signal transmission as unidirectional impose constraints that are not present in the physics of a system. Such constraints may be appropriate in special situations occurring in signal processing and electronics, mainly because such engineering systems have been on purpose designed to obtain unidirectional signal propagation. Natural physical and biological systems are not necessarily describable using such constraints. As a simple example, consider a hydraulic system composed of a water tank that takes as input a constant input flow through an input pipe and gives as output the pressure p at its output pipe. Once we connect to the output pipe another tank, the pressure p at the output pipe will change because the downstream tank will in turn apply a pressure to the upstream tank. Thus, the output behavior of the upstream tank changes when a downstream system is connected to its output. This phenomenon occurs also in electrical systems. The voltage at the output terminals of a voltage generator changes, due to a non-zero output impedance, when a load is applied to such output terminals. In these examples, the interconnection mechanism between an upstream system (the upstream tank or the voltage generator) and a downstream system (another tank or the electrical load) affects the dynamics of the internal state and thus of the output of the upstream system. We will model this phenomenon by a signal that travels from downstream to upstream, which we call retroactivity. The amount of such a retroactivity will change depending on the features of the interconnection and the downstream system. For example, if the aperture of the pipe connecting the two tanks is very small compared to the aperture of an output pipe of the downstream tank, the pressure p at the output of the upstream tank will not change much when the downstream tank is connected. Similarly, if the load applied to the voltage generator is very large compared to the output impedance of the voltage generator, the voltage at the output terminals will not change much when the load is added.
In the above examples, the input–output model of the isolated system does not take into account the interconnection mechanism that alters the output of the system once it is interconnected. We propose to directly model a system by taking into account its interconnection mechanism. That is, we add an input, called s, to the system to model any change in its dynamics that may occur upon interconnection with a downstream system. Similarly, we add to a system a signal r as another output, to model the fact that when such a system is connected downstream of another system, it will send upstream a signal that will alter the dynamics of the upstream system. More generally, we define a system S to have internal state x, two types of inputs (I), and two types of outputs (O): an input ‘u' (I), an output ‘y' (O), a retroactivity to the input ‘r' (O), and a retroactivity to the output ‘s' (I) (Figure 1). We represent such a system S by the equations
Figure 1.
A system S with input and output signals. The red signals denote signals originating by retroactivity upon interconnection.
 |
in which f, Y, and R are arbitrary functions and the signals x, u, s, r and y may be scalars or vectors. In this formalism, we define the input–output model of the isolated system as the one in equation (1) without r in which we have also set s=0. In practice, it is simpler to model the isolated system first, and only later model the interconnection mechanism to obtain model (1). Let Si be a system with inputs ui and si and with outputs yi and ri. Let S1 and S2 be two systems with disjoint sets of internal states. We define the interconnection of an upstream system S1 with a downstream system S2 by simply setting y1=u2 and s1=r2. For interconnecting two systems, we require that the two systems do not have internal states in common. For example, in the case of transcriptional components, this would mean that the two transcriptional components express different protein species; in the case of electrical circuits, this would mean that the two circuits do not share common electrical parts except for the ones that establish the interconnection mechanism.
It has been proposed by previous authors (Saez-Rodriguez et al, 2004, 2005) that the occurrence of retroactivity, that is, having non-zero signals r and s, depends on the specific choice of input u and output y. In particular, it has been proposed to deal with retroactivity by choosing (if they exist) specific u and y for the components that will result in zero signals s and r. Often, and particularly in the context of gene transcriptional networks such as those analyzed in more detail in this paper, it is not clear whether such choices are possible. We thus deal with retroactivity in a different way in this work, assuming that input and output choices u and y have been made, and posing the problem as one of understanding when the retroactivity signals s and r are, or can be made to be, small.
An abstract general formulation of the question, which links it to the disturbance rejection problem in control theory, is possible (See Supplementary information); however, for concreteness, we focus in this paper on what form the retroactivity signals s and r take for transcriptional networks, and we provide an operative measure of the effect of retroactivity s on the dynamics of the upstream system. We finally propose insulation devices as modules that have zero r and that attenuate the effect of s, which can be placed between an upstream system and a downstream one to insulate them.
Retroactivity in gene transcriptional networks
In the previous section, we have defined retroactivity as a general concept modeling the fact that when an upstream system is input–output connected to a downstream one, its dynamic behavior can change. In this section, we focus on transcriptional networks and show what form the retroactivity takes.
A transcriptional network is composed of a number of genes that express proteins that then act as transcription factors for other genes. Such a network can be generally represented as nodes connected by directed edges. Each node represents a gene and each arrow from node z to node x indicates that the transcription factor encoded in z, denoted Z, regulates gene x (Alon, 2007). In this paper, we model each node x of the network as an input–output module taking as input the transcription factors that regulate gene x and as output the protein expressed by gene x, denoted X. This is not the only possible choice for delimiting a module: one could in fact let the messenger RNA (mRNA) or the RNA polymerase flow along the DNA (as suggested by Endy, 2005) play the role of input and output signals. A directed edge between nodes z and x indicates that protein Z binds to the operator sites of gene x to alter (repress or activate) the expression of the latter. We denote by X the protein, by X (italics) the average protein concentration, and by x (lower case) the gene expressing protein X. A transcriptional component that takes as input protein Z and gives as output protein X is shown in Figure 2 in the dashed box. The activity of the promoter controlling gene x depends on the amount of Z bound to the promoter. If Z=Z(t), such an activity changes with time. We denote it by k(t). By neglecting the mRNA dynamics, we can write the dynamics of X as
Figure 2.
The transcriptional component takes as input u protein concentration Z and gives as output y protein concentration X. The transcription factor Z binds to operator sites on the promoter. The red part belongs to a downstream transcriptional block that takes protein concentration X as its input.
 |
in which δ is the decay rate of the protein. We refer to equation (2) as the isolated system dynamics. For the current study, the mRNA dynamics can be neglected because we focus on how the dynamics of X changes when we add downstream systems to which X binds. As a consequence, also the specific form of k(t) is not relevant. Now, assume that X drives a downstream transcriptional module by binding to a promoter p with concentration p (the red part of Figure 2). The reversible binding reaction of X with p is given by
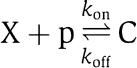 |
in which C is the complex protein promoter and kon and koff are the binding and dissociation rates of the protein X to the promoter site p. Since the promoter is not subject to decay, its total concentration pTOT is conserved so that we can write p+C=pTOT. Therefore, the new dynamics of X is governed by the equations
 |
in which the terms in the box represent the retroactivity to the output, that is, s=koffC−kon(pTOT−C)X, while the second of equation (3) describes the dynamics of the input stage of the downstream system driven by X. Then, we can interpret s as being a mass flow between the upstream and the downstream system. When s=0, the first of equation (3) reduces to the dynamics of the isolated system given in equation (2). Here, we have assumed that X binds directly to the promoter p. The case in which a signal molecule is needed to transform X to the active form that then binds to p can be treated in a similar way by considering the additional reversible reaction of X binding to the signal molecule. The end result of adding this reaction is the one of having similar terms in the box of equation (3) involving also the signaling molecule concentration.
How large is the effect of the retroactivity s on the dynamics of X and what are the biological parameters that affect it? We focus on the retroactivity to the output s. We can analyze the effect of the retroactivity to the input r on the upstream system by simply analyzing the dynamics of Z in the presence of its binding sites p0 in Figure 2 in a way similar to how we analyze the dynamics of X in the presence of the downstream binding sites p. The effect of the retroactivity s on the behavior of X can be very large (Figure 3). This is undesirable in a number of situations in which we would like an upstream system to ‘drive' a downstream one as is the case, for example, when a biological oscillator has to time a number of downstream processes. If, due to the retroactivity, the output signal of the upstream process becomes too low and/or out of phase with the output signal of the isolated system (as in Figure 3), the coordination between the oscillator and the downstream processes will be lost. We next propose a procedure to obtain an operative quantification of the effect of the retroactivity on the dynamics of the upstream system.
Figure 3.
Simulation results for the system in equation (3). Here, k(t)=0.01(1+sin(ωt)) with ω=0.005, kon=10, koff=10, δ=0.01, pTOT=100, X(0)=5. The choice of protein decay rate (in min−1) corresponds to a half-life of about 1 h. The frequency of oscillations is chosen to have a period of about 12 times the protein half-life in accordance to what is experimentally observed in the synthetic clock of Atkinson et al (2003). All simulation results were obtained by using MATLAB (Simulink), with variable-step ODE solver ODE23s. The green plot (solid line) represents X(t) originating by the isolated system in equation (2), while the blue plot (dashed line) represents X(t) obtained by the interconnected system of equation (3). Both transient and permanent behaviors are different.
Quantification of the effect of the retroactivity to the output
In this section, we propose a general approach for providing an operative quantification of the effect of the retroactivity to the output on the dynamics of the upstream system. This approach can be generally applied whenever there is a separation of timescales between the dynamics of the output of the upstream module and the dynamics of the input stage of the downstream module. This separation of timescales is usually encountered in transcriptional networks. In fact, the dynamics of the input stage of a downstream system is governed by the reversible binding reaction of the transcription factor with the operator sites. These reactions are often on the timescales of a second and thus are fast compared to the timescales of transcription and translation (often of several minutes) (Alon, 2007). These determine, in turn, the dynamics of the output of a transcriptional module. Such a separation of timescales is encountered even when we extend a transcriptional network to include as interconnection mechanisms between transcriptional modules protein–protein interactions (often with a subsecond timescale; Shen-Orr et al, 2002), as encountered in signal transduction networks. In this section, we illustrate our approach for quantifying the retroactivity effects in the case of transcription networks, while in section Design 2: amplification through phosphorylation we show, through an example, how the same approach can be employed for mixed transcription and signal transduction systems.
We quantify the difference between the dynamics of X in the isolated system (equation (2)) and the dynamics of X in the connected system (equation (3)) by establishing conditions on the biological parameters that make the two dynamics close to each other. This is achieved by exploiting the difference of timescales between the protein production and decay processes and its binding and unbinding process to the promoter p. By virtue of this separation of timescales, we can approximate system (3) by a one-dimensional system describing the evolution of X on the slow manifold (Kokotovic et al, 1999). This reduced system takes the form:
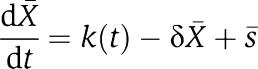 |
where [Xmacr] is an approximation of X and [sumacr] is an approximation of s, which can be written as 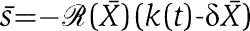 ). If
). If 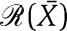 is zero, then also [sumacr]=0 and the dynamics of [Xmacr] becomes the same as the one of the isolated system (2). Since [Xmacr] approximates X, the dynamics of X in the full system (3) is also close to the dynamics of the isolated system (2) whenever
is zero, then also [sumacr]=0 and the dynamics of [Xmacr] becomes the same as the one of the isolated system (2). Since [Xmacr] approximates X, the dynamics of X in the full system (3) is also close to the dynamics of the isolated system (2) whenever  . The factor
. The factor 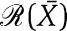 provides then a measure of the retroactivity effect on the dynamics of X. It is also computable as a function of measurable biochemical parameters and of the signal X traveling across the interconnection, as we next illustrate.
provides then a measure of the retroactivity effect on the dynamics of X. It is also computable as a function of measurable biochemical parameters and of the signal X traveling across the interconnection, as we next illustrate.
Consider again the full system in equation (3), in which the binding and unbinding dynamics are much faster than protein production and decay, that is, koff≫k(t), koff≫δ (Alon, 2007), and kon=koff/kd with kd=O(1). Even if the second equation goes to equilibrium very fast compared to the first one, the above system is not in ‘standard singular perturbation form' (Kokotovic et al, 1999). To explicitly model the difference in timescales between the two equations of system (3), we introduce a parameter ∈, which we define as ∈=δ/koff. Since koff≫δ, we also have that ∈≪1. Substituting koff=δ/∈ and kon=δ/(∈kd) in system (3), we obtain the system
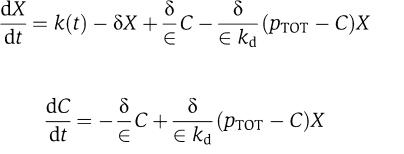 |
in which the singular perturbation parameter ∈ appears in both equations. We can take the above system to standard singular perturbation form by performing the change of variable y=X+C, in which y physically corresponds to the total concentration of protein X. Then, the system in the new variables becomes
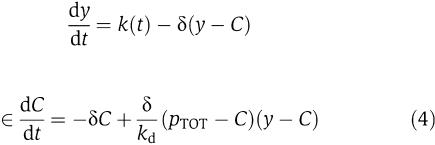 |
which is in standard singular perturbation form. This means, as some authors recently proposed (Ciliberto et al, 2007), that y (total concentration of protein) is the slow variable of the system (3) as opposed to X (concentration of free protein). We can then obtain an approximation of the dynamics of X in the limit in which ∈ is very small, by setting ∈=0. By setting ∈=0 in the second equation of (4), we obtain the manifold on which the dynamics of the system is governed by the slow variable dynamics. Such a manifold is called the slow manifold (Kokotovic et al, 1999) (The dynamics of (4) restricted to the slow manifold are a good approximation of the dynamics of system (4) only if the slow manifold is asymptotically stable, that is, only if the trajectories of the system tend to the slow manifold. It can be shown that this is the case (see Supplementary information). For a variable x involved in system (4), we denote by [xmacr] the value of the variable x once we have set ∈=0 in system (4). Let 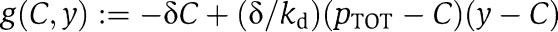 and let [Cmacr ]=γ([ymacr]) be the smallest root of g([Cmacr ], [ymacr])=0. The function γ([ymacr]) provides the steady-state value of C as a function of y and [Cmacr ]=γ([ymacr]) defines the slow manifold. Substituting the steady-state value of C as function of y in the first one of (4) leads to the reduced model
and let [Cmacr ]=γ([ymacr]) be the smallest root of g([Cmacr ], [ymacr])=0. The function γ([ymacr]) provides the steady-state value of C as a function of y and [Cmacr ]=γ([ymacr]) defines the slow manifold. Substituting the steady-state value of C as function of y in the first one of (4) leads to the reduced model
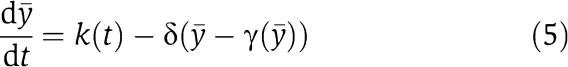 |
Given any solution of equation (5), we define [Cmacr ](t)=γ([ymacr](t)), and we let [Xmacr](t)=[ymacr](t)−[Cmacr ](t), which satisfies the following differential equation:
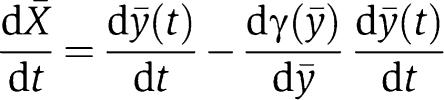 |
and which, since [Xmacr]=[ymacr]−[Cmacr ], finally leads to
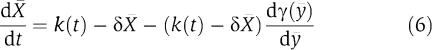 |
After a fast transient (established by the binding and unbinding dynamics of X to p), the signal y(t) is well approximated by [ymacr](t), and the signal C(t) is well approximated by [Cmacr ](t). Therefore, X(t)=y(t)−C(t) is also well approximated by [Xmacr](t)=[ymacr](t)−[Cmacr ](t). The smaller the ∈, the better is the approximation. Since [Xmacr] well approximates X, conditions for which the dynamics of equation (6) is close to the dynamics of the isolated system (2) also guarantee that the dynamics of X given in system (3) is close to the dynamics of the isolated system.
The difference between the dynamics in equation (6) (the connected system after a fast transient) and the dynamics in equation (2) (the isolated system) is zero when the term 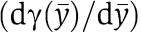 in equation (6) is also zero. We thus consider the factor
in equation (6) is also zero. We thus consider the factor 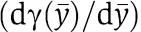 as a quantification of the effect of the retroactivity s after a fast transient in the approximation in which ∈≈0. We can also interpret the factor
as a quantification of the effect of the retroactivity s after a fast transient in the approximation in which ∈≈0. We can also interpret the factor 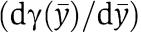 as a percentage variation of the dynamics of the connected system with respect to the dynamics of the isolated system at the quasi-steady state. We next determine the physical meaning of such a factor by calculating a more useful expression that is a function of key biochemical parameters. By using the implicit function theorem, one can compute the following expression for
as a percentage variation of the dynamics of the connected system with respect to the dynamics of the isolated system at the quasi-steady state. We next determine the physical meaning of such a factor by calculating a more useful expression that is a function of key biochemical parameters. By using the implicit function theorem, one can compute the following expression for 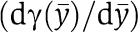 :
:
in which one can verify that 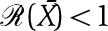 (see Supplementary information). The expression
(see Supplementary information). The expression 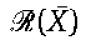 quantifies the effect of the retroactivity to the output on the dynamics of X after a fast transient, when we approximate X with [Xmacr] in the limit in which ∈≈0. The retroactivity effect is thus small if the affinity of the binding sites p is small (kd large) or if the signal X(t) is large enough compared to pTOT. Thus, the expression of
quantifies the effect of the retroactivity to the output on the dynamics of X after a fast transient, when we approximate X with [Xmacr] in the limit in which ∈≈0. The retroactivity effect is thus small if the affinity of the binding sites p is small (kd large) or if the signal X(t) is large enough compared to pTOT. Thus, the expression of 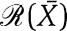 provides an operative quantification of the effects of retroactivity: such an expression can in fact be evaluated once the association and dissociation constants of X to p are known, the concentration of the binding sites pTOT is known, and the range of operation of the signal [Xmacr](t) that travels across the interconnection is also known.
provides an operative quantification of the effects of retroactivity: such an expression can in fact be evaluated once the association and dissociation constants of X to p are known, the concentration of the binding sites pTOT is known, and the range of operation of the signal [Xmacr](t) that travels across the interconnection is also known.
Note that the quantification of the effects of the retroactivity to the input r on the dynamics of Z can be obtained in exactly the same way as it has been obtained for s:
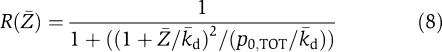 |
in which [kmacr]d=k−/k+ (here, k+ and k− are the binding and dissociation rates of Z to the promoter sites p0) and 1/[kmacr]d is the affinity of Z to its target sites p0 (with total concentration p0,TOT) on the promoter controlling gene x. Therefore, for a large-scale gene transcriptional network, one does not need to re-perform the above theoretical analysis for the larger system to obtain the measure of the retroactivity 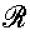 at an interconnection. In fact, the algebraic expression of
at an interconnection. In fact, the algebraic expression of  in terms of the biochemical parameters at each individual interconnection is the same independently of the size of the network. One only needs to evaluate such an expression at all interconnections to measure the amount of the retroactivity effect on the dynamics of an upstream module. In this respect, the proposed approach for the quantification of the effect of the retroactivity at interconnections is scalable.
in terms of the biochemical parameters at each individual interconnection is the same independently of the size of the network. One only needs to evaluate such an expression at all interconnections to measure the amount of the retroactivity effect on the dynamics of an upstream module. In this respect, the proposed approach for the quantification of the effect of the retroactivity at interconnections is scalable.
For simplifying notation, we will omit in the sequel the bar from the variables as X(t)≈[Xmacr](t) after a fast initial transient when ∈ is small. In the case in which X binds to a number N of different downstream binding sites, the analysis that we have proposed holds unchanged. In particular, equations (6) and (5) should be replaced by
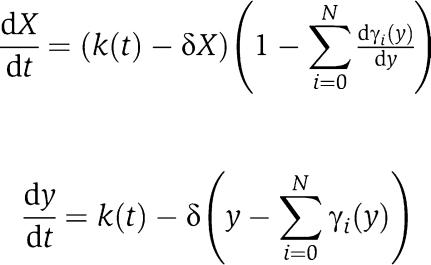 |
respectively, in which one can still show that 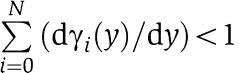 .
.
Electrical analogy. The analysis that we have performed holds generally for non-steady-state situations, in which protein concentrations change with time even in the permanent behavior. To point out a nice analogy between the biochemical retroactivity and the electrical retroactivity, consider the case of a voltage source, which could be A/C (alternate current) or D/C (direct current), powering a load. For simplicity, consider the D/C case, in which a voltage generator has a constant voltage Vgen and output impedance Ro. Apply a downstream load resistance R to the voltage generator. The voltage resulting at the load will not be Vgen because of the internal resistance, but it will be given by V = Vgen/(1 + Ro/R). The fact that Vgen≠V is a typical example of retroactivity in electrical circuits: the effective voltage applied to the load depends on the downstream load applied to the generator and on the output resistance of the generator component. Let us consider now the simplified case in which protein X is very stable so that its total concentration XTOT is conserved. Upon addition of a downstream binding site p with concentration p, one can compute the amount of free concentration of protein X. This is given by X = XTOT/(1 + p/kd and it is different from XTOT, which would be the free protein concentration without downstream binding sites. Then we can say that the retroactivity decreases as p/kd decreases. In the case in which R is very large, the input current I=Vgen/(R+Ro) to the downstream load is very small. As a consequence, the voltage drop across the internal impedance Ro is small and Vgen≈V. Similarly, a small number of binding sites p causes a small ‘flow' of protein X to the downstream sites. Therefore, we can say that p in the biochemical example plays a role similar to 1/R in the electrical example: if both are small, the ‘flow' (of charged particles or of proteins) toward the downstream system is small and retroactivity decreases. Furthermore, even if p, or 1/R, is large, the retroactivity can be decreased by having 1/kd (the affinity of the binding), or Ro, small enough. Hence, the affinity of protein X to the sites in the downstream component plays a role similar to the output resistance of the voltage generator in the electrical system.
Attenuation of the retroactivity
Consider a system S as the one shown in Figure 1 that takes u as input and gives y as output. We would like to design it in such a way that (a) the retroactivity r to the input is very small; (b) the effect of the retroactivity s to the output on the internal dynamics of the system is very small independently of s itself; and (c) its input–output relationship is about linear. Such a system is said to enjoy the insulation property and will be called an insulation component or insulation device (the meaning that we consider for ‘insulator' is different from the meaning that such a word is attributed in eukaryotic transcription). Indeed, such a system will not affect an upstream system because r≈0 and it will keep the same output y independently of any connected downstream system. In electronics, amplifiers enjoy the insulation property by virtue of the features of the OPAMP that they employ (Schilling and Belove, 1968). The concept of amplifier in the context of a biochemical network has been considered before in relation to its robustness and insulation property from external disturbances (Sauro and Kholodenko, 2004; Sauro and Ingalls, 2007). In this paper, we revisit the amplifier mechanism in the context of gene transcriptional networks with the objective of mathematically and computationally proving how suitable biochemical realizations of such a mechanism can attain properties (a), (b), and (c).
Retroactivity to the input
In electronic amplifiers, r is very small because the input stage of an OPAMP absorbs almost zero current. This way, there is no voltage drop across the output impedance of an upstream voltage source. Equation (8) quantifies the effect of retroactivity on the dynamics of Z as a function of biochemical parameters that characterize the interconnection mechanism. These parameters are the affinity of the binding site 1/[kmacr]d, the total concentration of such binding site p0,TOT, and the level of the signal Z(t). Therefore, to reduce the effect of r on Z, we can choose parameters such that (8) is small. A sufficient condition is to choose [kmacr]d large (low affinity) and p0,TOT small, for example. Having small value of p0,TOT and/or low affinity implies that there is a small ‘flow' of protein Z toward its target sites. Thus, we can say that a low retroactivity to the input is obtained when the ‘input flow' to the system is small. This interpretation establishes a nice analogy to the electrical case, in which low retroactivity to the input is obtained, as explained above, by a low input current. Such an interpretation can be further carried to the hydraulic example. In such an example, if the input flow to the downstream tank is small compared, for example, to the output flow of the downstream tank, the output pressure of the upstream tank will not be affected by the connection. Therefore, the retroactivity to the input of the downstream tank will be small.
Retroactivity to the output
In electronic amplifiers, the effect of the retroactivity to the output s on the amplifier behavior is reduced to almost zero by virtue of a large (theoretically infinite) amplification gain of the OPAMP and an equally large negative feedback mechanism that regulates the output voltage. Genetic realization of amplifiers has been previously proposed (see for example, Rubertis and Davies, 2003). However, such realizations focus mainly on trying to reproduce the layout of the device instead of implementing the fundamental mechanism that allows it to properly work as an insulator. Such a mechanism can be illustrated in its simplest form by Figure 4a, which is very well known to control engineers. For simplicity, we have assumed in such a diagram that the retroactivity s is just an additive disturbance. The reason why for large gains G the effect of the retroactivity s to the output is negligible can be verified through the following simple computation. The output y is given by
Figure 4.
(A) The basic feedback–amplification mechanism by which amplifiers attenuate the effect of the retroactivity to the output s. (B) An alternative representation of the same mechanism of (A), which will be employed to design biological insulation devices.
which leads to
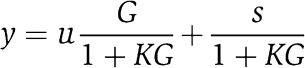 |
As G grows, y tends to u/K, which is independent of the retroactivity s.
We employ this general mechanism to attenuate the effect of the retroactivity to the output. A successful attenuation implies that the signal X(t) generated by the connected system will be very close to the signal X(t) generated by the isolated system. Consider the equivalent representation of the amplifier mechanism shown in Figure 4B. Such a representation allows us to decouple the design of a large amplification gain G from the design of a similarly large negative feedback gain G′. In the sequel, we consider the approximated dynamics of equation (6) for X. Let us thus assume that we can apply a gain G to the input k(t) and a negative feedback gain G′ to X with G′=KG. This leads to the new differential equation for the connected system (6) given by
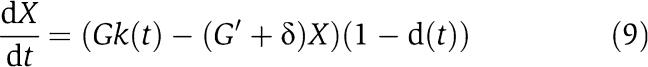 |
in which we have defined 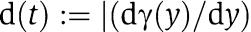 , where y(t) is given by the reduced system
, where y(t) is given by the reduced system 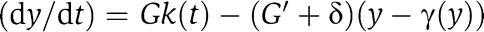 . It can be shown (see Supplementary information for the mathematical details) that as G and thus as G′ grow, the signal X(t) generated by the connected system (9) becomes close to the solution X(t) of the isolated system
. It can be shown (see Supplementary information for the mathematical details) that as G and thus as G′ grow, the signal X(t) generated by the connected system (9) becomes close to the solution X(t) of the isolated system
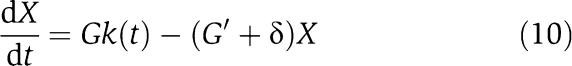 |
that is, the presence of the disturbance term d(t) will not significantly affect the time behavior of X(t). Since d(t) is a measure of the retroactivity effect on the dynamics of X, such an effect is thus attenuated by employing large gains G and G′. How can we obtain a large amplification gain G and a large negative feedback G′ in a biological insulation component? This question is addressed in the following section, in which two different realizations are presented and compared with each other.
Biological realizations of an insulation component
In the previous section, we have proposed a general mechanism to create an insulation component. In particular, we have specified how one can alter the biological features of the interconnection mechanism to have low retroactivity to the input r and we have shown a general method to attenuate the retroactivity to the output s. Such a method consists of a large amplification of the input and a large negative output feedback. The insulation component will be inserted in place of the transcriptional component of Figure 2. This will guarantee that the system generating Z, an oscillator, for example, will maintain the same behavior as in isolation and also that the downstream system that accepts X as its input will not alter the behavior of X. The net result of this is that the oscillator generating signal Z will be able to time downstream systems with the desired phase and amplitude independently of the number and the features of downstream systems. In this section, we determine two possible biological mechanisms that can be exploited to obtain a large amplification gain to the input Z of the insulation component and a large negative feedback on the output X of the insulation component. Both mechanisms realize the negative feedback through enhanced degradation. The first design realizes amplification through transcriptional activation, while the second design through phosphorylation of a protein that is in abundance in the system.
Design 1: amplification through transcriptional activation
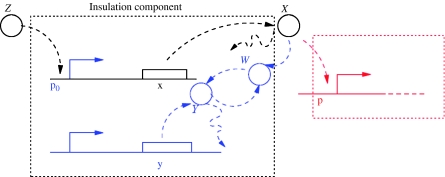
In this design, we obtain a large amplification of the input signal Z(t) by having promoter p0 (to which Z binds) as a strong, non-leaky, promoter. The negative feedback mechanism on X relies on enhanced degradation of X. Since this must be large, one possible way to obtain an enhanced degradation for X is to have a protease, called Y, be expressed by a strong constitutive promoter. The protease Y will cause a degradation rate for X, which is larger if Y is more abundant in the system. This design is schematically shown in Figure 5.
Figure 5.
The parts that design 1 affects are highlighted in blue. In particular, a negative feedback occurring through post-translational regulation and a promoter that produces a large signal amplification are the central parts of this design. The red part indicates the downstream component that takes as input the concentration of protein X.
To investigate whether such a design realizes a large amplification and a large negative feedback on X as needed, we analyze the full input–output model for the block in the dashed box of Figure 5. In particular, the expression of gene x is assumed to be a two-step process, which incorporates also the mRNA dynamics. Incorporating these dynamics in the model is relevant for the current study because they may contribute to an undesired delay between the Z and X signals. The reaction of the protease Y with protein X is modeled as the two-step reaction
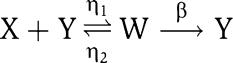 |
which can be found in standard references (see for example, Arkin et al, 1998). The input–output system model of the insulation component that takes Z as an input and gives X as an output is given by the following equations
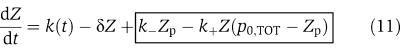 |
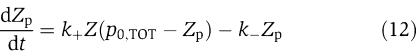 |
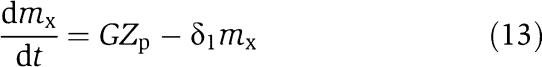 |
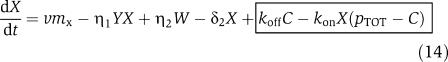 |
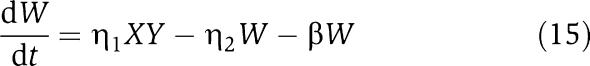 |
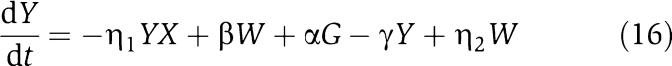 |
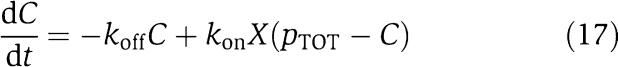 |
in which we have assumed that the expression of gene z is controlled by a promoter with activity k(t). These equations will be studied numerically and analyzed mathematically in a simplified form. The variable Zp is the concentration of protein Z bound to the promoter controlling gene x, p0,TOT is the total concentration of the promoter p0 controlling gene x, mX is the concentration of mRNA of X, C is the concentration of X bound to the downstream binding sites with total concentration pTOT, γ is the decay rate of the protease Y. The value of G is the production rate of X mRNA per unit concentration of Z bound to the promoter controlling x; the promoter controlling gene y has strength αG, for some constant α, and it has the same order of magnitude strength as the promoter controlling x. The terms in the box in equation (11) represent the retroactivity r to the input of the insulation component in Figure 5. The terms in the box in equation (14) represent the retroactivity s to the output of the insulation component of Figure 5. The dynamics of equations (11), (12), (13), (14), (15), (16) and (17) without s (the elements in the box in equation (14)) describe the dynamics of X with no downstream system.
We mathematically explain why system (11–17) allows to attenuate the effect of s on the X dynamics. Equations (11) and (12) simply determine the signal Zp(t) that is the input to equations (13), (14), (15), (16) and (17). For the discussion regarding the attenuation of the effect of s, it is not relevant what the specific form of signal Zp(t) is. Let then Zp(t) be any bounded signal ν(t). Since equation (13) takes ν(t) as an input, we will have that mX=Gnumacr(t), for a suitable signal numacr(t). Let us assume for the sake of simplifying the analysis that the protease reaction is a one-step reaction, that is, 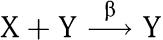 . Therefore, equation (16) simplifies to
. Therefore, equation (16) simplifies to 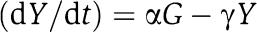 and equation (14) simplifies to
and equation (14) simplifies to 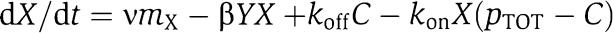 . If we consider the protease to be at its equilibrium, we have that Y(t)=αG/γ. As a consequence, the X dynamics becomes
. If we consider the protease to be at its equilibrium, we have that Y(t)=αG/γ. As a consequence, the X dynamics becomes
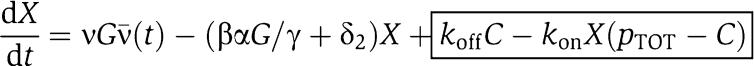 |
with C determined by equation (17). By using the same singular perturbation argument employed in the previous section, we obtain that the dynamics of X will be after a fast transient approximately given by
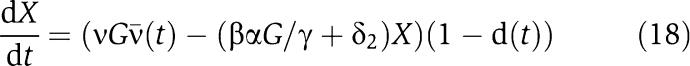 |
in which 0<d(t)<1 is the effect of the retroactivity s. Then, as G increases, X(t) becomes closer to the solution of the isolated system
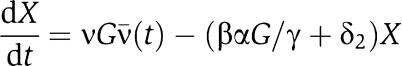 |
as explained in section Attenuation of the retroactivity (see the Supplementary Information for the mathematical details).
We now turn to the question of minimizing the retroactivity to the input r because its effect can alter the input signal Z(t). To decrease r, we guarantee that the retroactivity measure given in equation (8) is small. This is seen to be true if ([kmacr]d+Z)2/(p0,TOT[kmacr]d) is very large, in which 1/[kmacr]d=k+/k− is the affinity of the binding site p0 to Z. Since after a short transient, Zp=(p0,TOTZ)/([kmacr]d+Z), for Zp not to be a distorted version of Z, it is enough to ask that [kmacr]d≫Z. This, combined with the requirement that ([kmacr]d+Z)2/(p0,TOT[kmacr]d) is very large, leads to the requirement p0,TOT/[kmacr]d≪1. Summarizing, for not having distortion effects between Z and Zp and small retroactivity r, we need that
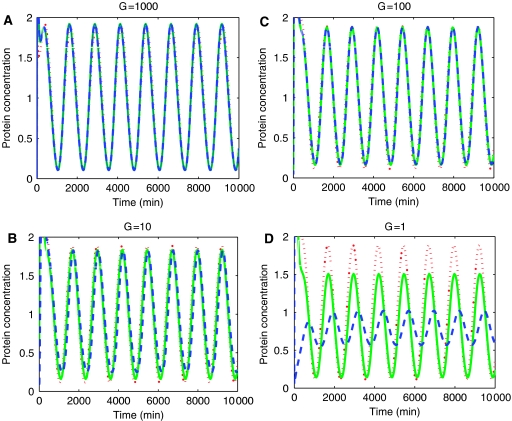
Simulation results. Simulation results are presented for the insulation system of equations (11), (12), (13), (14), (15), (16) and (17) as the mathematical analysis of such a system is only valid under the approximation that the protease reaction is a one-step reaction. In all simulations, we consider protein decay rates to be 0.01 min−1 to obtain a protein half-life of about 1 h. We consider always a periodic forcing k(t)=0.01(1+sin(ωt)), in which we assume that such a periodic signal has been generated by a synthetic biological oscillator. Therefore, the oscillating signals are chosen to have a period that is about 12 times the protein half-life in accordance to what is experimentally observed in the synthetic clock of Atkinson et al (2003). All simulation results were obtained by using MATLAB (Simulink), with variable-step ODE solver ODE23s. For large gains (G=1000, G=100), the performance considerably improves compared to the case in which X was generated by a plain transcriptional component accepting Z as an input (Figure 3). For lower gains (G=10, G=1), the performance starts to degrade for G=10 and becomes not acceptable for G=1 (Figure 6). Since we can view G as the number of transcripts produced per unit time (1 min) per complex of protein Z bound to promoter p0, values G=100 and 1000 may be difficult to realize in vivo, while the values G=10 and 1 could be more easily realized. The values of the parameters chosen in Figure 6 are such that [kmacr]d≫Z and p0,TOT≪[kmacr]d. This is enough to guarantee that there is small retroactivity r to the input of the insulation device independently of the value of the gain G, according to relations (19). The poorer performance of the device for G=1 is therefore entirely due to poor attenuation of the retroactivity s to the output.
Figure 6.
Design 1: results for different gains G. In all plots, red (dotted line) is the input Z to the insulation device, green (solid line) is the output X of the insulation device in isolation (without the downstream binding sites p), blue (dashed line) is the output X of the insulation device when downstream sites p are present. In all plots, k(t)=0.01(1+sin(ωt)), pTOT=100, koff=kon=10, δ=0.01, and ω=0.005. The parameter values are δ1=0.01, p0,TOT=1, η1=η2=β=γ=0.01, k_=200, k+=10, α=0.1, δ2=0.1, ν=0.1, and G=1000, 100, 10, and 1. The retroactivity to the output is not well attenuated for values of the gain G=1 and the attenuation capability begins to worsen for G=10.
Design 2: amplification through phosphorylation
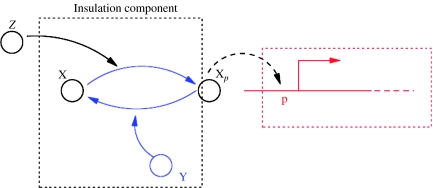
In this design, the amplification of Z is obtained by having Z activate the phosphorylation of a protein X, which is available in the system in abundance. That is, Z is a kinase for a protein X. The phosphorylated form of X, called Xp, binds to the downstream sites, while X does not. A negative feedback on Xp is obtained by having a phosphatase Y activate the dephosphorylation of protein Xp. Protein Y is also available in abundance in the system. This mechanism is depicted in Figure 7. A similar design has been proposed by Sauro and Ingalls (2007) and Sauro and Kholodenko (2004), in which a MAPK cascade as well as a negative feedback loop that spans the length of the MAPK cascade is considered as a feedback amplifier. Our design is much simpler as it involves only one phosphorylation cycle and does not require the additional feedback loop. In fact, we realize a strong negative feedback by the action of the phosphatase that converts the active protein form Xp to its inactive form X. This negative feedback, whose strength can be tuned by varying the amount of phosphatase in the system, is enough to mathematically and computationally show that the desired insulation properties are satisfied.
Figure 7.
The dashed box contains the insulation device. The blue parts highlight the mechanism that provides negative feedback and amplification. Negative feedback occurs through a phosphatase Y that converts the active form Xp back to its inactive form X. Amplification occurs through Z activating the phosphorylation of X.
We consider two different models for the phosphorylation and dephosphorylation processes. A one-step reaction model is initially considered to illustrate what biochemical parameters realize the input gain G and the negative feedback G′. Then, we turn to a more realistic two-step model to perform a parametric analysis and numerical simulation. The one-step model that we consider is the one of Heinrich et al (2002):
and
We assume that there is plenty of protein X and phosphatase Y in the system and that these quantities are conserved. The conservation of X gives X+Xp+C=XTOT, in which X is the inactive protein, Xp is the phosphorylated protein that binds to the downstream sites p, and C is the complex of the phosphorylated protein Xp bound to the promoter p. The Xp dynamics can be described by the first equation in the following model
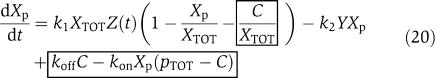 |
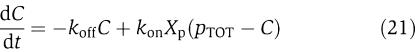 |
The boxed terms represent the retroactivity s to the output of the insulation system of Figure 7. For a weakly activated pathway (Heinrich et al, 2002), Xp≪XTOT. Also, if we assume that the concentration of total X is large compared to the concentration of the downstream binding sites, that is, XTOT≫pTOT, equation (20) is approximately equal to
Denote G=k1XTOT and G′=k2Y. Exploiting again the difference of timescales between the Xp dynamics and the C dynamics, after a fast initial transient, the dynamics of Xp can be well approximated by
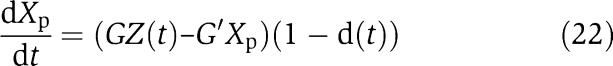 |
in which 0<d(t)<1 is the effect of the retroactivity s to the output after a short transient. Therefore, for G and G′ large enough, Xp(t) tends to the solution Xp(t) of the isolated system 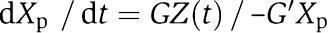 , as explained in section Attenuation of the retroactivity (see the Supplementary information for the mathematical details). As a consequence, the effect of the retroactivity to the output s is attenuated by increasing k1XTOT and k2Y enough. That is, to obtain large input and feedback gains, one should have large phosphorylation–dephosphorylation rates and/or a large amount of protein X and phosphatase Y in the system. This reveals that the values of the phosphorylation–dephosphorylation rates cover an important role toward the realization of the insulation property of the module of Figure 7.
, as explained in section Attenuation of the retroactivity (see the Supplementary information for the mathematical details). As a consequence, the effect of the retroactivity to the output s is attenuated by increasing k1XTOT and k2Y enough. That is, to obtain large input and feedback gains, one should have large phosphorylation–dephosphorylation rates and/or a large amount of protein X and phosphatase Y in the system. This reveals that the values of the phosphorylation–dephosphorylation rates cover an important role toward the realization of the insulation property of the module of Figure 7.
We next consider a more complex model for the phosphorylation and dephosphorylation reactions and perform a parametric analysis to highlight the roles of the various parameters for attaining the insulation properties. In particular, we consider a two-step reaction model such as those in Huang and Ferrell (1996). According to this model, we have the following two reactions for phosphorylation and dephosphorylation, respectively:
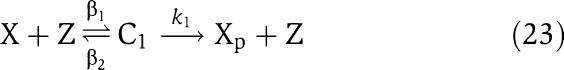 |
and
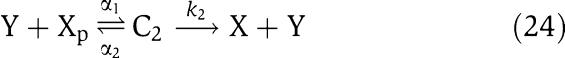 |
in which C1 is the (protein X/kinase Z) complex and C2 is the (phosphatase Y/protein Xp) complex. Additionally, we have the conservation equations YTOT=Y+C2, XTOT=X+Xp+C1+C2+C, because proteins X and Y are not degraded. Therefore, the differential equations modeling the insulation system of Figure 7 become
 |
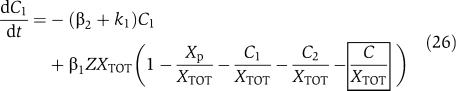 |
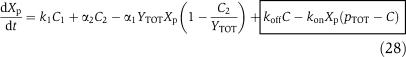 |
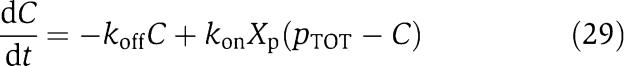 |
in which the expression of gene z is controlled by a promoter with activity k(t). The terms in the large box in equation (25) represent the retroactivity r to the input, while the terms in the small box in equation (25) and in the boxes of equations (26) and (28) represent the retroactivity s to the output. We assume that XTOT≫pTOT so that in equations (25) and (26) we can neglect the term C/XTOT because C<pTOT. Also, phosphorylation and dephosphorylation reactions in equations (23) and (24) can occur at a much faster rate (on the timescale of a second; Kholodenko et al, 2000) than protein production and decay processes (on the timescale of minutes; Alon, 2007). Choosing XTOT and YTOT sufficiently large, the separation of timescales between equation (25) and equations (26), (27), (28), and (29) can be explicitly modeled by letting ∈=δ/koff, kon=koff/kd, and by defining the new rate constants b1=β1XTOT∈/δ, a1=α1YTOT∈/δ, b2=β2∈/δ, a2=α2∈/δ, ci=∈ki/δ. Letting z=Z+C1 (the total amount of kinase) be the slow variable, we obtain the system in the standard singular perturbation form
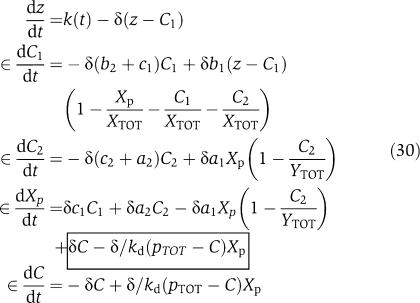 |
in which the boxed terms represent the retroactivity to the output s. We then compute the dynamics on the slow manifold by letting ∈=0. When we set ∈=0, the terms due to the retroactivity s vanish. This means that if the internal dynamics of the insulation device evolve on a timescale that is much faster than the dynamics of the input signal Z, then (provided we also have XTOT≫pTOT) the retroactivity s to the output has no effect on the dynamics of Xp at the quasi-steady state. This is a crucial feature of this design. Letting γ=(β2+k1)/β1 and 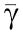 =(α2+k2)/α1, setting ∈=0 in the third and fourth equations of (30) the following relationships can be obtained:
=(α2+k2)/α1, setting ∈=0 in the third and fourth equations of (30) the following relationships can be obtained:
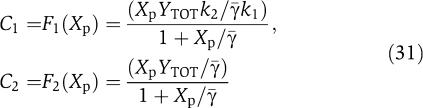 |
Using expressions (31) in the second of equation (30) with ∈=0 leads to
Assuming for simplicity that Xp≪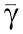 , we obtain that
, we obtain that 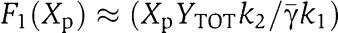 and that
and that 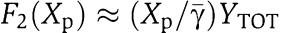 . As a consequence of these simplifications, equation (32) leads to
. As a consequence of these simplifications, equation (32) leads to
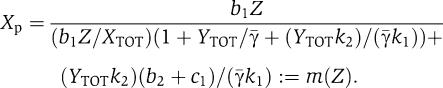 |
In order not to have distortion from Z to Xp, we require that
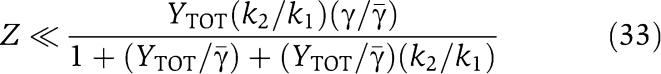 |
so that 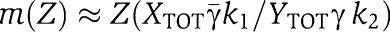 and therefore we have a linear relationship between Xp and Z with gain from Z to Xp given by
and therefore we have a linear relationship between Xp and Z with gain from Z to Xp given by 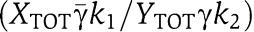 . In order not to have attenuation from Z to Xp, we require that the gain is greater than or equal to one, that is,
. In order not to have attenuation from Z to Xp, we require that the gain is greater than or equal to one, that is,
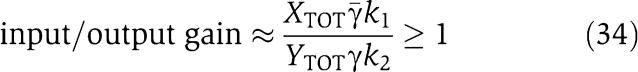 |
Requirements (33), (34), and Xp≪ are enough to guarantee that we do not have nonlinear distortion between Z and Xp and that Xp is not attenuated with respect to Z. To guarantee that the retroactivity r to the input is sufficiently small, we need to quantify the retroactivity effect on the Z dynamics due to the binding of Z with X. To achieve this, we proceed as in section Quantification of the effect of the retroactivity to the output by computing the Z dynamics on the slow manifold, which gives a good approximation of the dynamics of Z if ∈≈0. Such a dynamics is given by
are enough to guarantee that we do not have nonlinear distortion between Z and Xp and that Xp is not attenuated with respect to Z. To guarantee that the retroactivity r to the input is sufficiently small, we need to quantify the retroactivity effect on the Z dynamics due to the binding of Z with X. To achieve this, we proceed as in section Quantification of the effect of the retroactivity to the output by computing the Z dynamics on the slow manifold, which gives a good approximation of the dynamics of Z if ∈≈0. Such a dynamics is given by
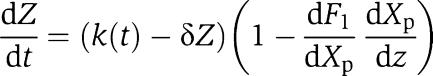 |
in which 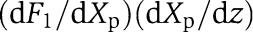 measures the effect of the retroactivity r to the input on the Z dynamics. Direct computation of dF1/dXp and of (dX/dZ) along with Xp≪
measures the effect of the retroactivity r to the input on the Z dynamics. Direct computation of dF1/dXp and of (dX/dZ) along with Xp≪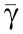 and with (33) leads to
and with (33) leads to 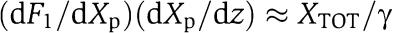 , so that to have small retroactivity to the input, we require that
, so that to have small retroactivity to the input, we require that
 |
In conclusion, for having attenuation of the effect of the retroactivity to the output s, we require that the timescale of the phosphorylation–dephosphorylation reactions is much faster than the production and decay processes of Z (the input to the insulation device) and that XTOT≫pTOT, that is, the total amount of protein X is in abundance compared to the downstream binding sites p. To obtain also a small effect of the retroactivity to the input, we require that γ≫XTOT as established by relation (35). This is satisfied if, for example, kinase Z has low affinity to bind with X. To keep the input–output gain between Z and Xp close to one (from equation (34)), one can choose XTOT=YTOT, and equal coefficients for the phosphorylation and dephosphorylation reactions, that is, γ=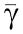 and k1=k2.
and k1=k2.
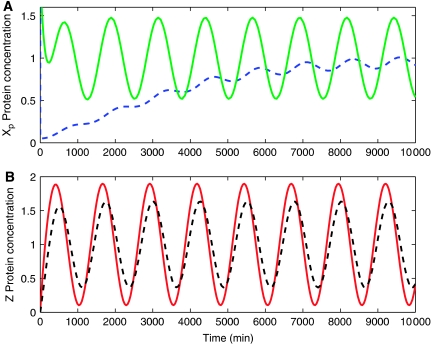
Simulation results. System in equations (25), (26), (27), (28) and (29) was simulated with and without the downstream binding sites p, that is, with and without, respectively, the terms in the small box of equation (25) and in the boxes in equations (26) and (28). This is performed to highlight the effect of the retroactivity to the output s on the dynamics of Xp. The simulations validate our theoretical study that indicates that when XTOT≫pTOT and the timescales of phosphorylation–dephosphorylation are much faster than the timescale of decay and production of the protein Z, the retroactivity to the output s is very well attenuated (Figure 8A). Similarly, the time behavior of Z was simulated with and without the terms in the large box in equation (25), that is, with and without X to which Z binds, to verify whether the insulation component exhibits retroactivity to the input r. In particular, the accordance of the behaviors of Z(t) with and without its downstream binding sites on X (Figure 8B) indicates that there is no substantial retroactivity to the input r generated by the insulation device. This is obtained because XTOT≪γ as indicated in equation (35), in which 1/γ can be interpreted as the affinity of the binding of X to Z. Our simulation study also indicates that a faster timescale of the phosphorylation–dephosphorylation reactions is necessary, even for high values of XTOT and YTOT, to maintain perfect attenuation of the retroactivity to the output s and small retroactivity to the output r. In fact, on slowing down the timescale of phosphorylation and dephosphorylation, the system looses its insulation property (Figure 9). In particular, the attenuation of the effect of the retroactivity to the output s is lost because there is not enough separation of timescales between the Z dynamics and the internal device dynamics. The device also displays a non-negligible amount of retroactivity to the input because the condition γ≪XTOT is not satisfied anymore.
Figure 8.
Simulation results for system in equations (25–29). In all plots, pTOT=100, koff=kon=10, δ=0.01, k(t)=0.01(1+sin(ωt)), and ω=0.005. (A, B), k1=k2=50, α1=β1=0.01, β2=α2=10, and YTOT=XTOT=1500. (A) The signal Xp(t) without the downstream binding sites p is in green (solid line), while the same signal with the downstream binding sites p is in blue (dashed line). The small error shows that the effect of the retroactivity to the output s is attenuated very well. (B) The signal Z(t) without X to which Z binds is in red (solid), while the same signal Z(t) with X present in the system (XTOT=1500) is in black (dashed line). The small error confirms a small retroactivity to the input. The values of the complexes concentrations C1 and C2 oscillate about 0.4, so they are comparable to the values of Xp.
Figure 9.
In all plots, pTOT=100 and koff=kon=10, δ=0.01, k(t)=0.01(1+sin(ωt)), and ω=0.005. Phosphorylation and dephosphorylation rates are slower than the ones in Figure 8, that is, k1=k2=0.01, while the other parameters are left the same, that is, α2=β2=10, α1=β1=0.01, and YTOT=XTOT=1500. (A) The signal Xp(t) without the downstream binding sites p is in green (solid line), while the same signal with the downstream binding sites p is in blue (dashed line). The effect of the retroactivity to the output s is dramatic. (B) The signal Z(t) without X in the system is in red (solid line), while the same signal Z(t) with X in the system is in black (dashed line). The device thus also displays a large retroactivity to the input r.
Discussion
The notion of retroactivity has generally been defined as a signal traveling back from a downstream system to its upstream system(s) upon interconnection. Retroactivity is the quantity that changes the behavior of an upstream system when its output is connected to downstream clients. ‘Downstream' and ‘upstream' refer to the direction in which we imagine a signal traveling, i.e. from its source to its clients. This retroactivity definition models all the effects that may change the input–output behavior of a module when it is input–output connected to other modules. The definition is general, and can be formulated as a type of disturbance rejection problem in control theory (see Supplementary Information). More specifically, we suggest a concrete approach to the quantification of the amounts of retroactivity (section Quantification of the effect of the retroactivity to the output), which applies to all systems in which the dynamics internal to a module and in particular the dynamics of the output of a module evolves on a slower timescale when compared to the dynamics of the interconnection process. That is, it applies whenever the dynamics of the output stage of a module is much slower than the dynamics of the input stage of a downstream module. This property is satisfied in a number of systems, and in particular in gene transcriptional networks. In fact, the internal dynamics of a module involves protein production and decay processes (generally with a timescale of minutes; Alon, 2007), which are slow when compared to the dynamics of protein–protein binding/unbinding (often with a subsecond timescale; Shen-Orr et al, 2002) and to the dynamics of protein promoter binding/unbinding (with a timescale of a second; Alon, 2007). Using this approach for transcriptional networks, we have provided a formula (expression (7)), which is a measure of retroactivity and holds at the steady state, during the transient, and during permanent non-constant behavior, such as oscillatory behavior. Such a formula is operationally useful because it can be evaluated just on the basis of measurable biochemical parameters such as association and dissociation rates, concentration of binding sites, and dynamic range of the (time varying) concentration of the protein interconnecting two modules. For large-scale transcriptional networks, such a formula does not need to be re-derived from scratch for each interconnection. In fact, expression (7) is the same for every interconnection in the network: one only needs to evaluate it for every interconnection, once the biochemical parameters and signal values characterizing the interconnection under consideration are known. In section Quantification of the effect of the retroactivity to the output, the quantification of the retroactivity effect has been obtained for transcriptional networks in which module interaction occurs at the transcriptional level. This procedure can be applied to obtain a formula similar to equation (7) also for the case of signal transduction networks, in which interconnection between modules occurs through protein–protein interaction. While we have left the general derivation of the retroactivity effect for signal transduction networks to future work, we have shown how to derive it in the example of insulation device involving phosphorylation cycles.
For transcriptional networks, sufficient conditions for a small retroactivity are low affinity of the regulatory protein to its binding sites in the downstream system and/or having a small number of binding sites in the downstream system compared to the amount of protein. These two conditions imply, using the electrical analogy, a small flow (or current) through the interconnection from an upstream system to a downstream one. Numerous natural genetic systems, however, utilize small numbers of regulatory molecules, such that the concentration of the binding sites, relative to the number of regulatory protein molecules, is not negligible (Reitzer and Magasanik, 1983; Yildirim and Mackey, 2003). It is thus often the case that retroactivity is not negligible, and hence modular analysis of the behavior of the interconnection is not possible. Instead of considering the interconnection of the two components as a larger module or considering a different input–output partitioning of the network to minimize retroactivity of the interconnection as proposed by several researchers (see for example, Bruggeman et al, 2002; Saez-Rodriguez et al, 2005), we have proposed (from a synthetic biology perspective) to place suitable insulation devices between components so that the behavior of the components is not altered upon interconnection. This allows modular analysis. In general, for the correct (according to some criterion) functioning of a natural system, it is not necessary that the retroactivity at each interconnection is small, because parts of the system may have been finely tuned to work well with each other in a specific interconnection configuration. From the point of view of understanding a large natural system or from the point of view of building one from small modules, it is desirable to have low retroactivity at the interconnections or to have some way to attenuate its effect (especially for the synthetic case). Whether there are naturally occurring systems that have been designed so that retroactivity at the interconnections is small (or has a small effect) and whether this insulation feature is necessary for the functioning of that specific system are challenging questions, which we plan to explore in the future.
We have illustrated a general solution to the problem of attenuating retroactivity upon interconnection of modules. This general solution, inspired by the design of electronic non-inverting amplifiers, relies on large input signal amplification and on a similarly large output negative feedback. It has been considered before in the context of signaling networks by Sauro and Kholodenko (2004) and Sauro and Ingalls (2007). In this paper, we have proposed two concrete realizations of such a general solution for gene transcriptional components and have mathematically and computationally shown the properties of these realizations with respect to retroactivity. The first solution (design 1), relies on a strong, non-leaky, promoter for input amplification and on an enhanced protein degradation through a protease as a negative feedback mechanism. The second solution (design 2) relies on protein phosphorylation as amplification mechanism and on protein dephosphorylation as a negative feedback mechanism. From the point of view of synthetic biology, the analysis performed for both designs provides a number of conditions on the biochemical parameters of the proposed device, which have to be satisfied for obtaining an insulator. Such conditions can be checked in practice once the biochemical parameters are known and thus they provide a design guideline for fabrication.
The remarkable ability of the phosphorylation–dephosphorylation cycle (design 2) to provide insulation relies on the relatively rapid timescale of these reactions in comparison to the protein production and decay processes. This was mathematically shown by the employment of singular perturbation analysis and confirmed by simulation study (Figures 8 and 9). By comparison, the relatively slower rates of protein synthesis and decay in design 1 limit the insulation capacity, unless the gain of the system is so large that it may be hardly realizable in vivo (Figure 6). It is not surprising that protein phosphorylation and dephosphorylation constitutes the most common type in signal transduction systems in nature. It has long been realized that the metabolic cost of phosphoryl groups is low, relative to the cost of protein synthesis and degradation and to other types of covalent protein modification, that the rates of protein phosphorylation and dephosphorylation can be very rapid (Kholodenko et al, 2000), and that the chemical stabilities of several types of phosphorylated amino acids are suitable for in vivo signaling functions (Voet and Voet, 2004). Here, we argue that another beneficial feature of covalent modification as a signal transduction system is an inherent capacity to provide insulation and thus to increase the modularity of the system in which it is placed.
Materials and methods
All simulations are performed in MATLAB (Simulink), Version 7.0.1, with variable step ODE solver ODE23s. Simulink models are available upon request.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Peter Woolf and Richard M. Murray for discussions that were very relevant to the development of this work. We also thank the reviewers for providing additional references and for their suggestions that improved the content and the readability of the paper. This work was supported in part by NSF grant DMS-0614371, in part by grant R01GM063642 from the NIGMS, and in part by the Rackham Faculty Research Grant from University of Michigan.
References
- Acerenza L, Sauro H, Kacser H (1989) Control analysis of time-dependent metabolic systems. J Theor Biol 151: 423–444 [DOI] [PubMed] [Google Scholar]
- Alon U (2003) Biological networks: the tinkerer as an engineer. Science 301: 1866–1867 [DOI] [PubMed] [Google Scholar]
- Alon U (2007) An Introduction to Systems Biology. Design Principles of Biological Circuits. Chapman-Hall/CRC Taylor & Francis, Boca Raton, FL [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2: 1–14 10.1038/msb4100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH (1998) Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149: 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthagiri AR, Lauffenburger DA (2000) Bioengineering models of cell signaling. Annu Rev Biomed Eng 2: 31–53 [DOI] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Meyers JT, Ninfa AJ (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113: 597–607 [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV, Hoek JB, Kholodenko BN (2002) Modular response analysis of cellular regulatory networks. J Theor Biol 218: 507–520 [PubMed] [Google Scholar]
- Ciliberto A, Capuani F, Tyson JJ (2007) Modeling networks of coupled enzymatic reactions using the total quasi-steady state approximation. PLoS Comput Biol 3: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demin O, Westerhoff H, Kholodenko B (1999) Control analysis of stationary fixed oscillations. J Phys Chem B 103: 10695–10710 [Google Scholar]
- Endy D (2005) Foundations for engineering biology. Nature 438: 449–452 [DOI] [PubMed] [Google Scholar]
- Fell D (1992) Metabolic control analysis a survey of its theoretical and experimental development. Biochem J 286: 313–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L, Hopfield J, Leibler S, Murray A (1999) From molecular to modular cell biology. Nature 402: 47–52 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Neel BG, Rapoport TA (2002) Mathematical models of protein kinase signal transduction. Mol Cell 9: 957–970 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Schuster S (1996) The Regulation of Cellular Systems. New York: Chapman & Hall [Google Scholar]
- Hofmeyr SJ (1997) Anaerobic energy metabolism in yeast as a supply–demand system. In New Beer in an Old Bottle: Eduard Buchner and the Growth of Biochemical Knowledge, Cornish-Bowden A (ed), pp 225–242. Valencia, Spain: Universitat de Valencia [Google Scholar]
- Hofmeyr SJ, Cornish-Bowden A (2000) Regulating the cellular economy of supply and demand. FEBS Lett 476: 47–51 [DOI] [PubMed] [Google Scholar]
- Huang CF, Ferrell JE (1996) Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA 93: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls B, Sauro H (2003) Sensitivity analysis of stoichiometric networks: an extension of metabolic control analysis to non-steady state trajectories. J Theor Biol 222: 23–36 [DOI] [PubMed] [Google Scholar]
- Jacob F (1977) Evolution and tinkering. Science 196: 1161–1166 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Brown GC, Hoek JB (2000) Diffusion control of protein phosphorylation in signal transduction pathways. Biochem J 350: 901–907 [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW, Gerhart JC (2005) The Plausibility of Life: Resolving Darwin's Dilemma. New Haven and London: Yale University Press [Google Scholar]
- Kokotovic P, Khalil HK, O'Reilly J (1999) Singular Perturbation Methods in Control. PHILADELPHIA: SIAM, Classics in Applied Mathematics, vol 25 [Google Scholar]
- Kremling A, Saez-Rodriguez J (2007) Systems biology—an engineering perspective. J Biotechnol 129: 329–351 [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA (2000) Cell signaling pathways as control modules: complexity for simplicity? Proc Natl Acad Sci USA 97: 5031–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason O, Verwoerd M (2006) Graph theory and networks in biology. Tech. Rep., http://arxiv.org/abs/q-bio.MN/ 0604006 [DOI] [PubMed]
- Papin JA, Reed JL, Palsson BO (2004) Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem Sci 29: 641–647 [DOI] [PubMed] [Google Scholar]
- Polderman JW, Willems JC (2007) Introduction to Mathematical Systems Theory. A Behavioral Approach, 2nd edn. New York: Springer-Verlag [Google Scholar]
- Reitzer LJ, Magasanik B (1983) Isolation of nitrogen assimilation regulator NRI, the product of the glnG gene of Escherichia coli. Proc Natl Acad Sci USA 80: 5554–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubertis GD, Davies SW (2003) A genetic circuit amplifier: design and simulation. IEEE Trans Nanobioscience 2: 239–246 [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Kremling A, Conzelmann H, Bettenbrock K, Gilles ED (2004) Modular analysis of signal transduction networks. IEEE Contr Syst Mag 24: 35–52 [Google Scholar]
- Saez-Rodriguez J, Kremling A, Gilles E (2005) Dissecting the puzzle of life: modularization of signal transduction networks. Comput Chem Eng 29: 619–629 [Google Scholar]
- Sauro H (2004) The computational versatility of proteomic signaling networks. Curr Proteomics 1: 67–81 [Google Scholar]
- Sauro HM, Ingalls B (2007) MAPK cascades as feedback amplifiers. Tech. Rep., http://arxiv.org/abs/0710.5195
- Sauro HM, Kholodenko BN (2004) Quantitative analysis of signaling networks. Prog Biophys Mol Biol 86: 5–43 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1976) Biochemical Systems Analysis. A Study of Function and Design in Molecular Biology. Reading, MA: Addison-Wesley [Google Scholar]
- Schilling DL, Belove C (1968) Electronic Circuits: Discrete and Integrated. New York, McGraw Hill [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA (2002) The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA 99: 5890–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E (1998) Mathematical Control Theory. New York: Springer-Verlag [Google Scholar]
- Voet D, Voet JG (2004) Biochemistry, 3rd edn. Hoboken, NJ: John Wiley and Sons [Google Scholar]
- Willems J (1999) Behaviors, latent variables, and interconnections. Systems, Control and Information 43: 453–464 [Google Scholar]
- Yildirim N, Mackey MC (2003) Feedback regulation in the lactose operon: a mathematical modeling study and comparison with experimental data. Biophys J 84: 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information