Abstract
One of the most widely used transgenic animal models in biology is Drosophila melanogaster, the fruit fly. Chemical information from this exceedingly small organism is usually accomplished by studying populations to attain sample volumes suitable for standard analysis methods. This paper describes a direct sampling technique capable of obtaining 50–300 nL of hemolymph from individual Drosophila larvae. Hemolymph sampling performed under mineral oil and in air at 30 s intervals up to 120 s after piercing larvae revealed that the effect of evaporation on amino acid concentrations is insignificant when the sample was collected within 60 s. Qualitative and quantitative amino acid analyses of obtained hemolymph were carried out in two optimized buffer conditions by capillary electrophoresis with laser-induced fluorescence detection after derivatizing with fluorescamine. Thirteen amino acids were identified from individual hemolymph samples of both wild-type (WT) control and the genderblind (gb) mutant larvae. The levels of glutamine, glutamate, and taurine in the gb hemolymph were significantly lower at 35%, 38%, and 57% of WT levels, respectively. The developed technique that samples only the hemolymph fluid is efficient and enables accurate organism-level chemical information while minimizing errors associated with possible sample contaminations, estimations, and effects of evaporation compared to the traditional hemolymph-sampling techniques.
Drosophila melanogaster, commonly known as the fruit fly, is an extensively used model species especially in genetics, neuroscience, and developmental biology. The major reasons for its popularity as a transgenic model is its relative biological simplicity, its sequenced genome, and the ease of breeding and maintaining large stocks in the laboratory. The importance of Drosophila as a neurodegenerative disease model in understanding disease pathogenesis and drug development has been highlighted recently.1-4 Amino acids such as glutamate (Glu), glutamine (Gln), and taurine (Tau) are known as important neurotransmitters or may play related functions in neurodegenerative disorders, and Drosophila has also been used in investigating tau-related neurodegenerative diseases.5-8 Chemical information including amino acids of Drosophila samples has drawn much attention over the last 4 decades.3,9-13 A recent discovery of a cystine–glutamate transporter (xCT) protein mutant has demonstrated significant behavioral effects and differential Glu receptor localizations compared to the control genotypes highlighting the importance of the determination of hemolymph amino acids of Drosophila larvae.13 Analytical methods to address the basal chemical content in Drosophila individuals are important characterization tools, and such techniques may also allow better insight into phenotypic anatomy and behavior of important transgenic animals. However, the small size of the organism has limited the analyses that can be performed to obtain chemical content information from individuals, and therefore, populations of fruit flies have been employed to obtain samples for chemical analyses.9,12,13
Most commonly, several fruit flies or larvae are homogenized together to obtain a large enough sample, about 100 μL, for the chemical analyses through high-performance liquid chromatography (HPLC).9 Due to the importance of attaining organism-level chemical information a recent study described a homogenization method for individual fly-head samples for capillary electrophoresis (CE) separations.14 In contrast to HPLC, the small volume compatibility of capillary electrophoresis along with features such as high separation efficiency, fast analysis, and low operation costs have made it a powerful separation technique, popular especially for biological sample analyses.15,16
The method of homogenization, however, is not ideal for fruit-fly hemolymph sampling. Hemolymph is a distinct biological fluid such that homogenization of tissue leads to undesirable dilution and contamination from the cellular material of the whole organism and from the contents of the gastrointestinal tract. There may be alterations for in vivo hemolymph amino acids by release of the protease enzymes from lysed cells. Also, the traditional homogenization technique is time-consuming and the addition of chemical treatments such as methanol and trichloroacetic acid could alter the native chemical information. In contrast to homogenization, Augustin et al.13 extracted hemolymph from a population by incubating larvae with pierced cuticles in a saline solution.13 They compared the hemolymph Glu levels of a control genotype and a mutant genotype that aided in elucidating the effect of an altered glutamate–cystine exchange protein gene.13 However, this quantitation method is problematic because the exact amount of leaked-hemolymph is not known and also the larval incubation in saline solution may alter the amino acid content.
This paper describes the development of a sampling technique to obtain in vivo hemolymph samples of 50–300 nL volumes from individual Drosophila larvae. The amino acid content is determined qualitatively and quantitatively via capillary electrophoresis with the laser-induced fluorescence detection (CE-LIF). The significance of hemolymph evaporation during open-air sampling has been assessed and compared to no evaporation methods. The developed sampling and analysis method that allows the comparison of individual fruit-fly hemolymph samples is shown for two fruit-fly genotypes.
EXPERIMENTAL SECTION
Materials
All chemicals used were of analytical grade or better. HPLC-grade acetone, mineral oil, sodium tetraborate decahydrate, cesium chloride, histamine dihydrochloride, and γ-amino-n-butyric acid (GABA) were purchased from Sigma (Sigma-Aldrich, St. Louis, MO). All other amino acids, fluorescamine, sodium dodecyl sulfate (SDS), and sodium hydroxide were obtained from Fisher Scientific (Itasca, IL). A US Filter Purelab Plus purification system (Lowell, MA) was used to obtain ultrafiltered-deionized water for preparing all solutions. Untreated 50 μm i.d. fused-silica capillary (360 μm o.d.) was purchased from Biotaq Inc. (Gaithersburg, MD), and 250 μm and 20.6 mm i.d. Tygon tubes were purchased from Cole-Parmer (Vernon Hills, IL).
A 50 mM borate stock solution prepared with sodium tetraborate decahydrate was further diluted to prepare the 20 mM borate run buffer. The pH of the borate buffer was adjusted to 9.1 with 0.5 M NaOH. Stock solutions containing 100 mM SDS were prepared by dissolving SDS in 20 mM borate buffer. The optimized MEKC run buffer contained 30 mM CsCl and 70 mM SDS in 20 mM borate solution. All buffer solutions were sonicated for 5 min prior to use. Fluorescamine was dissolved in acetone to prepare the 15 mg L−1 solution for derivatization. All standard amino acid solutions were prepared in the 20 mM borate buffer solution and diluted appropriately with the run buffer to the desired concentrations.
Sampling Probe Construction
A 60 cm long, 250 μm i.d. Tygon-tube piece was inserted into an 8 cm long 20.6 mm i.d. Tygon-tube housing such that about 1 cm of the 250 μm Tygon tube was cleared from one end (Figure 1A). The other end of the 250 μm Tygon tube was connected to a vacuum pump (Barnant Co., Barrington, IL), a regulator (Squire-Cogswell, Gurnee, IL), and a digital pressure gauge (Vacuubrand DVR 2, Wertheim, Germany). An electrode holder (World Precision Instruments Inc., FL) was used to hold both Tygon tubes as shown in Figure 1A. The whole sampling probe was then mounted on a three-dimensional micropositioner (model Tauruser-B, World Precision Instruments Inc., FL) that facilitated the movement of sampling probe into focus on the surface of a dissection plate placed under a dissection microscope (Leica MZ6, Heerbrugg, Switzerland).
Figure 1.
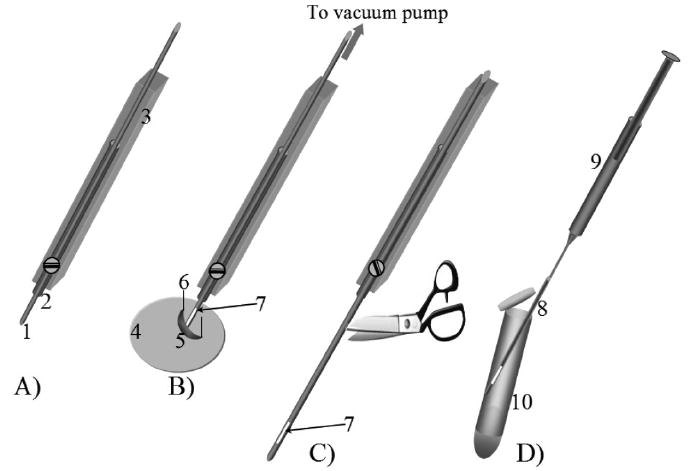
Schematic diagram of the sampling technique. (A) Sampling probe: 1, 250 μm i.d. Tygon tube; 2, 20.6 mm i.d. Tygon tube housing; 3, electrode holder. (B) Hemolymph sampling: 4, microdissection plate; 5, larvae; 6, microdissection pin; 7, hemolymph. (C) Sample-containing Tygon pulled out to cut: 7, hemolymph sample. (D) Transferring the hemolymph sample: 8, Tygon piece with the hemolymph sample; 9, 1 mL syringe connected to the Tygon piece; 10, 250 μL centrifuge tube with the required volume of borate buffer for dilution.
Sampling and Derivatization
Third instar Drosophila wild-type (WT) larvae, based on Oregon R strain, and genderblind (gb) mutants were reared on standard cornmeal–agar medium and maintained in the Department of Biology at UIC. Individual larva were placed on a paper towel and cleaned by rinsing with a saline solution followed by air-drying for 2 min. The dry larva was carefully pinned to a microdissection dish at its anterior and posterior ends with microdissection pins (Figure 1B), and a narrow incision was made through the cuticle with a third pin while visually observing through a dissection microscope. The leaking hemolymph was colleted into the 250 μm Tygon tube (Figure 1B) by applying a vacuum of 15 Torr below the atmospheric pressure. The 250 μm Tygon tube with the sample was then pulled out until 4 cm was cleared from the larger i.d. Tygon tube housing and cut carefully (Figure 1C). The volume of the collected sample, in the Tygon tube, was determined by measuring the fluid length as described by Kottegoda et al.17 where 1 mm of fluid distance is calculated to be 50 nL. As shown in Figure 1D a 1 mL syringe (Becton Dickinson and Co., Franklin, NJ.) was connected to the sample-carrying Tygon tube piece to transfer the hemolymph into a 250 μL centrifuge tube (Fisher Scientific, Itasca, IL) that contained the required volume of 20 mM borate buffer for dilution. The diluted hemolymph sample was then frozen at −20 °C until analysis. An amino acid standard mixture of known concentrations was also sampled using the above technique to assess the variations of the measured concentrations incurred during the sampling and the analyses processes. Equal volumes (4 μL) of fluorescamine solution and amino acid standards or diluted hemolymph were mixed together in a 250 μL microcentrifuge tube (Fisher Scientific, Itasca, IL) to derivatize the amino acids. Hemolymph samples collected after 180 s of piercing the larval body cavity were diluted 250, 500, and 750 times with 20 mM borate buffer and derivatized with 15 mg mL−1 fluorescamine solution to verify complete derivatization of amino acids. Three standard cornmeal–agar medium (fruit-fly food) samples were weighed and dissolved in borate buffer, and the filtrate was obtained for the analyses.
Assessing the Effect of Evaporation
All the third instar larvae employed to study the effect of evaporation were of the WT genotype. Only one hemolymph sample can be collected from each larva, and the shortest time interval between piercing the larval body cavity and sample collection was 30 s. Therefore, to assess the effect of evaporation on the hemolymph amino acid concentrations, samples were collected at 30 s intervals up to 2 min after piercing. Dissections were also done under mineral oil to prevent hemolymph evaporation. A cleaned–dry WT larva was immersed in a drop of mineral oil, pinned to the dissection plate, and the body cavity was pierced as described previously. Thirty seconds after the piercing, the leaked-hemolymph, which often forms a thin layer between the wiggling larvae and the dissection plate, is collected into the Tygon tube probe as explained in the previous section. The aqueous hemolymph and mineral oil are immiscible and distinguishable within the Tygon tube, and the length of hemolymph was measured to find its volume. The content of the whole Tygon tube was then transferred into a 250 μL centrifuge tube and diluted with the 20 mM borate buffer. After mixing and centrifuging, the sample was left to settle for about 10 min in the closed centrifuge tube. The aqueous phase was then collected by piercing the bottom of the centrifuge tube with a 1 mL syringe to withdraw the diluted sample.
Capillary Electrophoresis
The chemical analyses were carried out on a home-built CE system equipped with a commercial high-voltage power supply (Spellman, CZE 1000R, Hauppage, NY) and photomultiplier detector (H7421-50 Hamamutsu corp., Japan) operating with a diode laser (TECBL-10G-405, World Star Tech., Canada) at 405 nm. The excitation laser light was filtered through a narrow band-pass filter (Newport 10BPF10-410) followed by a broad-band (Edmund optics NT46-155) filter, and the fluorescence was separated from excitation light using a long-pass filter (Omega optical Inc. XF3088) and a broad-band filter (Edmund optics NT46-150). The high-voltage power supply, separation time, and the data acquisition were controlled through an interface board (E 6229, National Instruments, Austin, TX) by a custom LabView (National Instruments, Austin, TX) program. Untreated 50 μm i.d., 50 cm fused-silica capillary with a 1 cm detection window at the effective length of 36 cm was utilized for separation. Initially, the capillary was conditioned by rinsing with deionized water, followed by 0.5 M sodium hydroxide and deionized water, respectively. Prior to each injection, the capillary was rinsed with 0.5 M sodium hydroxide and then with run buffer. The derivatized samples or the standards were injected into the separation capillary by gravity for 15 s at a 15 cm displacement. The optimized applied potentials for the 20 mM borate and the MEKC run buffers were 30 and 19 kV, respectively. Each sample or the standard was analyzed in triplicate. The analyte peaks of the hemolymph were identified by spiking with 24 different amino acid standards. Quantification of amino acids was performed with external calibration curves of standard amino acids after verification with the method of standard additions.
Data Analyses
The raw data were imported to Microsoft Excel to plot electropherograms and to perform quantitation and statistical analysis. Statistical comparisons of peak heights, sample volumes, and amino acids concentrations carried out with Microsoft Excel involved initial testing of the variances of the two selected data sets with the F test. Then, depending upon the homoscedastic or heteroscedastic nature of the subjected data sets, two-tailed Student’s t test was performed on them with a confidence level of 95% (P < 0.05) to determine whether the means are significantly different from each other. Mean values reported are followed by the corresponding standard deviations (±).
RESULTS AND DISCUSSION
Identification of Amino Acids in Hemolymph of WT and gb Mutant Larvae
The average hemolymph volume collected from individual third instar larvae was 174 nL (±81, n = 80), and the average weight of the used larvae was 1.39 mg (±0.15). Previous studies have shown differences in chemical contents for different stages of the Drosophila life cycle.9 The size range for our studies shows that the third instar Drosophila larvae were carefully selected for the analyses. The variation in the volume of collected hemolymph is likely depended on the variation of incision of the cuticle. The larval body cavity was pierced only once to obtain leaked-hemolymph, and the samples were collected within the desired time interval (30, 60, 90, and 120 s). There was no attempt to keep the sample volumes consistent. The leaked-hemolymph volumes were also independent of the genotypes (P < 0.05) with the average WT and gb mutant sample volumes as 178 nL (±84, n = 69) and 165 nL (±43, n = 12), respectively. Sample handling, especially those with lower absolute volumes, was facilitated by 250-fold dilution immediately after the collection. Figure 2A is an electropherogram of 250-fold diluted WT hemolymph obtained with a 20 mM borate buffer (pH 9.1). Nine individual amino acids were resolved, and the electropherogram also shows a number of unidentified peaks including a large peak migrating after aspartate (Asp). Separation of hemolymph from the gb mutant larvae in the 20 mM borate buffer is shown in Figure 2B, and for the both larvae types the qualitative peak patterns were the same.
Figure 2.
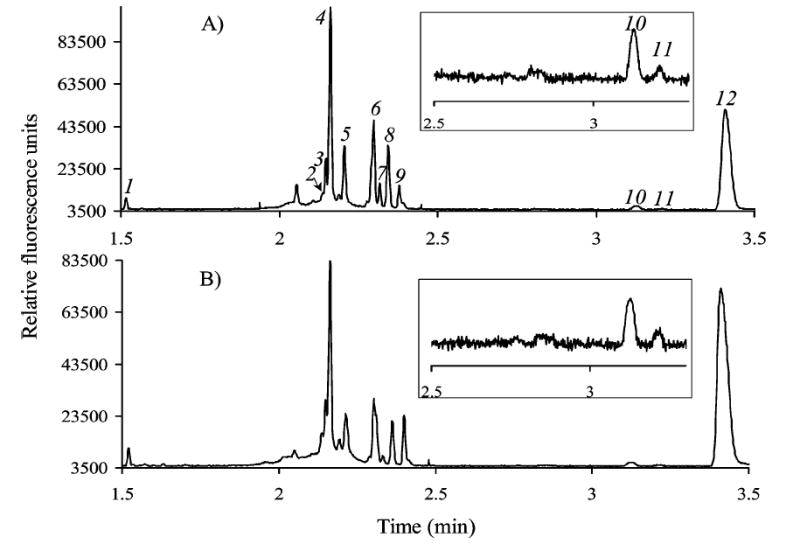
Electropherograms for separation of fluorescamine-labeled amino acids in hemolymph of (A) wild-type and (B) gb mutants in 20 mM borate buffer (pH 9.1) at 30 kV in a 50 cm bare fused-silica capillary with 50 μm i.d. and 360 μm o.d. 1, arginine; 2, tyrosine; 3, histidine; 4, glutamine; 5, asparagine and threonine; 6, alanine and serine; 7, taurine; 8, lysine; 9, glycine; 10, glutamate; 11, aspartate; 12, unknown.
It has been demonstrated that univalent cations with smaller hydrodynamic radii can be utilized to resolve hydrophobic amino acids in buffers with anionic surfactants.18 In this study Cs+ is used as the univalent modifier for the MEKC buffer system. Figure 3 shows the separation of WT hemolymph in the MEKC run buffer that resolved leucine from phenylalanine and serine from alanine that comigrated in the 20 mM borate buffer. In the MEKC run buffer GABA comigrated with threonine, and three amino acids, asparagine, isoleucine, and methionine, migrated together. Arginine (Arg), Gln, glycine (Gly), Glu, and histidine were resolved in both run buffers. The separation of gb hemolymph in the MEKC run buffer showed similar results. Previous studies based on pooled homogenized fruit-fly samples and pooled hemolymph samples have reported the presence of the above amino acids.9,11,19 It is noted that the large unknown peak that eluted after Asp remained a single component for all the samples in all utilized buffers. This peak may be a peptide component. Other studies of homogenized Drosophila samples have also reported peptide peaks in the acidic region.9,10
Figure 3.
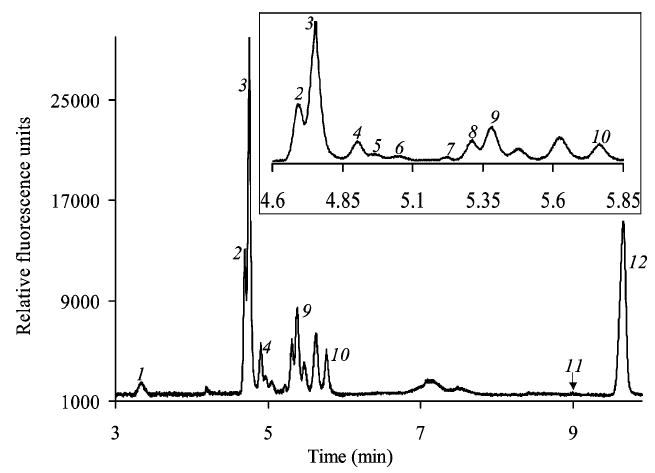
Electropherogram for separation of fluorescamine-labeled amino acids of wild-type hemolymph in 20 mM borate, 70 mM SDS, and 30 mM CsCl, MEKC run buffer (pH 9.1) at 19 kV in a 50 cm bare fused-silica capillary with 50 μm i.d. and 360 μm o.d. 1, arginine; 2, histidine; 3, glutamine; 4, asparagine, isoleucine, and methionine; 5, GABA and threonine; 6, leucine; 7, phenylalanine; 8, serine; 9, alanine; 10, glycine; 11, glutamate; 12, unknown.
Analyses of more concentrated hemolymph samples such as 50- and 100-fold diluted did not reveal amino acids other than the ones identified from the 250-fold diluted samples. The separation resolution and speed were not improved with the pH 9.5 run buffers as has been reported by others for fluorescamine-labeled amino acids.20-22 A higher potential could be applied for the hemolymph separations in the 20 mM borate buffer to reduce separation time to 3.5 min compared to that of the MEKC buffer separation which took 9.9 min. Therefore, the 20 mM borate buffer was employed for the quantitation of the seven major amino acids of hemolymph.
Analyses of fly food samples in both buffers showed that it does not contain Tau and the component that eluted after Asp for the hemolymph samples. The observed concentrations of Arg, Glu, Gln, lysine (Lys), Gly, and Asp in fly food were about 3 orders of magnitude lower compared to those of previously published hemolymph amino acid levels.9,13 Although these findings suggested that the effect of food contamination of hemolymph would not contribute significantly to hemolymph amino acid levels, larval body cavity was pierced carefully, by visual observation through the dissection microscope, to avoid damaging its gastrointestinal tract, or other internal organs.
Sample Dilution and Fluorescamine Concentration
Previously reported hemolymph amino acid levels were in the millimolar range9,13 and Figures 2 and 3 show unidentified peaks those could be due to other proteins or peptides of hemolymph. Hence, the amount of fluorescamine utilized was assessed to explore the extent of derivatization. The average net peak heights of Arg, Glu, and Gln for the 250-fold diluted samples in relative fluorescence units (RFU) were 1.3 × 104, 8.4 × 103, and 6.0 × 104, respectively, with a relative standard deviation below 5%. Reactions performed with the same mass of derivatizing reagent with 500- and 700-fold diluted samples showed peak heights that were about 51% and 68% less, respectively, than the 250-fold diluted hemolymph peaks. The decreasing ratio of the peak heights correlated with the dilution factors suggesting that the extent of derivatization is about equivalent in all samples. Therefore, 15 mg mL−1 fluorescamine was used with 250-fold diluted hemolymph for further study. Quantitation via calibration curves is relatively simple and was used in this study. The method of standard additions was compared to the external calibration curves, and no significant matrix effects were observed for the amino acids with 250-fold diluted samples.
Effect of Evaporation on the Amino Acid Concentrations
The effect of evaporation on the hemolymph amino acid concentrations was assessed due to the fact that nanoscale sample volumes (174 ± 81 nL) were collected while they were exposed to the atmosphere. Shown in Table 1 are the average net peak heights for six amino acids obtained from electropherograms for samples collected 30, 60, 90, and 120 s after piercing the larval body cavity. The average values for the 30 s interval, the shortest possible sample collection period, is based on hemolymph samples from 17 individual larvae, and the average value for each other time period is based on 10 individual animals. The average peak heights of Arg, Glu, Gly, Lys, and Tau showed a significant increase of over 31% (P < 0.05) when the time gap between piercing and sampling was increased from 30 s to 90 s and above (Table 1). A significant increase of 36% in the Gln peak height (P < 0.05) was observed when the time gap was increased up to 120 s. However, Table 1 shows no significant difference in the mean peak heights of the 30 and 60 s time interval samples (P < 0.05) with respect to all six amino acids. These results suggest that the effect of evaporation on the amino acid concentrations of the hemolymph is insignificant when the sample collection is completed before 90 s of piercing. Accordingly, hemolymph was collected within a very short time period, 30 s, after piercing the larvae. The possible effect of evaporation during the first 30 s of the sampling process was also assessed due to the extremely small sample volume. Figure 4 shows the average amino acid concentrations for individual hemolymph samples obtained from 10 larvae while they were under mineral oil and for 17 individual samples while they were exposed to atmosphere. In both approaches the sampling process commenced 30 s after making the narrow cut in the larval body cavity for hemolymph withdrawal to minimize the hemolymph exposure to atmosphere in dry sampling. The average concentrations obtained from two sampling techniques were not significantly different for Gln, Tau, Lys, and Gly suggesting that there is no significant effect of evaporation compared to the samples collected under zero evaporation.
Table 1.
Average Net Peak Heights with the Standard Deviations (±) for Six Amino Acids Obtained from the Electropherograms of the Samples Collected 30, 60, 90, and 120 s after Piercing the Larval Body Cavitya
| time since piercing the body cavity (s)
|
||||
|---|---|---|---|---|
| 30b | 60c | 90c | 120c | |
| Arg | 6.3 (±2.0) × 103 | 6.5 (±2.3) × 103 | 8.4 (±2.8) × 103 d | 1.0 (±0.4) × 104 d |
| Glu | 1.5 (±0.5) × 103 | 1.7 (±0.7) × 103 | 2.7 (±1.6) × 103 d | 3.0 (±1.4) × 103 d |
| Gly | 1.6 (±0.3) × 104 | 1.6 (±0.3) × 104 | 2.1 (±0.3) × 104 d | 2.3 (±0.7) × 104 d |
| Lys | 1.9 (±0.7) × 104 | 2.5 (±1.1) × 104 | 2.7 (±1.0) × 104 d | 3.0 (±1.4) × 104 d |
| Tau | 1.4 (±0.3) × 104 | 1.4 (±0.6) × 104 | 1.9 (±0.7) × 104 d | 2.1 (±1.1) × 104 d |
| Gln | 8.8 (±2.8) × 104 | 9.5 (±4.0) × 104 | 1.1 (±0.4) × 105 | 1.2 (±0.4) × 105 d |
Separation conditions are the same as in Figure 2.
n (number of larvae) = 17.
n = 10.
Significantly different compared to the 30 s time period at P < 0.05.
Figure 4.
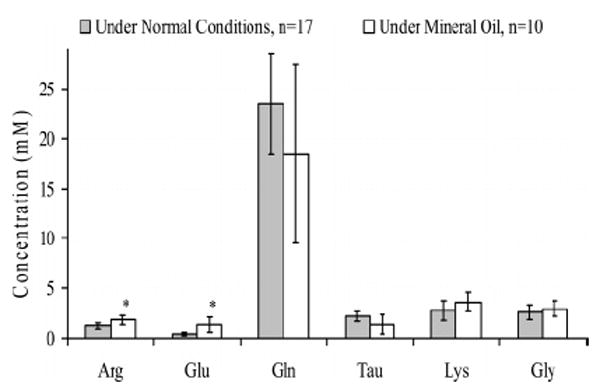
Amino acid concentrations of hemolymph samples collected 30 s after piercing the larvae under the “normal” open-air conditions and those collected while the larvae were under mineral oil. * Significantly different at P < 0.05.
However, Arg and Glu concentrations from the samples collected under mineral oil were significantly greater compared to those collected while the larvae were exposed to the atmosphere. Accordingly, the observed difference for Arg and Glu levels between the two methods cannot be a consequence of hemolymph evaporation in the open-air sampling technique. It was noted that larvae wriggle considerably when immersed in mineral oil, which could be due to suffocation or stress. Previous studies have shown that the stress level of the animals correlate with the variation of amino acid levels such as Glu,23,24 which also is a neurotransmitter. Although there may be a number of possibilities, the elevated hemolymph Arg levels could be due to the wriggling as arginine phosphate is the phosphagen of insects.25 Further studies are underway to better understand the effect of larval stress on the amino acid levels of its hemolymph.
Quantification of Amino acids in the WT and gb Mutant Larvae
Although the results of the qualitative analyses of hemolymph of WT and gb mutant larvae were the same, the electropherograms (Figure 2) show different peak heights especially for amino acids such as Gln and Tau. Table 2 shows the average amino acid concentrations for WT and gb genotypes based on 17 and 10 individual hemolymph samples, respectively. The levels of Tau, Glu, and Gln of the gb mutant larvae were significantly lower at 57%, 38%, and 35% of WT levels, respectively (P < 0.05). The gb mutants used in this study are based on a novel Drosophila xCT gene, and the genetic elimination of gb is discovered to reduce extracellular Glu concentrations indicating its regulation by the xCT transporters for the first time.13 The ratio of Glu levels between the mutant and WT larvae types, 0.61, of this study is similar to the previously reported Glu ratio, 0.62, for the mutant and the “precise excision” genotype based on samples from populations of larvae through HPLC analyses.13 Both WT and precise excision are control genotypes. However, the absolute Glu level, 1.72 mM, reported by Augustin et al.13 for the precise excision larvae is higher compared to the 0.44 mM reported for the samples collected in air and closer to those of samples collected under mineral oil, 1.32 mM, from WT larvae. The sampling technique of Augustin et al.13 required immersion of larvae in a saline solution for 5 min; it is possible that there are some similarities to mineral oil immersion. The WT and gb genotypes were not significantly different in terms of Arg, Lys, Gly, and Asp (Table 2).
Table 2.
Average Concentrations (mM ± Standard Deviation) of Eight Amino Acids in Hemolymph of Wild-Type and Genderblind (gb) Mutant Larvae Based on Samples from Individual Organisms (n)a
| wild-type (n = 17) | gb mutants (n = 10) | |
|---|---|---|
| Arg | 1.22 ± 0.29 | 1.28 ± 0.24 |
| Glu | 0.44 ± 0.16 | 0.27 ± 0.11b |
| Gln | 23.5 ± 5.0 | 15.1 ± 4.1b |
| Tau | 2.17 ± 0.52 | 0.94 ± 0.25b |
| Lys | 2.76 ± 1.02 | 2.61 ± 0.71 |
| Gly | 2.57 ± 0.76 | 2.68 ± 0.60 |
| Asp | 0.25 ± 0.20 | 0.20 ± 0.07 |
Separation conditions are the same as in Figure 2.
Significantly different at P < 0.05.
In vitro sampling was carried out with six trials of standards showing an average sample volume of 227 ± 59 nL. The observed levels for Arg, Asp, Glu, and Tau were 100 (±14)%, 108 (±12)%, 100 (±7)%, and 106 (±12)%, respectively, of the expected levels of 1.00, 0.25, 0.35, and 1.20 mM, respectively. This analysis is helpful in determining the extent of the hemolymph concentration variations due to the sampling and analytical processes so that the amino acid variations among individual organisms can be better assessed and understood. Although, the hemolymph amino acid levels reported in this study include variations incurred during the sampling and analyses processes, the variations shown in Table 2 and the box and whisker plots in Figure 5 suggest there are dominant animal-to-animal amino acid level variations. There are significant variations in the ranges of amino acid levels between the genotypes that well exceed that seen with the in vitro sampling. One obvious trend is that the median value (horizontal line in the box) is found to be closer to the lower quartile (bottom of box) for the gb mutants compared to WT larvae. Importantly, this may be a reflection of the number of animals that are studied, n = 10 for gb versus n = 17 for WT, and suggests that an averaged value approach does not clearly reflect the variations observed. Moreover, the range observed (total y-extent of box and whiskers) tends to be larger for the WT animals. An observed similarity is the upper/lower quartile range (height of box) for amino acids for all animals with the exceptions of Tau and Asp where the upper/lower quartile range is much smaller. The ability to study individual animals provides the ability to explore population distributions that are not available with multianimal collection methods employed in previous studies. Accordingly, this sampling technique also allows us to better understand the animal-to-animal chemical variations to study disease models and their behavioral aspects.
Figure 5.
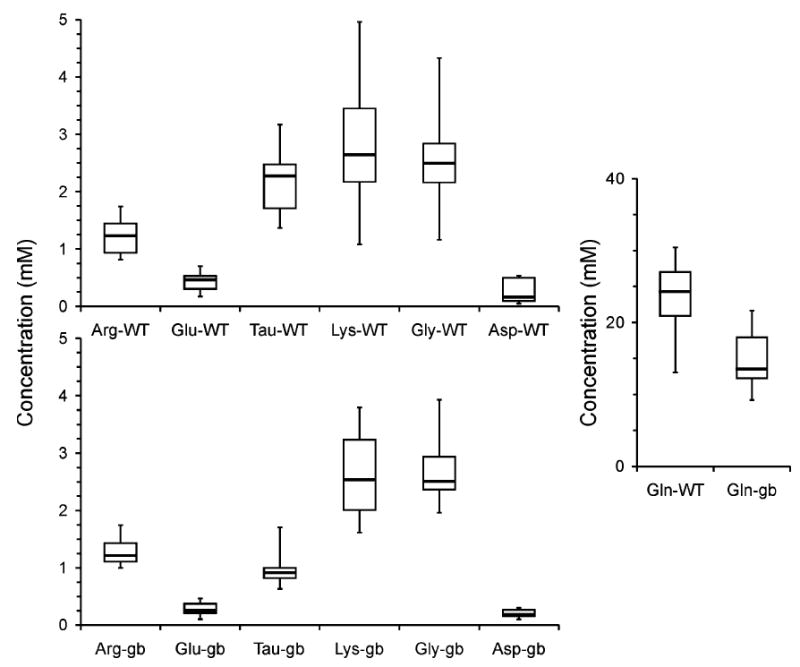
Box and whisker plots showing the variations of seven amino acids for the wild-type (WT, n = 17) and genderblind (gb, n = 10) larvae. The top and the bottom of the box show the upper and the lower quartiles, respectively, and the horizontal line in the middle indicates the median of the corresponding distribution while the minimum and maximum observed values are the bars connected to the box.
CONCLUSION
The novel sampling technique described in this paper is capable of collecting nanoscale volumes of biological fluids from small individual organisms. This new low-cost organism-level sampling technique efficiently collected in vivo hemolymph samples from individual Drosophila larvae for chemical analyses while minimizing the possible contaminations and alterations to the samples collected under the conventional homogenization and pooled sampling techniques. The absolute amino acid levels of the individual hemolymph samples were determined using CE-LIF after assessing the affects of evaporation. Samples collected under zero evaporation and at 30 s time intervals up to 120 s after piercing the larvae indicated the fact that effect of evaporation on the amino acid concentrations is insignificant when the hemolymph collection was completed within 60 s from piercing. More than 13 amino acids were separated in individual hemolymph samples of both WT and gb mutant larvae employing two optimized buffer conditions with CE-LIF after derivatizing with fluorescamine. The quantitation of amino acids showed significantly lower levels for Glu, Gln, and Tau for the gb mutants compared to the WT larvae. The distribution of observed single-animal amino acid concentrations for both genotypes exceeded the variability seen for in vitro sampling with standard amino acids and provides and indication of animal-to-animal variability. Further studies, employing the developed techniques, are underway to better understand the changes of amino acids in Drosophila hemolymph due to genetic modifications and stress conditions. The ability to study the hemolymph of individual fruit flies will allow improved chemical information from this important transgenic model in the study of basic neuroscience and human disease, and the developed technique can also be utilized for fluid sampling from other important small organs or organisms.
Acknowledgments
This work was supported by NIH MH 067971 (S.A.S.) and NIH NS 045628 Grants (D.E.F.).
References
- 1.Shulman JM, Shulman LM, Weiner WJ, Feany MB. Curr Opin Neurol. 2003;16:443–449. doi: 10.1097/01.wco.0000084220.82329.60. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S, Feany MB. Hum Mol Genet. 2004;13:2011–2018. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- 3.Robinson AS, Franz G, Atkinson PW. Insect Biochem Mol Biol. 2004;34:113–120. doi: 10.1016/j.ibmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Celotto AM, Palladino MJ. Mol Interventions. 2005;5:292–303. doi: 10.1124/mi.5.5.9. [DOI] [PubMed] [Google Scholar]
- 5.Shank RP, Aprison MH. Life Sci. 1981;28:837–842. doi: 10.1016/0024-3205(81)90044-8. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto K, Kimura H, Sakai Y. Brain Res. 1983;265:163–168. doi: 10.1016/0006-8993(83)91350-1. [DOI] [PubMed] [Google Scholar]
- 7.Feany MB, Dickson DW. Ann Neurol. 1996;40:139–148. doi: 10.1002/ana.410400204. [DOI] [PubMed] [Google Scholar]
- 8.Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, Feany MB. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 9.Chen PS, Hanimann FZ. Naturforsch. 1965;20b:307–312. [PubMed] [Google Scholar]
- 10.Chen PS, Buhler R. J Insect Physiol. 1969;16:615–627. doi: 10.1016/0022-1910(70)90095-8. [DOI] [PubMed] [Google Scholar]
- 11.Burnet B, Sang JH. Genetics. 1968;59:211–235. doi: 10.1093/genetics/59.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ream PJ, Suljak SW, Ewing AG, Han K. Anal Chem. 2003;75:3972–3978. doi: 10.1021/ac034219i. [DOI] [PubMed] [Google Scholar]
- 13.Augustin H, Grosjean Y, Chen K, Sheng Q, Featherstone DE. J Neurosci. 2007;27:111–123. doi: 10.1523/JNEUROSCI.4770-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell PR, Paxon TL, Han KA, Ewing AG. Anal Chem. 2005;77:6902–6908. doi: 10.1021/ac050963m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Righetti PG. Biopharm Drug Dispos. 2001;22:337–351. doi: 10.1002/bdd.276. [DOI] [PubMed] [Google Scholar]
- 16.Thongkhao-On K, Kottegoda S, Pulido JS, Shippy SA. Electrophoresis. 2004;25:2978–2984. doi: 10.1002/elps.200405941. [DOI] [PubMed] [Google Scholar]
- 17.Kottegoda S, Shaik I, Shippy SA. J Neurosci Methods. 2002;121:93–101. doi: 10.1016/s0165-0270(02)00245-5. [DOI] [PubMed] [Google Scholar]
- 18.McLaren DG, Boulat O, Chen DDY. Electrophoresis. 2002;23:1912–1920. doi: 10.1002/1522-2683(200206)23:12<1912::AID-ELPS1912>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Pierce VA, Mueller LD, Gibbs AG. J Exp Biol. 1999;202:2349–2358. doi: 10.1242/jeb.202.17.2349. [DOI] [PubMed] [Google Scholar]
- 20.Zacharis CK, Tempels FWA, Theodoridis GA, Voulgaropoulos AN, Underberg WJM, Somsen GW, De Jong GJ. J Chromatogr A. 2006;1132:297–303. doi: 10.1016/j.chroma.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 21.Ho YH, Wu HL. Electrophoresis. 2006;27:2300–2309. doi: 10.1002/elps.200500810. [DOI] [PubMed] [Google Scholar]
- 22.Dong Q, Jin W, Shan J. Electrophoresis. 2002;23:559–564. doi: 10.1002/1522-2683(200202)23:4<559::AID-ELPS559>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Milakofsky L, Hare TA, Miller JM, Vogel WH. Life Sci. 1985;36:753–761. doi: 10.1016/0024-3205(85)90195-x. [DOI] [PubMed] [Google Scholar]
- 24.Sherman A, Gebhart GF. Neuropharmacology. 1974;13:673–675. doi: 10.1016/0028-3908(74)90057-4. [DOI] [PubMed] [Google Scholar]
- 25.Beis I, Newsholme EA. Biochem J. 1975;152:23–32. doi: 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]


