SUMMARY
During vertebrate cell division, chromosomes oscillate with periods of smooth motion interrupted by abrupt reversals in direction. These oscillations must be spatially constrained in order to align and segregate chromosomes with high fidelity, but the molecular mechanism for this activity is uncertain. We report here that the human kinesin-8, Kif18A, has a primary role in the control of chromosome oscillations. Kif18A accumulates as a gradient on kinetochore microtubules in a manner dependent on its motor activity. Quantitative analyses of kinetochore movements reveal that Kif18A reduces the amplitude of preanaphase oscillations and slows poleward movement during anaphase. Thus, the microtubule-depolymerizing kinesin, Kif18A, has the unexpected function of suppressing chromosome movements. Based on these findings we propose a molecular model in which Kif18A regulates kinetochore microtubule dynamics to control mitotic chromosome positioning.
INTRODUCTION
During mitosis chromosomes establish connections to mitotic spindle microtubules (MTs) via specialized protein complexes, called kinetochores, and subsequently translocate to the midzone of the bipolar spindle. This process, known as “congression”, is essential for preserving the fidelity of the genome as it ensures that sister-chromosomes will be segregated on either side of the cytokinetic furrow when it bisects the spindle midzone. Despite decades of study on this important process, the molecular mechanisms that allow chromosomes to find and remain at the spindle equator remain unclear.
Classic live imaging studies of congressing chromosomes established that bioriented chromosomes move at constant velocity and display abrupt changes in direction both before and after alignment (Rieder and Salmon, 1994; Skibbens et al., 1993). These oscillatory movements are surprisingly similar in unaligned and aligned chromosomes with the exception that unaligned chromosomes tend to exhibit more directional persistence towards the center of the spindle between reversals (Skibbens et al., 1993). Current models for congression attempt to explain the decrease in directional persistence near the metaphase plate as a result of the combined influence of kinetochore-microtubule (kMT) dynamic instability and away-from-pole forces generated along chromosome arms by MTs and chromokinesins (i.e., polar ejection forces) (Joglekar and Hunt, 2002; Khodjakov et al., 1999; Rieder and Salmon, 1994). While experimental evidence supports the idea that polar ejection forces vary with spindle position and thus could provide a positional cue (Ault et al., 1991; Cassimeris et al., 1994; Rieder et al., 1986; Rieder and Salmon, 1994), other data indicate that they are insufficient to explain congression (Kapoor and Compton, 2002). For example, abrogation of the polar ejection force via inhibition of chromokinesins does not abolish congression (Levesque and Compton, 2001). Furthermore, experimental removal of chromosome arms does not prevent kinetochores from aligning at the spindle equator (Brinkley et al., 1988). Taken together, these studies suggest that another mechanism, likely acting at kinetochores, provides spatial cues that are critical for alignment.
An alternative mechanism for congression revolves around an intriguing idea that kinetochores are “smart” and can sense their position within the spindle (Mitchison, 1989). In “smart” kinetochore models, the position-sensitive mechanism that determines when chromosomes change direction acts at kinetochores (Kapoor and Compton, 2002; Rieder and Salmon, 1994). While molecular evidence to support a mechanism that would permit kinetochores to monitor their spindle position is lacking, the motor proteins of the kinesin-8 subfamily have the potential to fulfill such a role. In vitro studies have established that kinesin-8s are plus-end directed motors that can depolymerize stable MTs specifically at their plus-ends in a length-dependent manner, suggesting a role in directly regulating MT dynamics and MT length in vivo (Gupta et al., 2006; Mayr et al., 2007; Pereira et al., 1997; Varga et al., 2006). Furthermore, genetic and siRNA-based studies indicate that kinesin-8 motors, including human Kif18A, are required for proper mitotic chromosome alignment (Gandhi et al., 2004; Garcia et al., 2002; Goshima and Vale, 2003; Mayr et al., 2007; West et al., 2002; Zhu et al., 2005). However, the question of how kinesin-8 motors functionally contribute to congression remains unanswered.
To investigate the mechanism by which Kif18A regulates chromosome congression we have used high-resolution live cell imaging combined with quantitative measurements of kinetochore movements. We report that Kif18A controls the persistent movement of chromosomes by both increasing the rate at which they make directional switches and slowing the velocity of their movement. Furthermore, Kif18A’s accumulation on kMTs and its ability to suppress oscillatory movements are dependent on its motor activity and vary within the spindle. Based on these discoveries, we propose a model for chromosome congression in which Kif18A forms a gradient along kMTs that directly regulates their length and dynamics to facilitate chromosome alignment at the spindle equator
RESULTS
Kif18A Forms a Motor-dependent Gradient at kMT Plus-ends
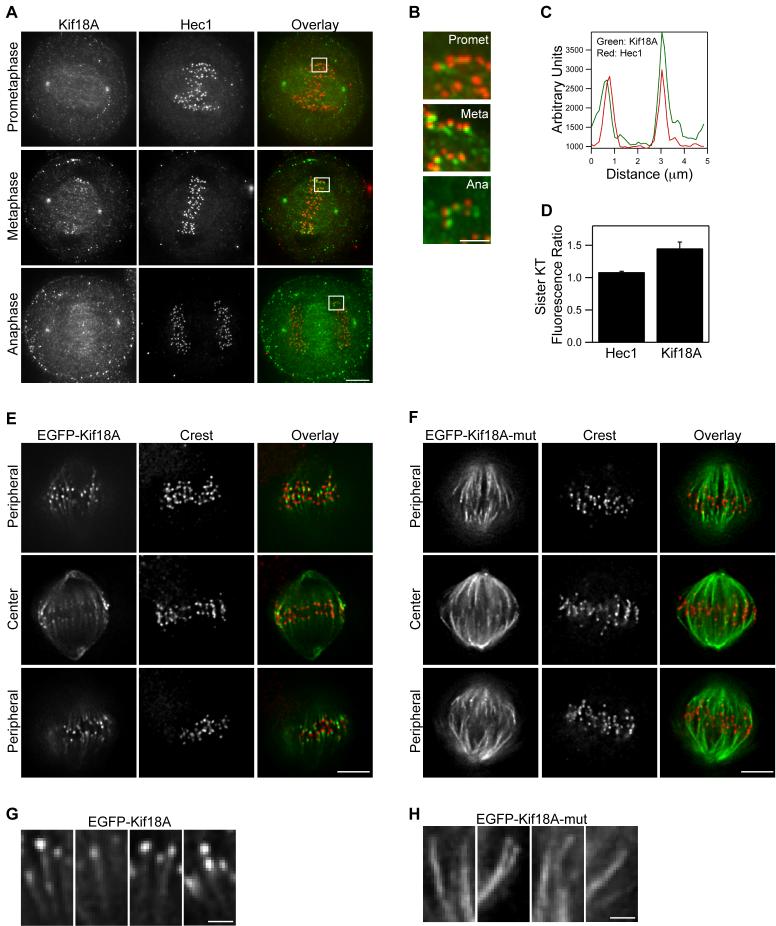
Endogenous Kif18A exhibits a dynamic localization to the plus-ends of kMTs (Figure 1A-B and Figure S1). The localization of Kif18A in mitotic HeLa cells was analyzed with anti-Kif18A antibodies and by expression of EGFP-Kif18A. Similar localization was observed with both approaches. In prometaphase cells the motor is found along spindle MTs and localizes near the outer-kinetochore marker Hec1 at only a subset of kinetochores (Figure 1A-D). In metaphase cells with aligned chromosomes, Kif18A localizes as a comet-like gradient along most if not all kMTs, and this localization requires Kif18A’s motor activity (Figure 1A-H and Figure S1). A mutant form of Kif18A (EFGP-Kif18A-mut), containing alanine substitutions in three conserved motor domain residues required for kinesin motility and kinesin-13 depolymerization (Moore et al., 2005; Woehlke et al., 1997), uniformly binds to spindle MTs and does not exhibit a gradient-like localization pattern (Figure 1E-H). During anaphase, Kif18A is still seen at kinetochores and additionally begins to accumulate in the spindle midzone (Figure 1A and Figure S1). Kif18A concentrates at the midbody during telophase and cytokinesis (Figure S1).
Figure 1. Kif18A displays dynamic, motor-dependent localization to the plus-ends of kinetochore microtubules.
(A) Mitotic HeLa cells in the indicated stages were fixed and stained with anti-Kif18A antibodies (green in overlay) and anti-Hec1 antibodies (red in overlay). Scale bar is 5 μm.
(B) Magnified views of the regions indicated by white boxes in prometaphase (promet), metaphase (meta) and anaphase (ana) cells in (A). Scale bar is 2 μm.
(C) Representative linescan across metaphase sister kinetochores in a HeLa cell stained with anti-Kif18A (green) and anti-Hec1 (red) antibodies. In all scans the peak of Kif18A fluorescence was distal to the peak of Hec1 fluorescence with respect to the centromere (n = 19 kinetochore pairs from 3 cells).
(D) The ratio of peak fluorescence intensity on sister kinetochores was calculated from metaphase cells co-stained for Hec1 and Kif18A by immunofluorescence. The peak intensity of Hec1 is equal between sister kinetochores while Kif18A is significantly higher on one sister kinetochore than the other (n = 19 kinetochore pairs from 3 cells; p = 0.001). Error bars indicate SEM.
(E and F) Metaphase HeLa cells expressing either EGFP-Kif18A (E) or EGFP-Kif18A-mutant (F) (green in overlay) and stained with human CREST serum to visualize kinetochores (red in overlay). Optical slices from the periphery and the center of each spindle are displayed. Scale bars are 5 μm.
(G and H) Magnified images of EGFP-Kif18A (G) or EGFP-Kif18A-mut (H) along kMTs. Scale bars are 1 μm.
Several additional features of Kif18A localization in metaphase cells were also observed. Analyses of metaphase sister kinetochore pairs co-immunostained with Hec1 and Kif18A antibodies revealed that the peak of Kif18A fluorescence is just distal to the peak of Hec1 fluorescence (Figure 1C), consistent with localization of Kif18A at kMT plus-ends. However, unlike Hec1 fluorescence, which is equal on both sister kinetochores, the peak intensity of Kif18A is significantly greater on one sister kinetochore relative to the other (Figure 1C-D). Analysis of optical sections through metaphase spindles revealed that the concentration of Kif18A, but not EGFP-Kif18A-mut, is greater on kMTs at the periphery of the spindle compared to those at the spindle interior closer to the pole-to-pole axis. Thus, the accumulation of Kif18A on kMT plus-ends varies within the spindle and this differential accumulation is motor-dependent (Figure 1E-F). Consistent with previous studies (Mayr et al., 2007), we found that Kif18A’s localization to kinetochores was also dependent on MTs, as Kif18A was not detected at kinetochores after depolymerization of MTs with nocodazole or vinblastine (Figure S2). Taken together, these data suggest Kif18A utilizes its plus-end directed motility to form a gradient along kMTs during metaphase and that accumulation is greater at the outer periphery of the spindle.
Kinetochore Oscillations are Sensitive to Alterations in Kif18A Expression
To address Kif18A’s role during chromosome congression, cells were either depleted of Kif18A by siRNA treatment or transfected with EGFP-Kif18A to increase Kif18A expression. Treatment with Kif18A-specific siRNAs resulted in the depletion of Kif18A from mitotic spindles and kinetochores (Figure S3). The amount of Kif18A that remained after siRNA depletion was below the level of detection for our antibody (>90% depletion, Figure S3C). Consistent with previous reports (Mayr et al., 2007; Zhu et al., 2005), we observed that depletion of Kif18A resulted in failed chromosome alignment (Figure S3A-B) and an increase in prometaphase cells (Figure S3D). In contrast, expression of EGFP-Kif18A did not cause a mitotic delay or disrupt chromosome alignment (Figure S3D).
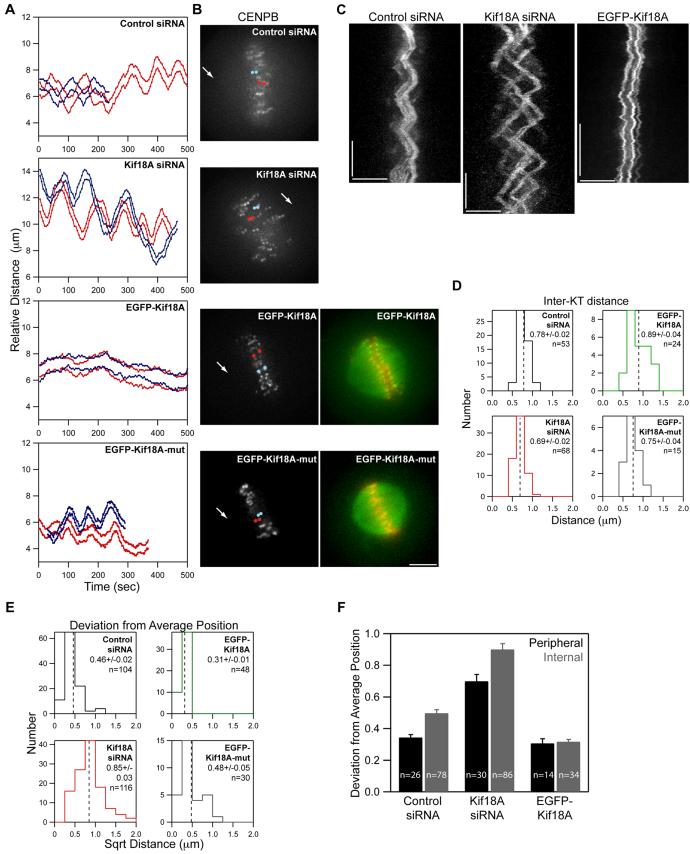
To address a role for Kif18A in regulating mitotic chromosome movements, HeLa cells co-expressing fluorescent kinetochore and centrosome markers were analyzed by time-lapse microscopy following transfection with siRNAs or EGFP-Kif18A. We observed that chromosomes in these cells were bioriented and made oscillatory movements around the equator of the spindle reminiscent of those observed in late prometaphase and metaphase control cells (Figure 2A-C; Supplemental movies 1-3). Bioriented kinetochores in these cells were also under tension, as determined by measuring the distance between sister kinetochores over time. While distributions of inter-kinetochore distance were similar in all cell types tested, alterations in Kif18A levels correlated with small but significant changes in the average inter-kinetochore distance (Figure 2D). Kif18A depletion reduced the average inter-kinetochore distance, consistent with previous measurements in fixed cells (Mayr et al., 2007; Zhu et al., 2005), while EGFP-Kif18A expression led to greater inter-kinetochore distance. However, the most striking effects induced by altering Kif18A expression were changes in oscillation amplitude. Oscillatory movements were dramatically increased in the absence of Kif18A and were suppressed in EGFP-Kif18A cells. These effects are evident in plots of individual kinetochore movements relative to spindle poles (Figure 2A) and in kymographs generated from CENP-B fluorescence (Figure 2C).
Figure 2. Kif18A affects the oscillatory movements of kinetochores.
(A) Distance versus time plots of two kinetochore pairs (red and blue lines) from each of the indicated cell types. Relative distance was calculated by measuring the separation between each kinetochore and one spindle pole. Images were collected every 5 seconds for control and Kif18A siRNA cells and every 2 seconds for EGFP-Kif18A and EGFP-Kif18A-mut cells.
(B) Still frames of CENP-B fluorescence in cells used to derive the distance versus time plots shown in (A). Arrows indicate position of pole used for relative distance measurements. Red and blue dots are overlayed on kinetochores that were tracked to generate the red and blue traces in (A) respectively. Two-color images of EGFP-Kif18A or EGFP-Kif18A-mut (green) with mRFP-CENP-B (red) were taken immediately after time-lapse imaging of kinetochore movements was stopped. Scale bar is 5 μm.
(C) Representative kymographs of CENP-B fluorescence from cells transfected with control siRNAs, Kif18A-specific siRNAs or EGFP-Kif18A. Vertical scale bars = 2 minutes; horizontal scale bars = 5 μm.
(D) Histograms displaying average inter-kinetochore distances for sister kinetochore pairs in live cells. The mean ± SEM is indicated for each distribution and the mean position is marked by a vertical dotted line. “n” indicates the number kinetochore pairs tracked from 10 (control and Kif18A siRNA), 7 (EGFP-Kif18A) or 4 (EGFP-Kif18A-mut) cells. The average inter-kinetochore distances measured from Kif18A siRNA and EGFP-Kif18A expressing cells are significantly different from those measured in control siRNA treated cells (p = 1.0 × 10-4 and p = 0.02 respectively). Average inter-kinetochore distances for EGFP-Kif18A expressing cells are also significantly different from EGFP-Kif18A-mut expressing cells (p = 0.02).
(E) Histograms displaying deviation from average position (DAP) calculations for the indicated cell types. The mean ± SEM is given for each distribution and the mean position is marked by a vertical dotted line. “n” indicates the number of kinetochores analyzed from the same data set used in (D). The DAPs for kinetochores in Kif18A-depleted cells and EGFP-Kif18A cells are significantly different from the DAP for kinetochores in control siRNA cells (p = 4.4 × 10-22 and 1.2 × 10-9 respectively). The DAP for EGFP-Kif18A kinetochores is also significantly different from the DAP for EGFP-Kif18A-mut kinetochores (p = 0.002).
(F) Average DAP measurements for kinetochores on the periphery of the spindle (peripheral) and along the pole-to-pole axis (internal) from cells treated with control or Kif18A siRNAs or over-expressing EGFP-Kif18A. Error bars are SEM; “n” indicates the number of kinetochores from the data set used in (E). DAPs for peripheral and internal kinetochores are significantly different in control and Kif18A-depleted cells (p = 5.9 × 10-7 and p = 7.0 × 10-4 respectively) but not in EGFP-Kif18A cells (p = 0.75).
In order to quantify changes in oscillatory movement in a population of kinetochores, we developed a new measurement that scores the deviation from the average position (DAP) for individually tracked bioriented kinetochores. The DAP for a kinetochore is a linear measure of oscillation amplitude that provides a means to quantitatively analyze kinetochore movements even when unambiguous determination of directional changes is difficult, such as in the suppressed oscillations observed in EGFP-Kif18A cells (for more information see Figure S4). These studies revealed that depletion of Kif18A significantly increased the DAP from 0.46 ± 0.02 μm to 0.85 ± 0.03 μm, while overexpression of Kif18A significantly reduced the DAP to 0.31 ± 0.01 μm (Figure 2E and Table 1). Therefore, the distance that kinetochores move from their average position during oscillations is increased approximately 2-fold in the absence of Kif18A and is reduced by approximately 30% in the presence of EGFP-Kif18A. Furthermore, the suppression of oscillatory movements is dependent on Kif18A’s motor activity, as expression of EGFP-Kif18A-mut did not affect oscillations (Figure 2A-C and E). These data indicate that Kif18A limits persistent movement of bioriented kinetochores in a motor-dependent fashion and that increased oscillation amplitude is likely the primary defect that leads to the unaligned chromosome phenotype seen in Kif18A-depleted cells.
Table 1.
Measurements of preanaphase kinetochore movements
| P vel | AP vel | Switch | Amp | DAP | IKD | |
|---|---|---|---|---|---|---|
| Control siRNA | 1.99 ± 0.07 | 1.81 ± 0.07 | 1.58 ± 0.05 | 1.21 ± 0.08 | 0.46 ± 0.02 | 0.78 ± 0.02 |
| Kif18A siRNA | 2.97 ± 0.09** | 2.67 ± 0.08** | 1.23 ± 0.11** | 2.28 ± 0.15** | 0.85 ± 0.03** | 0.69 ± 0.02** |
| EGFP-Kif18A | ND | ND | ND | ND | 0.31 ± 0.01** | 0.89 ± 0.04* |
| EGFP-Kif18A-mut | ND | ND | ND | ND | 0.48 ± 0.05 | 0.75 ± 0.04 |
Average measurements of poleward velocity (P vel), away from pole velocity (AP vel), switch rate (Switch), oscillation amplitude (Amp), deviation from average position (DAP) and inter-kinetochore distance (IKD) are given ± SEM.
indicates p ≤ 0.05
indicates p ≤ 0.01 when compared to controls.
In addition to suppressing oscillations, we observed that EGFP-Kif18A expression often led to uncoordinated sister kinetochore movements. Specifically, one kinetochore frequently attempted to move poleward while its sister remained stationary or also attempted poleward movement (see Supplemental movie 3). The transient increases in inter-kinetochore distance caused by these events are evident in our distribution of measurements as a shoulder above 1 μm and may explain the increase in average inter-kinetochore distance induced specifically by EGFP-Kif18A but not EGFP-Kif18A-mut expression (Figure 2D).
Oscillation Amplitude Correlates with Kif18A Accumulation at Kinetochores
Since the concentration of Kif18A is higher on kMTs at the periphery of the spindle compared to those nearer to the pole-to-pole axis (Figures 1E), we compared oscillatory movements of kinetochores based on their location within the spindle. In control siRNA treated cells, we found that oscillations of peripheral kinetochores were significantly reduced compared to those of kinetochores closer to the long axis of the spindle (Figure 2F) consistent with previous studies in PtK1 cells (Canman et al., 2002; Cimini et al., 2004). Depletion of Kif18A significantly increased the movements of peripheral and internal kinetochores, suggesting that Kif18A limits oscillatory movements of all kinetochores (Figure 2F). Interestingly, we also found that oscillations of peripheral and internal kinetochores were not significantly different in EGFP-Kif18A cells and that oscillations in these cells are comparable to those of peripheral kinetochores in control cells (Figure 4F). These data show that oscillation amplitude is inversely correlated with the concentration of Kif18A on kMTs.
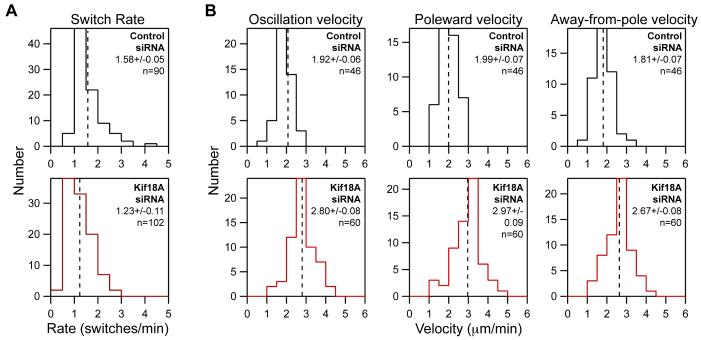
Figure 4. Kif18A regulates both the directional switch rate and velocity of kinetochore oscillations.
(A) Histograms showing the distribution of kinetochore directional switch rates measured in control and Kif18A siRNA cells. The mean ± SEM is given for each distribution and the mean position is marked by a vertical dotted line. An average switch rate was calculated for each kinetochore and “n” indicates the number of kinetochores analyzed from 10 control and 10 Kif18A siRNA cells. The two data sets are significantly different (p = 2.8 × 10-5).
(B) Histograms of average kinetochore velocities during oscillatory movements in control and Kif18A siRNA treated cells. The “oscillation velocity” distribution displays the average velocity for each kinetochore analyzed. “Poleward velocity” and “Away-from-Pole velocity” distributions include only velocities from movements made toward or away-from-the-pole that the kinetochore was attached to respectively. The mean ± SEM is given for each distribution and the mean position is marked by a vertical dotted line. “n” indicates the number of kinetochores analyzed from 5 control and 5 Kif18A siRNA cells. Kinetochore velocities in Kif18A-depleted cells are significantly different than those in control depleted cells (p = 2.8 × 10-15 for average oscillation velocity; p = 5.4 × 10-13 for poleward velocity; p = 4.2 × 10-12 for away-from-pole velocity).
The Effects of Kif18A on Chromosome Oscillations are Not Due to Changes in Spindle Length
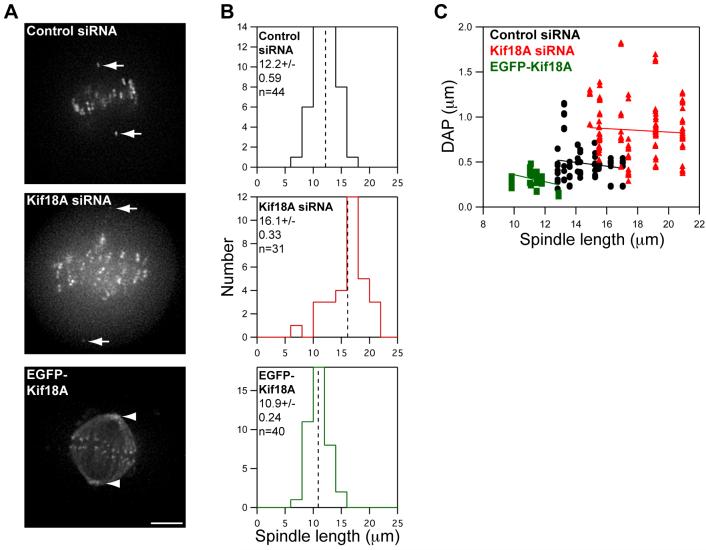
Consistent with studies in fixed cells (Mayr et al., 2007), we found that spindles in live cells depleted of Kif18A are longer on average than control spindles (Figure 3A-B). Polar ejection forces generated by each half spindle are believed to play a significant role in the regulation of chromosome oscillations during mitosis by generating away-from-pole forces along chromosome arms, which increase as chromosomes enter MT-dense regions near centrosomes (Cassimeris et al., 1994; Rieder et al., 1986; Rieder and Salmon, 1994). In principle, an increase in spindle length could indirectly lead to larger chromosome oscillations by positioning areas of MT density further from the spindle equator. However, analysis of kinetochore oscillations as a function of spindle length does not support this hypothesis (Figure 3C). Within a cell population there is little correlation between spindle length and oscillation amplitude. Furthermore, when oscillatory movements in control and Kif18A-depleted cells with spindles of similar length are compared (between 15 and 17 μm), oscillations are larger in the absence of Kif18A (Figure 3C). These data indicate that the effects of Kif18A on chromosome oscillations are not simply an indirect effect of changes in spindle length and suggest a direct role for Kif18A in controlling chromosome movements.
Figure 3. Kif18A’s effects on oscillations are not an indirect effect of changes in spindle length.
(A) Images of live HeLa cells expressing Venus-centrin and EGFP-CENP-B (control and Kif18A siRNA) or EGFP-Kif18A. Spindle lengths were determined by measuring the distance between Venus-centrin foci (arrows) or EGFP-Kif18A spindle pole labeling (arrowheads). Scale bar is 5 μm.
(B) Histograms of spindle lengths measured in live cells. The mean ± SEM is given for each distribution and the mean position is marked by a vertical dotted line. “n” indicates the number of cells analyzed. Spindle lengths in Kif18A-depleted and EGFP-Kif18A expressing cells are significantly different from those in control-depleted cells (p = 4.4 × 10-7 and 0.002 respectively).
(C) Scatter plot of deviation from average position (DAP) measurements as a function of spindle length. Black circles = control siRNA, red triangles = Kif18A siRNA and green squares = EGFP-Kif18A. Lines represent regression fits to each data set.
Kif18A Affects the Velocity and Switch Rate of Kinetochore Movements
Oscillation amplitude could be affected by alterations in two independent characteristics of chromosome movement: velocity and the frequency of directional switches. To calculate the switch rate for a kinetochore, we counted the number of times it changed directions during oscillatory movement and divided that number by the amount of time the kinetochore was filmed. On average, the kinetochore switch rate is reduced in cells depleted of Kif18A compared to controls (Figure 4A and Table 1). Kinetochores in control-depleted cells changed direction at an average rate of 1.58 ± 0.05 min-1, while those in Kif18A-depleted cells switched directions at an average rate of 1.23 ± 0.11 min-1. These data suggest that kinetochores in Kif18A-depleted cells undergo longer periods of persistent movement between turnarounds.
The velocity of oscillatory movements was also significantly increased in Kif18A-depleted cells relative to controls (Figure 4B and Table 1). Kinetochores in control-depleted cells moved at an average rate of 1.92 ± 0.06 μm/min while those in Kif18A-depleted cells oscillated at an average rate of 2.80 ± 0.08 μm/min. Furthermore, poleward and away-from-pole movements were equally affected (Figure 4B and Table 1). The increase in velocity in the absence of Kif18A is surprising since it implies that Kif18A, a MT depolymerizer, acts to slow chromosome movement in vivo. In contrast to our findings, another study reported that Kif18A depletion reduces the velocity of kinetochore movements (Mayr et al., 2007). The exact reason for these conflicting results is not clear, but we feel that the differences may be due to the use of low time resolution in the previous study, which would prevent accurate determination of directional chromosome speeds (see Experimental Procedures).
Our data indicate that both an increase in velocity and a decrease in directional switch frequency lead to a 2-fold increase in oscillation amplitude from 1.21 ± 0.08 μm to 2.28 ± 0.15 μm in Kif18A-depleted cells, consistent with the observed 2-fold change in DAP (Table 1). Thus, a combination of changes in switch rate and velocity quantitatively explains the increased movements observed in Kif18A-depleted cells.
Kif18A Affects the Velocity of Poleward Anaphase Movements
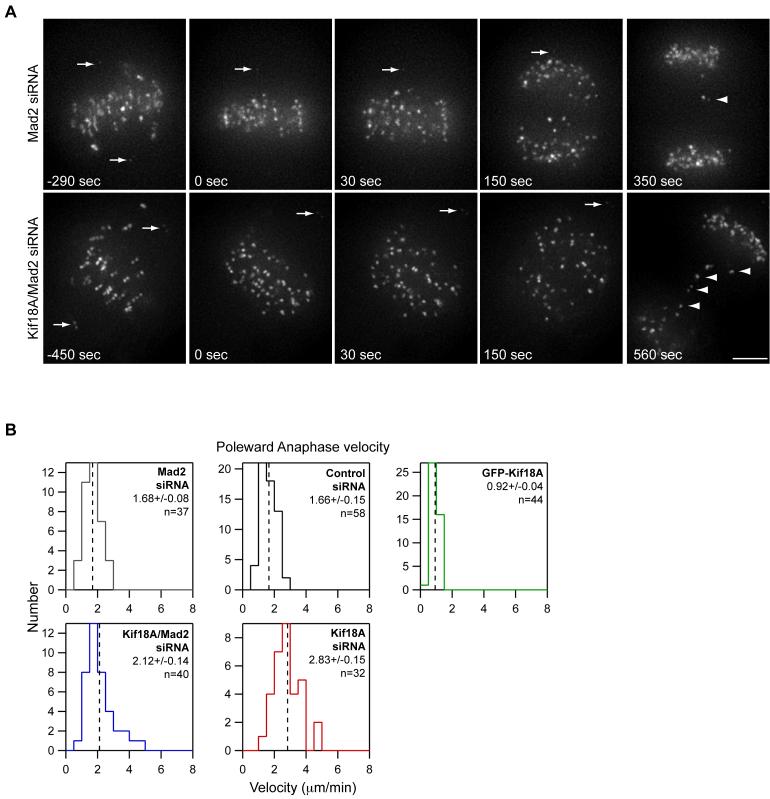
Since Kif18A is also present on kinetochores in anaphase cells (Figure 1A), we tested whether Kif18A affects the velocity of poleward chromosome movements during anaphase. Depletion of Kif18A results in a mitotic delay mediated by the spindle assembly checkpoint, and very few Kif18A-depleted cells go through anaphase (Mayr et al., 2007; Zhu et al., 2005) (Figure S3D). Kif18A-depleted cells, however, are able to progress through anaphase and exit mitosis when the checkpoint protein Mad2 is simultaneously depleted (Mayr et al., 2007) (Figure S3D). Therefore, we analyzed anaphase kinetochore movements in cells co-depleted of Mad2 and Kif18A. Consistent with previous studies, Mad2-depleted cells entered anaphase before completing congression (Canman et al., 2002; Meraldi et al., 2004). While most kinetochores segregated to the spindle poles normally, lagging chromosomes were frequently seen in cells depleted of Mad2 alone or co-depleted of Mad2 and Kif18A (Figure 5A; see supplemental movies 4 and 5). Anaphase A rates for non-lagging chromosomes increased approximately 20% from 1.68 ± 0.08 μm/min to 2.12 ± 0.14 μm/min in the absence of Kif18A (Figure 5B). However, we observed that loss of Kif18A function did not affect all kinetochores equally under these conditions and that only a subset of kinetochores in Mad2/Kif18A depleted cells moved faster than controls. This could be due to the increased frequency of merotelic attachments seen when cells are induced to enter anaphase precociously (Cimini et al., 2003).
Figure 5. Kif18A affects poleward movement during anaphase.
(A) Selected images from time-lapse analyses of Mad2-depleted and Kif18A/Mad2-co-depleted HeLa cells expressing EGFP-CENP-B and Venus-centrin. Time is given in seconds relative to anaphase sister kinetochore separation. Arrows mark the position of Venus-centrin labeled spindle poles. In both cells, the lower spindle pole is out of focus at the time of anaphase. Kinetochores on lagging chromosomes are marked by arrowheads. Cells were filmed at 5-second intervals; scale bar = 5 μm.
(B) Histograms of anaphase A velocities measured in control siRNA, Mad2 siRNA, Kif18A/Mad2 siRNA, Kif18A siRNA and EGFP-Kif18A cells. The mean ± SEM is given for each distribution and the mean position is marked by a vertical dotted line. “n” indicates the number of kinetochores analyzed from 3 cells (Kif18A siRNA, Mad2 siRNA and Kif18A/Mad2 siRNA) or 4 cells (control siRNA and EGFP-Kif18A). The average anaphase A velocities in Kif18A-depleted, Kif18A/Mad2-co-depleted and EGFP-Kif18A expressing cells are significantly different from anaphase A velocities in control cells (p = 4.9 × 10-9, p = 4.0 × 10-3 and p = 4.4 × 10-16 respectively). Anaphase A velocities in Mad2-depleted and Kif18A/Mad2-co-depleted cells are also significantly different (p = 0.01).
To further investigate Kif18A’s effects on anaphase kinetochore movements, we analyzed the small fraction of Kif18A siRNA treated cells that happened to enter anaphase without checkpoint knockdown. For these studies we chose cells with large pre-anaphase oscillations that initiated chromosome segregation without completing alignment, which are indications of Kif18A depletion. The average kinetochore poleward velocity in these cells was increased by approximately 40% to 2.83 ± 0.15 μm/min and importantly, the majority of kinetochores displayed increased speed relative to controls (Figure 5B and Supplemental movies 6 and 7). In contrast, expression of EGFP-Kif18A slowed anaphase A velocity by approximately 45% to 0.92 ± 0.04 μm/min (Figure 5B and Supplemental movie 8). Taken together, these results suggest that Kif18A acts as a governor to limit the rate of kinetochore movements during mitosis, a function that is quite unexpected considering its MT depolymerization activity in vitro (Mayr et al., 2007).
DISCUSSION
Metaphase chromosomes in vertebrate cells make oscillatory movements around the spindle equator, and the regulation of these movements is believed to be important for establishing and maintaining alignment (Kapoor and Compton, 2002; Rieder and Salmon, 1994; Skibbens et al., 1993). Our data indicate that Kif18A functions to limit these oscillatory movements and control chromosome alignment.
We show that during mitosis, Kif18A suppresses the amplitude of kinetochore oscillations in part by increasing the rate at which kinetochores change directions. Kinetochore movements in vertebrate cells are thought to depend primarily on kMT plus-end dynamics (Inoue and Salmon, 1995; Rieder and Salmon, 1994). Kif18A’s localization to kMT plus-ends and its in vitro MT depolymerizing activity suggest that it may directly modulate chromosome movements (Mayr et al., 2007). Directional switching involves both catastrophe and rescue of kMT plus-ends. Interestingly, Kip3p increases both the rescue and catastrophe frequencies of MTs in budding yeast (Gupta et al., 2006). A similar effect during mitosis would increase the frequency and suppress the amplitude of kinetochore oscillations, as we observed.
Kif18A-mediated changes in kinetochore oscillation amplitude were also due in part to a surprising and counter-intuitive effect on the velocity of chromosome movements. In the absence of Kif18A, kinetochore velocity was increased during pre-anaphase oscillations and anaphase. Conversely, overexpression of Kif18A slowed poleward anaphase movements. These data indicate that Kif18A functions to slow kinetochore velocity and, importantly, argue against the previously suggested idea that Kif18A produces the force that drives chromosome movements (Mayr et al., 2007).
Exactly how Kif18A affects kinetochore velocity is an interesting question that warrants further investigation. One possible explanation for the observed effects on kinetochore velocity is suggested by recent studies of the Drosophila kinesin-8 motor, Klp67A (Buster et al., 2007). In the absence of Klp67A, the rate of kMT flux, and thus the rate of kMT minus-end shortening, is increased, which leads to faster anaphase poleward movement. However, it is unclear whether changes in flux rate can fully explain the dramatic effects of altering Kif18A expression in human cells where flux makes only a minor contribution to the movement and alignment of chromosomes (Ganem and Compton, 2006). For example, flux in human cells accounts for 20% of chromosome poleward velocity (Ganem et al., 2005), so even if Kif18A overexpression completely suppressed flux, it would not be enough to explain the 45% decrease in velocity that we observed. Alternatively, Kif18A’s accumulation at the plus-ends of kMTs might affect the kinetics of tubulin addition and release. The measured effects of kinesin-8 motors on MT dynamics and chromosome movements in yeast, where MTs do not flux, support this hypothesis (Garcia et al., 2002; Gupta et al., 2006; Maddox et al., 2000; Mallavarapu et al., 1999; Pearson et al., 2003; West et al., 2002). Thus, based on current data we favor a model in which Kif18A affects kinetochore velocity through regulation of kMT plus-end dynamics, although effects on minus-end dynamics cannot be ruled out. Future work aimed at determining whether and how kinesin-8 motors directly modulate MT dynamics should help resolve this question.
Our studies also reveal that Kif18A forms a gradient on kMTs that is dependent on its motor activity, suggesting that Kif18A’s plus-end directed motility is required for the concentration of the motor at the plus-ends of kMTs. Interestingly, the extent of Kif18A’s accumulation at kMT plus-ends varies within the spindle, as it is more concentrated on kMTs at the spindle periphery. The absolute concentration of motor could be influenced by a variety of factors such as length, stability or numbers of MTs within the kinetochore fiber. Interestingly, studies of purified Kip3p reveal that its rate of in vitro depolymerization is proportional to MT length, which is correlated with the accumulation of a higher concentration of the motor at the plus-ends (Varga et al., 2006). This leads us to propose a model wherein Kif18A utilizes a combination of length-dependent plus-end accumulation and concentration-dependent modulation of kMT plus-end dynamics to control mitotic chromosome positioning. In our model, Kif18A protein will accumulate at the plus-end of a kMT as it lengthens beyond the midzone (the center of the graph in Figure 6), and will dissociate as the kMT shortens. Our observation that the concentration of Kif18A is higher on one sister kinetochore than the other is consistent with this idea. As Kif18A accumulates at the plus-end, it increases to a threshold beyond which the probability that a kMT will undergo catastrophe is high, and in turn increases the chance that a chromosome will switch from away-from-pole to poleward movement. Such a mechanism would limit persistent movement and restrict oscillations of bioriented chromosomes to a region around the spindle equator where kMTs connected to opposite spindle poles are of relatively equal length (Figure 6).
Figure 6. Model for Kif18A regulation of mitotic chromosome movements.

The concentration of Kif18A at kinetochores is proportional to kMT length and stability. Once Kif18A reaches a threshold level (dashed gray line) at kMT plus-ends, it increases the probability that the kMT will undergo catastrophe and therefore increases the probability that the kinetochore will change direction. In a control cell (gray diagonal lines) Kif18A restricts oscillatory movements to a region near the spindle equator where kMTs emanating from opposite poles are of relatively equal length (black bracket). Increasing the concentration of Kif18A in the cell by over-expressing EGFP-Kif18A (green diagonal lines) increases the accumulation of Kif18A on kMTs and further restricts movements (green bracket). Reducing the concentration of Kif18A by treating cells with Kif18A-specific siRNAs (red diagonal lines) prevents threshold accumulation of the motor and leads to larger kinetochore oscillations (red bracket).
This model, in which Kif18A regulates kMT plus-ends in a concentration-dependent manner, is consistent with our analyses of kinetochore movements. We observed that increasing the concentration of Kif18A in the cell leads to both an increase in the accumulation of Kif18A at kMT plus-ends and a reduction in oscillation amplitude (Figure 6). In this situation, kinetochores have relatively high levels of Kif18A the majority of the time, and thus kMTs may be strongly biased towards shortening. This, in turn, could cause sister kinetochores on bioriented chromosomes to initiate poleward movement simultaneously, reducing coordinated sister chromosome movement and transiently increasing inter-kinetochore distance as we observed when Kif18A was over-expressed. In contrast, decreasing Kif18A leads to larger kinetochore oscillations, perhaps by preventing threshold accumulation of Kif18A at kMT plus-ends (Figure 6). Quantitatively, however, oscillation amplitude does not seem to be solely dependent on Kif18A concentration since near complete depletion of the protein does not completely randomize chromosome distribution (e.g. >90% depletion only increases oscillation amplitude 2-fold). Therefore, other cues, such as polar ejection forces or tension-dependent mechanisms, may be acting in parallel.
The correlation between Kif18A’s localization and kinetochore oscillations in control cells is also consistent with this model. Kif18A accumulates to a greater extent on the kMTs at the periphery of the spindle and the oscillations of peripheral kinetochores are reduced compared to those attached to kMTs along the pole-to-pole axis. This phenomenon is not specific to HeLa cells as similar variations in kinetochore movements have been observed in PtK1 cells (Canman et al., 2002; Cimini et al., 2004). Interestingly, increasing the concentration of Kif18A in the cell suppresses all kinetochore movements to the level seen at the spindle periphery in control cells. The fact that increased Kif18A does not further limit peripheral chromosome movements suggests that endogenous Kif18A already suppresses chromosome movements maximally at the spindle periphery in control cells.
In conclusion, our data suggest a model in which length-dependent modulation of kMT dynamics by Kif18A provides a spatial cue to control chromosome oscillations and thereby facilitate accurate organization and segregation of chromosomes during cell division.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HeLa cells were cultured as previously described (Maney et al., 1998). HeLa cells were transfected with plasmid DNA by electroporation using a Nucleofector II (Amaxa) according to the manufacturer’s instructions. Cells were transfected with siRNA using oligofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions. For Kif18A-depletion, cells were transfected with 60 μM each of siRNAs targeting the Kif18A sequences GCCAAUUCUUCGUAGUUUU and GCAGCUGGAUUUCAUAAA (Ambion). Treatment with this combination of siRNAs or with either siRNA alone at 120 μM produced indistinguishable effects. For Mad2-depletion cells were transfected with 60 μM each of siRNAs targeting the sequences GGAUGACAUGAGGAAAAUA and GCGUGGCAUAUAUCCAUCU (Ambion). For control depletions cells were transfected with 120 μM (for Kif18A single knockdown experiments) or 240 μM (for Kif18A/Mad2 double knockdown experiments) of Negative Control siRNA #1 (Ambion). Control siRNA treatment did not alter chromosome alignment or kinetochore movements relative to untreated control cells (data not shown).
Construction of DNA Plasmids
EGFP-CENP-B (pGFPCPB1) was constructed by PCR amplification of codons 1-167 of the C. griseus CENP-B gene (a kind gift from M. Valdivia) and subcloning into the EcoRI and Xba sites of pEGFP-C1 (Clontech). To prepare mRFP-CENP-B (pMX234) the EGFP gene of pEGFP-N1 (Clontech) was replaced by PCR-amplified mRFP1.0 (Campbell et al., 2002) and codons 1-167 of CENP-B to generate an EcoRI-NotI fragment bearing the mRFP-CENP-B fusion. EGFP-Kif18A was constructed by PCR amplification of codons 1-898 of the human Kif18A gene and subcloning into the EcoRI and Not1 sites of pEGFP-C1 (Clontech). Site directed mutagenesis was used to change H304, R308 and K311 to alanine in the EGFP-Kif18A-mutant. Venus-centrin was a kind gift from Benjamin Major and Randall Moon.
Immunofluorescence, Deconvolution and Linear Protein Mapping
HeLa cells were fixed as previously described (Maney et al., 1998). For drug treatments, cells were treated with 20 μM nocodazole (Sigma), 10 μM vinblastine (Sigma) or an equal volume of DMSO (Sigma) for 30 minutes prior to fixation. Antibodies against the c-terminus of Kif18A were raised in rabbits against a GST-tagged polypeptide containing amino acids 593-898 of Kif18A and then affinity purified. Cells were labeled with the following primary antibodies: mouse-anti-Hec1 (1:500, Abcam Inc.), rabbit-anti-Kif18A (1:50), mouse-anti-alpha-tubulin (1:50, Sigma) or human-CREST serum (1:50, a kind gift from Bill Brinkley) for 1 hour at room temperature. Anti-mouse, anti-rabbit and anti-human secondary antibodies conjugated to fluorescein or rhodamine (Jackson Laboratories) were used at 1:50 for 1 hour at room temperature. Stained cells were mounted in Vectashield with DAPI (Vector). Cells were imaged on a Nikon upright microscope equipped with a CCD camera and a 60× 1.4 NA lens (Nikon) or a Deltavision system equipped with a CCD camera and 60× 1.4 NA lens (Olympus). Selected images were deconvolved using a Deltavision image processing workstation (Applied Precision). The linescan function in the Softworx program (Applied Precision) was used to determine the spatial relationships and peak fluorescent values of Hec1 and Kif18A from immunofluorescent images.
SDS-PAGE and Western Blot
HeLa cells were lysed in 1X Laemmli sample buffer 24 or 48 hours after addition of siRNA. Lysates were briefly sonicated, boiled for 10 min and separated on 4-12% acrylamide gradient gels by SDS-PAGE. Proteins were transferred to nitrocellulose membrane and analyzed by Western Blot with polyclonal anti-Kif18A antibodies (1:100) and monoclonal anti-GAPDH antibodies (1:1000, Calbiochem). Proteins were visualized by chemoluminescence and GAPDH signals were quantified with the gel analyzer function in ImageJ.
Live Cell Imaging
HeLa cells were cultured in MEMalpha (Life Technologies) medium with 10% FBS (Hyclone) at 37°C and 5% CO2 on 35mm2 glass coverslip dishes coated with poly-l-lysine (MatTek Corp.) for 48 hours after DNA transfection and 24-36 hours after siRNA transfection before analysis by time-lapse microscopy. Prior to filming the cells were switched to 37°C CO2-independent media (Life Technologies). Cells were imaged with a Deltavision RT system (Applied Precision) equipped with a CCD camera, 60× 1.42 NA lens (Olympus) and a 37°C environmental chamber (Applied Precision). Z-stacks containing five focal planes with 0.5 μm spacing were acquired at intervals of 2, 5 or 20 seconds. Some cells were also imaged using a CARV (BD Biosciences) spinning disc confocal system attached to a TE2000 inverted microscope (Nikon) equipped with a ORCA ER (Hamamatsu) camera and a Plan Apo 60× 1.4 NA lens (Nikon). CARV images were collected at 5-second intervals using a single focal plane. Cells were maintained at 37°C with a thermoelectric stage.
Quantification of Kinetochore Movements
Based on the similarities between kinetochore movements in Kif18A-depleted and EGFP-Kif18A expressing cells to those of late prometaphase and metaphase control cells, kinetochore movements during these stages in controls were quantified. For all preanaphase measurements, the chosen kinetochores were bioriented, under tension and moving around the equator of the spindle. Kinetochore and spindle pole movements were tracked using maximum intensity projection movies from live cell imaging experiments and the Manual Tracking plug-in for ImageJ (NIH). Kinetochore movement parameters, spindle lengths and inter-kinetochore distances were quantified from tracking data using Igor Pro 6.0 software (Wavemetrics). All velocity measurements were made by linear regression analysis of kinetochore distance versus time plots. Velocity measurements during oscillatory movements were only made from cells filmed at time intervals of 5 or 2 seconds because kinetochores filmed at 20 second intervals frequently changed directions between data points making velocity measurements artifactually low (kinetochores change directions approximately every 40 seconds in HeLa cells based on our switch rate measurements). Average oscillation amplitudes were calculated by dividing average velocities by average switch rates. Statistical comparisons between data sets were performed using two-tailed t-tests assuming unequal variances. In cases where multiple measurements were made from the same kinetochore over time, an average value was calculated. The reported p-values are from comparisons of these kinetochore averages where the number of events is taken as the number of kinetochores analyzed. Indications of significance (p ≤ 0.05) from statistical tests using average values from each cell analyzed were consistent with those reported with the exception of inter-kinetochore distance changes in Kif18A-depleted or EGFP-Kif18A cells, which were not significantly different when compared in this manner.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Greg Martin (Keck Center for Neural Imaging) for assistance with live imaging, to Andrew Franck for programming assistance, and to Julia Chang for technical assistance with drug treatment experiments. We thank Chad Pearson, and the members of the Wordeman and Asbury labs for invaluable discussions and for critical reading of the manuscript. A National Institutes of Health Grant (GM69429) to L. Wordeman, grants from the Searle Scholars Program (06-L-111) and the Packard Fellowship for Science and Engineering to Charles Asbury, and a Ruth L. Kirschstein National Research Service Award (GM778572) to J. Stumpff supported this work. The Center for Cell Dynamics is an NIH Center for Excellence (GM066050).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ault JG, DeMarco AJ, Salmon ED, Rieder CL. Studies on the ejection properties of asters: astral microtubule turnover influences the oscillatory behavior and positioning of mono-oriented chromosomes. J Cell Sci. 1991;99(Pt 4):701–710. doi: 10.1242/jcs.99.4.701. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Zinkowski RP, Mollon WL, Davis FM, Pisegna MA, Pershouse M, Rao PN. Movement and segregation of kinetochores experimentally detached from mammalian chromosomes. Nature. 1988;336:251–254. doi: 10.1038/336251a0. [DOI] [PubMed] [Google Scholar]
- Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Salmon ED, Fang G. Inducing precocious anaphase in cultured mammalian cells. Cell Motil Cytoskeleton. 2002;52:61–65. doi: 10.1002/cm.10032. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Salmon ED. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J Cell Sci. 1994;107(Pt 1):285–297. doi: 10.1242/jcs.107.1.285. [DOI] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. Functional roles of poleward microtubule flux during mitosis. Cell Cycle. 2006;5:481–485. doi: 10.4161/cc.5.5.2519. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Hunt AJ. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J. 2002;83:42–58. doi: 10.1016/S0006-3495(02)75148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Compton DA. Searching for the middle ground: mechanisms of chromosome alignment during mitosis. J Cell Biol. 2002;157:551–556. doi: 10.1083/jcb.200202073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Gabashvili IS, Rieder CL. “Dumb” versus “smart” kinetochore models for chromosome congression during mitosis in vertebrate somatic cells. Cell Motil Cytoskeleton. 1999;43:179–185. doi: 10.1002/(SICI)1097-0169(1999)43:3<179::AID-CM1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007 doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Chromosome alignment at mitotic metaphase: balanced forces or smart kinetochores? In: D. F. M. Warner JR, editor. Cell Movement Volume 2: kinesin, dynein and microtubule dynamics. Alan R. Liss, Inc; New York: 1989. pp. 421–430. [Google Scholar]
- Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxsey SJ, Goldstein LS. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol. 1997;136:1081–1090. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional Analysis of Human Microtubule-based Motor Proteins, the Kinesins and Dyneins, in Mitosis/Cytokinesis Using RNA Interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.