Abstract
β-Catenin/T-cell factor (Tcf) signaling is constitutively active in the majority of human colorectal cancers, and there are accompanying changes in Bcl-2 expression. Similarly, 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP)-induced colon tumors in the rat have increased β-catenin and elevated Bcl-2. To examine the possible direct transcriptional regulation of rat Bcl-2 by β-catenin/Tcf, we cloned and characterized the corresponding promoter region and found 70.1% similarity with its human counterpart, BCL2. Bcl-2 promoter activity was increased in response to LiCl and exogenous β-catenin, including oncogenic mutants of β-catenin found in PhIP-induced colon tumors. Protein/DNA arrays identified E2F1, but not β-catenin/Tcf, as interacting most strongly with the rat Bcl-2 promoter. Exogenous E2F1 increased the promoter activity of rat Bcl-2, except in mutants lacking the E2F1 sites. As expected, β-catenin induced its downstream target c-Myc, as well as E2F1 and Bcl-2, and this was blocked by siRNA to c-Myc or E2F1. These findings suggest an indirect pathway for Bcl-2 overexpression in PhIP-induced colon tumors involving β-catenin, c-Myc and E2F1.
Keywords: β-catenin, Bcl-2, c-Myc, E2F1, wnt signaling, colorectal cancer
Introduction
Most human colorectal cancers have mutations in the adenomatous polyposis coli (APC) gene or β-catenin gene (CTNNB1), leading to increased β-catenin protein expression and activation of downstream β-catenin/T-cell factor (Tcf) target genes (Behrens, 2005). These cancers also exhibit marked changes in the expression of Bcl-2 family proteins (Bedi et al., 1995).
Similar findings have been reported in the colon tumors from animals treated with 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP) and 2-amino-3-methylimidazo(4,5-f)quinoline (IQ), which are dietary agents that the US National Toxicology Program has classified as ‘reasonably anticipated to be human carcinogens’. Indeed, there are a number of similarities between human and rat colon tumors with respect to β-catenin/Tcf signaling and the expression of Bcl-2 family proteins. First, as in the human situation, PhIP-and IQ-induced colon tumors in the rat contain mutations in Apc or Ctnnb1, but not in both of these genes (Dashwood et al., 1998). Second, these mutations stabilize β-catenin through inhibition of phosphorylation, ubiquitination and proteasome degradation, leading to increased expression of β-catenin protein (Al-Fageeh et al., 2004). Third, β-catenin/Tcf target genes frequently are overexpressed, including c-Myc (Blum et al., 2001, 2003). Fourth, during the progression from normal colonic mucosa to adenoma and carcinoma there is an increase in anti-apoptotic Bcl-2 protein and loss of proapoptotic Bax (Hayashi et al., 1996).
These findings allowed us to ask whether changes in apoptosis-related proteins, and specifically Bcl-2, might be directly related to β-catenin/Tcf signaling. Cloning and characterization studies ruled out the rat Bcl-2 gene as a direct target of β-catenin, but c-Myc and the transcription factor E2F1 were identified as intermediates in the upregulation of rat Bcl-2.
Results
The ratio of Bcl-2/Bax mRNA is increased in PhIP-induced colon tumors containing β-catenin mutations
Previous work showed elevated Bcl-2 and decreased Bax protein levels in heterocyclic amine-induced colon tumors (Hayashi et al., 1996); thus, we first examined the expression of Bcl-2 and Bax mRNA in six PhIP-induced colon tumors. Results are summarized in Table 1 in terms of the Bcl-2/Bax ratio, together with the mutation status of β-catenin, as determined by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) analysis. Codons 32 and 34 of Ctnnb1 are known ‘hot spots’ for heterocyclic amineinduced mutation (Dashwood et al., 1998; Blum et al., 2003), and three PhIP-induced colon tumors had β-catenin GGA→GAA (G34E) or GGA→GTA (G34 V) mutations. Although such mutations do not substitute critical Ser/Thr residues directly, they nonetheless stabilize the β-catenin protein by interfering with the relative extent of Ser33 phosphorylation by glycogen synthase kinase-3β (GSK-3β) and subsequent ubiquitination/proteasome degradation (Al-Fageeh et al., 2004). No β-catenin mutations were detected in three other colon tumors (Table 1). Interestingly, tumors with β-catenin mutations had an 8-fold increase in Bcl-2 versus Bax expression, whereas tumors that were wild-type for β-catenin had no marked changes in Bcl-2 or Bax. This suggested that β-catenin might upregulate Bcl-2 gene expression through direct or indirect mechanisms.
Table 1.
Bcl-2/Bax expression and β-catenin mutation status in PhIP-induced colon tumors
| Tumor ID # | Relative Bcl-2 mRNA expressiona | Relative Bax mRNA expressiona | Bcl-2/Bax ratio | Ctnnb1 mutation statusb |
|---|---|---|---|---|
| 98-01-18 | 4.0 | 0.5 | 8 | GGA→GAA (G34E) |
| 98-02-16 | 0.5 | 1.0 | 0.5 | None detected |
| 98-02-18 | 4.0 | 0.5 | 8 | GGA→GAA (G34E) |
| 98-02-20 | 4.0 | 0.5 | 8 | GGA→GTA (G34 V) |
| 98-02-22 | 1.0 | 1.0 | 1 | None detected |
| 98-02-23 | 1.0 | 1.0 | 1 | None detected |
Abbreviations: PhIP, 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine.
Tumor versus adjacent normal-looking tissue.
detected by PCRbased single strand conformation polymorphism analysis and direct sequencing (Dashwood et al., 1998), data not presented. Ctnnb1, gene designation for rat β-catenin.
Cloning andcharact erization of the rat Bcl-2 5′-flanking sequence
To investigate whether Bcl-2 might be a direct β-catenin/Tcf target gene in the rat, we cloned a 1745 bp fragment of the 5′-flanking region of rat Bcl-2, also containing part of exon 1 (see GenBank accession no. AF531426). Analysis using MatInspector (Genomatix) revealed putative DNA-binding sites for E2F1, NFκB, AP-2, SP1, c-Myb, SMAD, GATA, PAX3 and other transcription factors. Interestingly, four putative Tcf binding sites also were predicted.
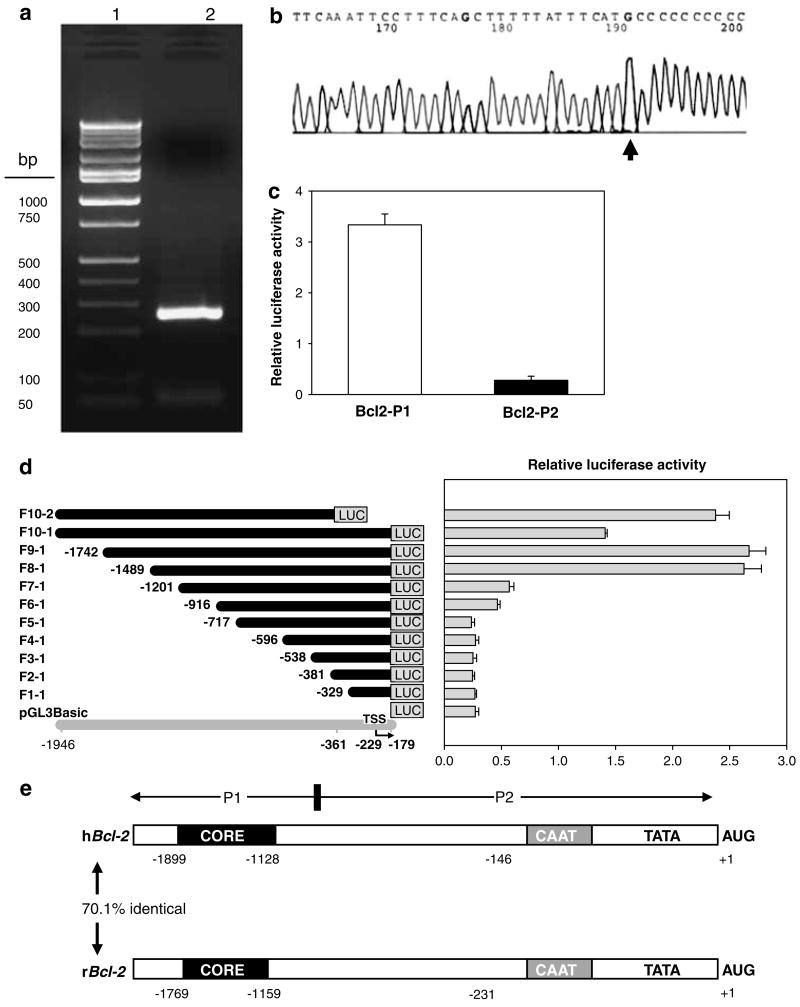
In 5′-RACE experiments, fresh poly(A)+RNA (Ambion, Austin, TX, USA) consistently generated a single 260-bp product and no larger or smaller fragments, suggesting only full-length poly(A)+RNAs were amplified (Figure 1a). Moreover, following gel purification and sequence analysis (Figure 1b), the transcription start site was readily assigned to a C nucleotide 231 bp upstream of the translation start site ATG, with a similar genomic organization as human Bcl-2 (Figure 1e). Indeed, hBcl-2 and rBcl-2 share 70.1% similarity, and the core promoter region of rBcl-2 has 85% identity with hBcl-2, containing several GC boxes, and CAAT and TATA box motifs located close to the AUG translation start site, as in hBcl-2 (Harigai et al., 1996).
Figure 1.
Characterization of the rat Bcl-2 promoter. (a) Product from 5′-rapid amplification of cDNA ends (5′-RACE) on a 2% agarose gel stained with ethidium bromide. In 5′-RACE, fresh poly(A)+RNA (Ambion) was used to ensure a high percentage of full-length transcripts, which consistently generated a single 260 bp PCR product (lane 2; lane 1=marker ladder). Degradation of full-length poly(A)+RNAs was discounted based on the lack of shorter or longer 5′-RACE products in repeat experiments, and DNA sequencing (GenBank accession no. AF531426) indicated a common 5′ end with the major TSS assigned to a C nucleotide 231 bp upstream of the translation start site (b), consistent with the human BCL-2 counterpart. (c) Bcl-2 promoter-luciferase reporter construct containing promoter region P1 had >10-fold higher activity than region P2 when transfected into HEK293 cells and luciferase activities were measured in whole cell lysates 48 h post-transfection. (d) Deletion analysis. Left panel: schematic view of Bcl-2 promoter-luciferase reporter constructs; gray line, position in genomic DNA sequence; bent arrow, transcription start site (TSS); black lines, extent of genomic DNA attached to the luciferase (LUC) gene. Promoter fragments were designated as F1-1 through F10-2, as shown at left. Right panel: Bcl-2 promoter activity in HEK293 cells. Transcriptional activity was evaluated by transient transfection, using the constructs shown in the left panel. Cells were extracted using 1×Reporter lysis buffer, 48 h post-transfection. Values represent the mean±s.d. of three independent transfections for each construct, normalized for transfection efficiency using pSV-β-Gal, as reported before (Li et al., 2004). (e) Genomic organization of human BCL-2 (hBCL-2) and rat Bcl-2 (rBcl-2) promoters, with numbering relative to the translation start codon (AUG). CORE, core promoter region.
In human embryonic kidney (HEK) 293 cells (Figure 1c), transient transfection of a reporter fragment containing −1945 to −906 of rat Bcl-2 (designated hereafter as ‘P1’) showed 10 times higher promoter activity than the fragment between −905 and −1 (referred to hereafter as ‘P2’). Sequential deletion analysis showed that deletion at position −1201 (F7-1) or shorter resulted in significant loss of Bcl-2 promoter activity (Figure 1d). However, F8-1, F9-1 and F10-2 had high promoter activity, indicating that the core promoter is located between −1201 and −1489 bp. Interestingly, F10-1 had lower promoter activity than F9-1 and F10-2, suggesting that negative regulatory element(s) might exist at each end of the Bcl-2 promoter.
Identification of transcription factors bound to the rat Bcl-2 promoter
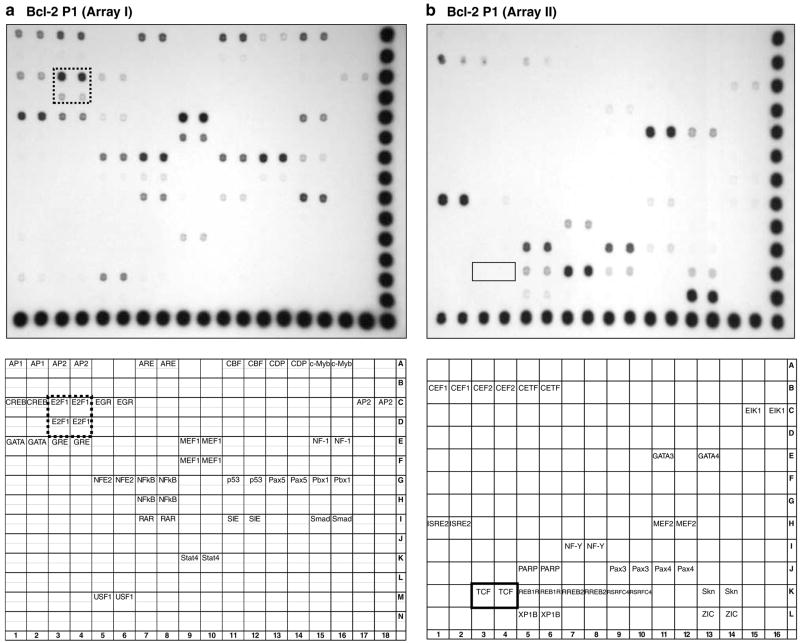
Nuclear proteins bound to the rat Bcl-2 promoter were purified by DNA pull-downs (Zeng et al., 2003), and they were identified using protein/DNA arrays, as reported before (Li et al., 2004). In Array I (Figure 2a), strong signals were obtained for E2F1 (dotted box), NFκB, MEF1, CBF, USF1, c-Myb, NF-1, Pax5 and Smad3/4. There also were signals for AP-1, AP-2, p53, GATA and CREB. In Array II, strong signals were detected for GATA3, ISRE, PARP, RREB2, Pax3 and ZIC (Figure 2b). However, no Tcf signal was observed (Figure 2b, box), even though four putative Tcf sites were predicted using MatInspector software.
Figure 2.
Transcription factors bound to the rat Bcl-2 promoter. Nuclear proteins bound to a 1040 bp (−1945 to −906) fragment of rBcl-2 promoter region P1 were pulled down from nuclear extracts of HEK293 cells and transcription factors were identified using TranSignal protein/DNA Array I (a) and Array II (b) (Panomics), as reported (Li et al., 2004). Top panels, hybridization signals; bottom panels, transcription factor identities. Box, Tcf signal; dotted box, E2F1 signal.
Subsequently, mobility-shift assays were performed with 32P-labeled oligos containing one of the four putative Tcf sites, in vitro translated Tcf4 (or Lef1), and nuclear extracts from HEK 293 cells. Promising bands initially were observed, but excess cold oligo failed to produce the expected competition, and no supershift was seen with Tcf4 or Lef1 antibodies (data not shown). Taken together with the protein/DNA analyses (Figure 2b), we concluded that the putative Tcf sites in rat Bcl-2 were not authentic.
E2F1 is a transcriptional activator of rat Bcl-2
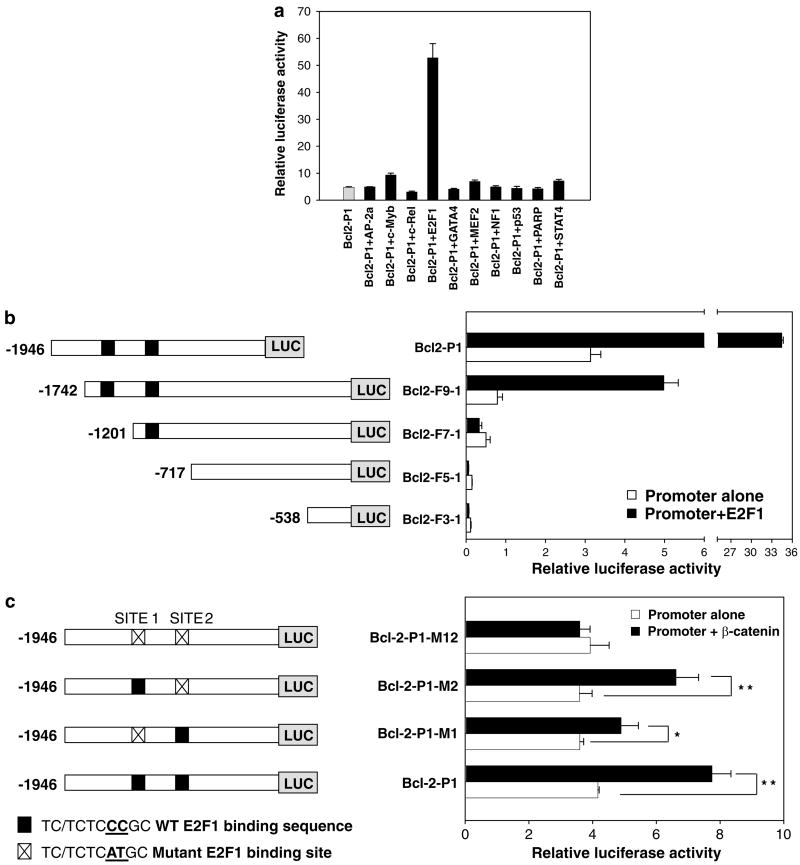
Because no authentic Tcf sites were found, 10 different transcription factors with affinity for the rat Bcl-2 promoter in protein/DNA arrays (Figure 2) were tested for their effects on Bcl2-P1. A striking response was seen with E2F1, which increased promoter activity >10-fold (Figure 3a). Reporter assays were performed with deletion constructs containing two, one, or zero E2F sites in the rat Bcl-2 promoter (Figure 3b). Exogenous E2F1 increased reporter activities 7- to 11-fold with constructs containing both E2F sites, but there was no effect with constructs containing one or no E2F sites. Site-directed mutagenesis targeted at both E2F sites completely abolished the induction of Bcl-2 promoter activity by exogenous β-catenin, and the response was markedly attenuated by mutation of site 1, but not site 2 (Figure 3c). Thus, site 1 appears to be more important for β-catenin- or E2F1-induced activation of the Bcl-2 promoter.
Figure 3.
Confirmation of E2F1 sites in the rat Bcl-2 promoter. (a) HEK293 cells were transfected with Bcl-2-P1 plus constructs expressing transcription factors identified in protein-DNA arrays (Figure 2). pSV-β-Gal was used as internal control. Cell lysates were analyzed for luciferase and β-Gal activities 48 h post-transfection. Luciferase activity was normalized to β-Gal to obtain the relative luciferase activity. E2F-1 binding sites were confirmed using (b) deletion analyses and (c) site-directed mutagenesis. Left, diagram of Bcl-2 promoter-luciferase constructs. Right, Bcl-2 promoter activities in the presence and absence of exogenous (b) E2F1 or (c) β-catenin. Data, mean±s.d., n=3. WT, wild-type. *P<0.05; **P<0.01.
β-Catenin upregulates Bcl-2 promoter activity
Although Bcl-2 was excluded as a direct target of β-catenin, owing to the lack of authentic Tcf sites (see above), we observed induction of Bcl-2-P1 reporter activity after forced expression of β-catenin. Thus, in HEK293 cells, overexpression of wild-type β-catenin by transient transfection increased the reporter activity twofold (Figure 4a), and LiCl, a well-known inhibitor of GSK-3β that stabilizes endogenous β-catenin (Aberle et al., 1997; Al-Fageeh et al., 2004), also increased Bcl-2 promoter activity. Immunoblotting of whole cell lysates confirmed that β-catenin and Bcl-2 proteins were increased by exogenous β-catenin and LiCl treatment (Figure 4b). Rather than using other means to increase endogenous wild-type β-catenin, such as soluble Wnt3a (Park et al., 2006), we examined oncogenic mutants found in PhIP-induced colon tumors, namely D32N, G34E and Δ45 (Figure 4c). Each β-catenin mutant activated Bcl-2 reporter activity more effectively than wild-type β-catenin, and there was a concomitant increase in Bcl-2 and β-catenin protein expression in cell lysates.
Figure 4.
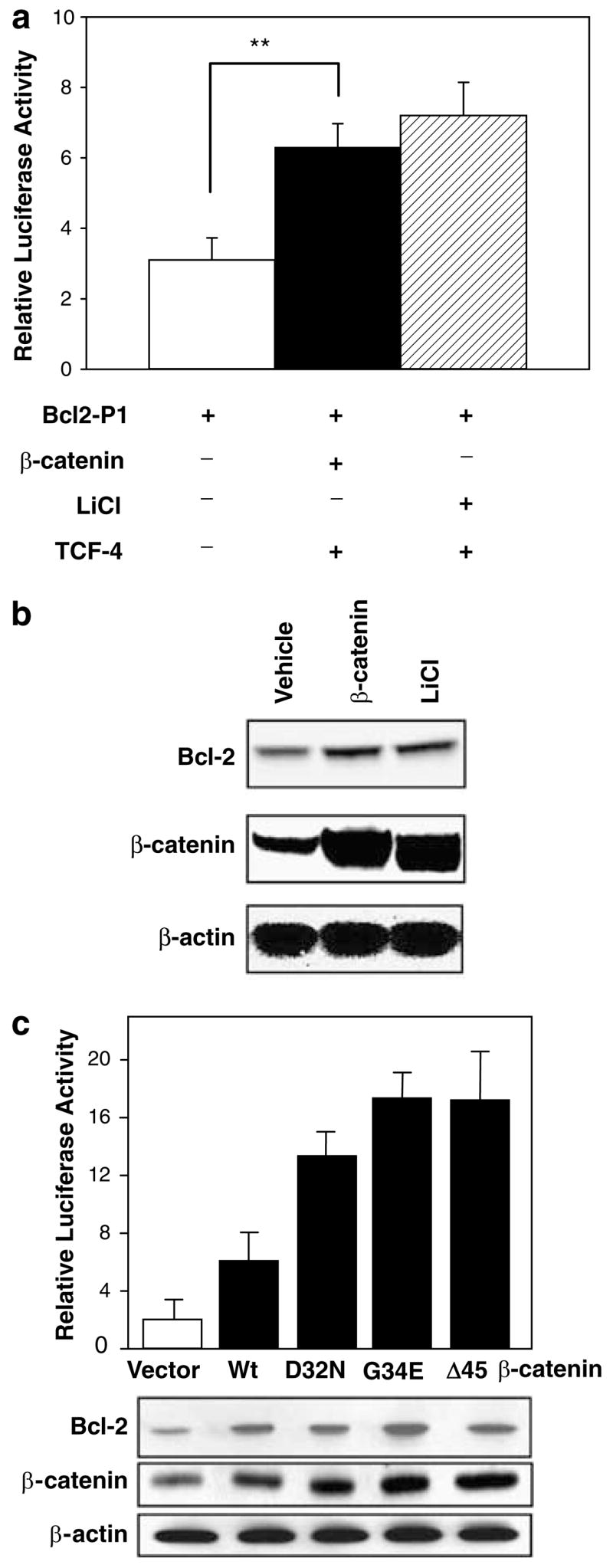
Regulation of Bcl-2 promoter activity by β-catenin. (a) Induction of Bcl-2 promoter activity by forced expression of β-catenin. HEK293 cells were transfected with Bcl-2-P1 promoter-luciferase construct alone, or Bcl-2-P1 plus a construct that overexpressed wild-type β-catenin; pSV-β-Gal was used as internal control. Alternatively, endogenous β-catenin was overexpressed with 30mM LiCl. (b) Immunoblot of whole cell lysates showing increased Bcl-2 and β-catenin following exogenous β-catenin or LiCl treatment. (c) Induction of Bcl-2 promoter–reporter activity by wild-type (Wt) β-catenin and β-catenin mutants D32N, G34E and Δ45, with concomitant changes in β-catenin and Bcl-2 protein expression in whole cell lysates.
β-Catenin upregulates c-Myc, E2F1 andhence Bcl-2
Because c-Myc is a well-known direct target of β-catenin/Tcf signaling (He et al., 1998), we next used siRNA to knockdown c-Myc. Overexpression of β-catenin induced rat Bcl-2 promoter activity (Figure 5a, upper panel), and increased protein expression of c-Myc, E2F1 and Bcl-2 (Figure 5a, lower panel). Reduction of c-Myc by siRNA blocked β-caten-independent induction of rat Bcl-2 promoter activity, as well as E2F1 and Bcl-2 protein expression. These data confirmed that c-Myc is not only a β-catenin/Tcf target, but also an E2F1 activator. Finally, using siRNA to knockdown E2F1 (Figure 5b), there was a corresponding attenuation of Bcl-2 promoter activity and Bcl-2 protein expression, with more cells undergoing apoptosis, as evidenced by an increased in cleaved (active) caspase-3.
Figure 5.
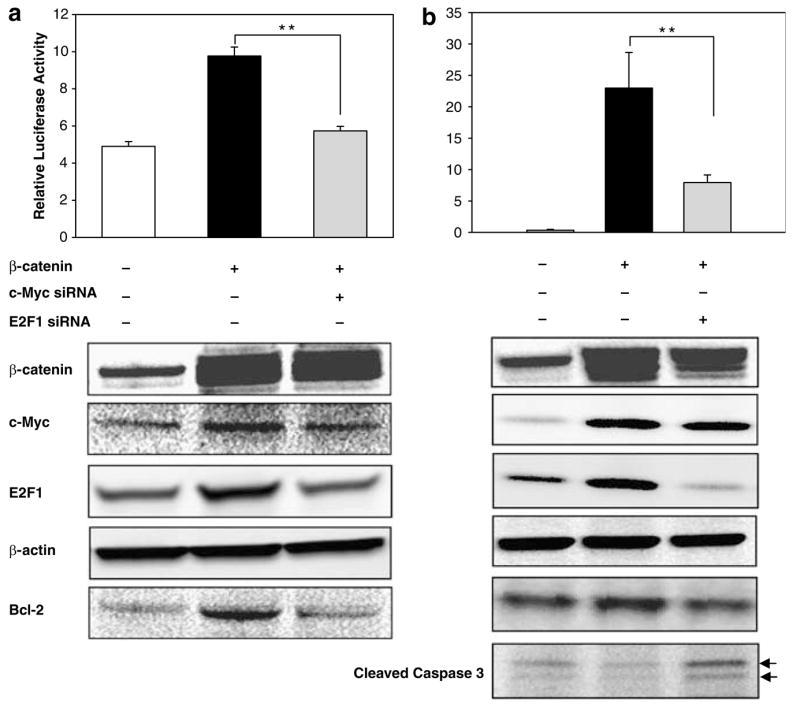
Knockdown of c-Myc or E2F1 attenuates β-catenin-dependent induction of Bcl-2 promoter activity and Bcl-2 protein expression. HEK293 cells were transfected with Bcl-2-P1, in the presence and absence of exogenous β-catenin, plus c-Myc siRNA or E2F1 siRNA. Luciferase and β-Gal activities were assayed 48 h post-transfection. Luciferase activity was normalized to β-Gal activity to obtain the Relative Luciferase Activity. Data, mean±s.d., n=3; **P<0.01. Corresponding whole cell lysates were immunoblotted for β-catenin, c-Myc, E2F1, β-actin, Bcl-2 and cleaved caspase 3, as indicated. Arrows, 19-kDa and 17-kDa bands indicative of cleaved (active) caspase 3.
Discussion
Elevated expression of β-catenin has been detected in colorectal and other cancers, resulting in constitutive activation of numerous β-catenin/Tcf target genes (Behrens, 2005). We reported on the high frequency of β-catenin mutations in PhIP- and IQ-induced rat colon tumors (Dashwood et al., 1998), the overexpression of β-catenin/Tcf targets such as c-Myc and c-Jun (Blum et al., 2001), and the elevated expression of Bcl-2 protein with loss of Bax (Hayashi et al., 1996). In the present investigation of PhIP-induced colon tumors, a striking concordance was found between the presence of β-catenin mutations and increased Bcl-2/Bax mRNA expression (Table 1), suggesting that β-catenin/Tcf might activate the rat Bcl-2 gene (and/or inhibit Bax) at the transcriptional level.
The Bcl-2 proto-oncogene is frequently expressed in human cancers, and Bcl-2 is regulated both transcriptionally and post-transcriptionally (Bedi et al., 1995; Harigai et al., 1996). We cloned and characterized the rat Bcl-2 promoter for the first time and found a similar genomic organization as the human counterpart, BCL-2 (Seto et al., 1988; Harigai et al., 1996), with a core promoter that shares 85% identity between the two species. We also identified transcription factors bound to the rat Bcl-2 promoter, and excluded several putative Tcf sites on the basis of the results from protein/DNA arrays (Figure 2) and mobility-shift assays (data not shown). Thus, the rat Bcl-2 gene does not appear to be a direct β-catenin/Tcf target.
Various transcription factors have been implicated as regulators of Bcl-2 (Wilson et al., 1996; Salomoni et al., 1997; Smith et al., 1998; Mayo et al., 1999; Pugazhenthi et al., 1999; Romero et al., 1999; Tamatani et al., 1999; Grossmann et al., 2000). Human BCL-2 contains an authentic E2F response element (Gomez-Manzano et al., 2001) and was identified as an E2F1 target gene by cDNA microarray analysis (Muller et al., 2001). We report here, for the first time, that E2F1 also directly regulates the rat Bcl-2 gene, with strong signals in protein/DNA arrays (Figure 2) and loss of promoter activity upon deletion of the E2F1 sites (Figure 3).
Rat Bcl-2 promoter activity was increased by LiCl treatment and by exogenous wild-type and mutant β-catenins. Lithium has been shown to stabilize β-catenin via inhibition of GSK-3β (Behrens, 2005), and increased Bcl-2 levels in rat frontal cortex, hippocampus and striatum, as well as in cultured retinal ganglion cells (Manji et al., 2000; Huang et al., 2003). Of particular interest, however, oncogenic mutants of β-catenin from PhIP-induced colon tumors strongly activated Bcl-2 promoter activity and Bcl-2 protein expression, supporting a link between increased β-catenin and Bcl-2 (Figure 4c).
c-Myc is a well-known β-catenin/Tcf target (He et al., 1998), and is strongly overexpressed in rat colon tumors both at the mRNA and protein level (Blum et al., 2001; Fujiwara et al., 2004). Interestingly, a recent report indicated that c-Myc-regulated microRNAs modulate E2F1 expression (O’Donnell et al., 2005). In our experiments, knockdown of c-Myc by siRNA blocked the induction of E2F1 and Bcl-2 by β-catenin, as well as inhibiting rat Bcl-2 promoter activity (Figure 5a), and knockdown of E2F1 by siRNA also attenuated Bcl-2 reporter activity and Bcl-2 protein expression (Figure 5b). We conclude that an indirect pathway exists between β-catenin and Bcl-2 in PhIP-induced colon tumors, in which mutations in β-catenin activate β-catenin/Tcf signaling, increase c-Myc, elevate E2F1 expression and enhance Bcl-2 expression. Further studies of this pathway are warranted, including the possible contribution of miroRNAs in PhIP-induced colon tumors and early lesions such as colonic aberrant crypts and dysplastic foci (Ochiai et al., 2003).
Materials and methods
Source of colon tumors
Colon tumors were from a study in which male F344 rats were treated with PhIP, as reported previously (Dashwood et al., 1998). At necropsy, one portion of each tumor was taken for histopathology, and other portions were frozen in liquid nitrogen for molecular analyses.
Competitive RT-PCR
Total RNA was isolated from colon tumors and adjacent normal looking tissue using RNeasy RNA isolation kit (Qiagen, Valencia, CA, USA). cDNAs were amplified with primers specific for Bcl-2 or Bax in the presence of serial dilutions of competitor DNA (Clontech, Palo Alto, CA, USA). Parallel reactions were run with primers and competitor DNA for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). PCR products were separated on 2% agarose gels, visualized by ethidium bromide staining and quantified on an AlphaImager 2200 (AlphaInnotech, San Leandro, CA, USA). Bcl-2 or Bax levels, normalized relative to GAPDH, were expressed for tumor versus normal looking tissue.
Mutation screening
PhIP-induced colon tumors and adjacent normal looking tissue were subjected to DNA isolation and PCR-based single strand conformation polymorphism (PCR-SSCP) analyses, using the experimental conditions reported previously (Dashwood et al., 1998).
Cloning of the 5′-flanking region of Bcl-2
The 5′-flanking region of the rat Bcl-2 gene was amplified using the Rat GenomeWalker kit (Clontech). The primary PCR was performed with Adapter Primer 1, supplied with the kit and gene-specific primer rBcl2P-GSP1 (5′-TGCATTCT TG GATGAAGGGGTGTCTT-3′). Subsequently, secondary PCR was performed with Adapter Primer 2 (supplied with the kit) and a nested gene-specific primer rBcl2P-GSP2 (5′-TCCCCCTTGGCATGAGATGCAGGAAAT-3′). The primary PCR was performed for 35 cycles at 98°C for 20 s and 65°C for 4 min, with an additional 10 min extension at 72°C after the final cycle. The nested PCR was run for 30 cycles, denaturing for 20 s at 98°C, annealing for 5 min at 68°C, and ending with a final extension for 10 min at 72°C. PCR products were subcloned into pGEM-T (Promega, Madison, WI, USA) and subjected to DNA sequencing in both directions on an ABI Prizm model 377 sequencer (Applied Biosystems, Bedford, MA, USA). Based on the new sequence, primers rBcl2P-GSP3 (5′-GGGAACGGGGACCAGAATCCTCTTCT-3′) and rBcl2P-GSP4 (5′-TTAAACTCCGAAGGGCCAATGCG TTTTC-3′) were used to obtain additional flanking sequence of Bcl-2.
5′-Rapid amplification of cDNA ends (5′-RACE)
The transcription start site was identified using 5′-RACE System Version 2.0 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. First strand cDNA was synthesized from 100 ng rat liver poly(A)+RNA using an antisense gene-specific primer RA-b2-GSP1 (5′-CCTCT GTGACAGCTTAT-3′). The cDNA was purified and an oligo-dC tail was added to the 3′end of the cDNA using terminal transferase TdT. Homopolymeric C tailed cDNA was then amplified by PCR using Abridged Anchor Primer (supplied with the kit) and a nested gene-specific primer RAb2- GSP2 (5′-CGGTTATCATACCCTGTTCTCCCGGCTT- 3′). PCR tubes were transferred from ice to a thermal cycler pre-equilibrated to 94°C, and after 2–3 min 35 cycles were performed of 30 s/94°C, 30 s/55°C and 60 s/72°C, with final extension at 72°C for 10 min. The PCR product was further amplified using nested primers AUAP (supplied with the kit) and RA-b2-GSP3 (5′-GAAGCTGCAGGTACCAATAGCA CTT-3′), and cycling parameters identical to the first round. 5′-RACE products were purified using Wizard PCR Preps DNA Purification System (Promega) and sequenced using primer RA-b2-GSP3.
Construction of Plasmids
Progressive deletion constructs of the rat Bcl-2 promoter, including Bcl2-P1 (−1945 to −906) and Bcl2-P2 (−905 to −1), were engineered by cloning PCR fragments between KpnI and XhoI sites of the reporter luciferase vector pGL3Basic (Promega); primer sequences are available upon request. Plasmids containing one or two mutant E2F1-binding sites were generated from pGL3-Bcl2-P1 by site-directed mutagenesis using the QuickChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). To generate pcDNA3.1-E2F1, an E2F1 expression construct, full-length human E2F1 cDNA was amplified by RT-PCR. The PCR products were cloned into pcDNA3.1(+) (Invitrogen, Carlsbad, CA, USA) between BamHI and EcoRV sites. According to the same methodology, plasmids expressing AP2α, c-Rel, c-Myb, GATA4, MEF2, NF-1, p53, PARP and STAT4 were constructed by subcloning the corresponding full-length cDNA into pcDNA3.1(+). The wild-type β-catenin cDNA construct pcDNAI/β-catenin was kindly provided by Hans Clevers and Marc van de Wetering. Oncogenic β-catenin mutants, generated by fragment switching, were as described before (Dashwood et al., 2002; Al-Fageeh et al. 2004; Dashwood et al., 2005). All constructs were confirmed by sequencing in both directions.
c-Myc and E2F1 knockdown by siRNA
Inhibition of c-Myc expression in HEK293 cells was performed using SureSilencing Human MYC siRNA and Antibody Kit (SuperArray Biosciences, Frederick, MO, USA). Cells were transfected with MYC-specific siRNA population using Lipofectamine2000 (Invitrogen) as recommended by the manufacturer. Non-specific siRNA was used as negative control. In subsequent experiments, E2F1 was knocked down using ON-TARGETplus SMARTpool human E2F1 siRNA from Dharmacon (Chicago, IL, USA).
Cell culture and transient transfection experiments
HEK293 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% horse serum (Invitrogen). Rat kidney epithelial (RK3E) cells were grown in DMEM supplemented with 10% bovine fetal serum (Invitrogen). Cultures were maintained at 37°C in a humidified 5% CO2-containing atmosphere. Transfection was performed using TransFast (Promega) or FuGENE 6 (Roche, Palo Alto, CA, USA) following manufacturer’s instructions, and cells were harvested 48 h after transfection. To inhibit GSK-3β and induce endogenous β-catenin, 30mM LiCl was added to culture medium; 30mM NaCl was used as control.
β-Galactosidase (β-Gal) and luciferase assays
β-Galactosidase and luciferase assays were performed as reported previously (Li et al., 2004).
Isolation of transcription factors bound to the rat Bcl-2 promoter
Transcription factors bound to the 1040 bp Bcl-2 promoter 1 (−1945 to −906) were purified by DNA pull-down assays according to the procedure described previously (Li et al., 2004). Bcl-2 promoter fragments were end labeled with biotin using Bio-16-dUTP (Enzo Life Sciences, Farmingdale, NY, USA) and the Klenow fragment of DNA polymerase I (Fermentas Inc., Hanover, MD, USA). Proteins were eluted on ice in 50 μl of TGED buffer containing 2M NaCl.
Protein/DNA array analyses
After DNA pull-down assays, TranSignal Protein/DNA Arrays I and II (Panomics, Redwood, CA, USA) were used to identify the transcription factors associated with the rat Bcl-2 promoter, as described before for the β-catenin gene Ctnnb1 (Li et al., 2004).
Western blotting
Whole cell lysates were prepared using Reporter Lysis buffer (Promega) and the protein concentration was determined as reported (Li and Dashwood, 2004; Li et al., 2004). Proteins were separated on 4–12% bis-tris gels (Novex, Invitrogen) and transferred to nitrocellulose membranes (Invitrogen), and after incubation with primary antibody followed by secondary antibody conjugated to horseradish peroxidase, detection was by Western Lighting Chemiluminescence Reagents Plus (Perkin Elmer Life Science, Boston, MA, USA).
Acknowledgments
Minako Nagao and Hideaki Inamori are gratefully acknowledged for their help in the studies with PhIP-induced colon tumors, which were supported by a fellowship from the Foundation for Promotion of Cancer Research, Tokyo, Japan. We thank Mark van de Wetering and Hans Clevers of University Hospital Utrecht, The Netherlands, for the wildtype β-catenin construct. This work was supported by NIH grants CA65525, CA80176 and CA90890, and by NIEHS center grant P30 ES00210.
References
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Fageeh M, Li Q, Dashwood WM, Myzak MC, Dashwood RH. Phosphorylation and ubiquitination of oncogenic mutants of β-catenin containing substitutions at Asp32. Oncogene. 2004;23:4839–4846. doi: 10.1038/sj.onc.1207634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi A, Pasricha PJ, Akhtar AJ, Barber JP, Bedi GC, Giardiello FM, et al. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55:1811–1816. [PubMed] [Google Scholar]
- Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem Soc Trans. 2005;33:672–675. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- Blum CA, Xu M, Orner GA, Fong AT, Bailey GS, Stoner GD, et al. β-Catenin mutation in rat colon tumors initiated by 1,2-dimethylhydrazine and 2-amino-3-methylimidazo[4,5-f]quinoline, and the effect of post-initiation treatment with chlorophyllin and indole-3-carbinol. Carcinogenesis. 2001;22:315–320. doi: 10.1093/carcin/22.2.315. [DOI] [PubMed] [Google Scholar]
- Blum CA, Tanaka T, Zhong X, Li Q, Dashwood WM, Pereira C, et al. Mutational analysis of Ctnnb1 and Apc in tumors from rats given 1,2-dimethylhydrazine or 2-amino-3- methylimidazo[4,5-f]quinoline: mutational ‘hotspots’ and the relative expression of β-catenin and c-jun. Mol Carcinogen. 2003;36:195–203. doi: 10.1002/mc.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashwood RH, Suzui M, Nakagama H, Sugimura T, Nagao M. High frequency of β-catenin (Ctnnb1) mutations in the colon tumors induced by two heterocyclic amines in the F344 rat. Cancer Res. 1998;58:1127–1129. [PubMed] [Google Scholar]
- Dashwood WM, Orner GA, Dashwood RH. Inhibition of β-catenin/Tcf activity by white tea, green tea, and epigallocatechin-3-gallate (EGCG): minor contribution of H2O2 at physiologically relevant EGCG concentrations. Biochem Biophys Res Commun. 2002;296:584–588. doi: 10.1016/s0006-291x(02)00914-2. [DOI] [PubMed] [Google Scholar]
- Dashwood WM, Carter O, Al-Fageeh M, Li Q, Dashwood RH. Lysosomal trafficking of β-catenin induced by the tea polyphenol epigallocatechin-3-gallate. Mutat Res. 2005;591:161–172. doi: 10.1016/j.mrfmmm.2005.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Ochiai M, Ohta T, Ohki M, Aburatani H, Nagao M, et al. Global gene expression analysis of rat colon cancers induced by a food-borne carcinogen, 2-amino-3-methylimidazo[4,5-f]quinoline. Carcinogenesis. 2004;25:1495–1505. doi: 10.1093/carcin/bgh155. [DOI] [PubMed] [Google Scholar]
- Gomez-Manzano C, Mitlianga P, Fueyo J, Lee HY, Hu M, Spurgers KB, et al. Transfer of E2F-1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61:6693–6697. [PubMed] [Google Scholar]
- Grossmann M, O’Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351–6360. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigai M, Miyashita T, Hanada M, Reed JC. A cis-acting element in the BCL-2 gene controls expression through translational mechanisms. Oncogene. 1996;12:1369–1374. [PubMed] [Google Scholar]
- Hayashi R, Luk H, Horio D, Dashwood RH. Inhibition of apoptosis in colon tumors induced in the rat by 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Res. 1996;56:4307–4310. [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Huang X, Wu DY, Chen G, Manji H, Chen DF. Support of retinal ganglion cell survival and axon regeneration by lithium through a Bcl-2-dependent mechanism. Invest Ophthalmol Vis Sci. 2003;44:347–354. doi: 10.1167/iovs.02-0198. [DOI] [PubMed] [Google Scholar]
- Li Q, Dashwood WM, Zhong X, Al-Fageeh M, Dashwood RH. Cloning of the rat β-catenin gene (Ctnnb1) promoter and its functional analysis compared with the Catnb and CTNNB1 promoters. Genomics. 2004;83:231–242. doi: 10.1016/j.ygeno.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Li Q, Dashwood RH. Activator protein-2α associates with adenomatous polyposis coli/β-catenin and inhibits β-catenin/T-cell factor transcriptional activity in colorectal cancer cells. J Biol Chem. 2004;269:45669–45675. doi: 10.1074/jbc.M405025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji HK, Moore GJ, Chen G. Lithium up-regulates the cytoprotective protein Bcl-2 in the CNS in vivo: a role for neurotrophic and neuroprotective effects in manic depressive illness. J Clin Psychiatry. 2000;61:82–96. [PubMed] [Google Scholar]
- Mayo MW, Wang C, Drouin SS, Madrid LV, Marshall AF, Reed JC, et al. WT1 modulates apoptosis by transcriptionally upregulating the bcl-2 proto-oncogene. EMBO J. 1999;18:3990–4003. doi: 10.1093/emboj/18.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai M, Ushigome M, Fujiwara K, Ubagai T, Kawamori T, Sugimura T, et al. Characterization of dysplastic aberrant crypt foci in the rat colon induced by 2-amino-3-methylimidazo[4,5-f]quinoline. Am J Pathol. 2003;163:1607–1614. doi: 10.1016/S0002-9440(10)63517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Park S, Gwak J, Cho M, Song T, Won J, Kim DE, et al. Hexachlorophene inhibits Wnt/β-catenin pathway by promoting Siah-mediated β-catenin degradation. Mol Pharmacol. 2006;70:960–966. doi: 10.1124/mol.106.024729. [DOI] [PubMed] [Google Scholar]
- Pugazhenthi S, Miller E, Sable C, Young P, Heidenreich KA, Boxer LM, et al. Insulin-like growth factor-1 induces bcl-2 promoter through a transcription factor camp-response element-binding protein. J Biol Chem. 1999;274:2829–2837. doi: 10.1074/jbc.274.39.27529. [DOI] [PubMed] [Google Scholar]
- Romero O, Martinez AC, Camonis J, Rebollo A. Aiolos transcription factor controls cell death in T cells by regulating Bcl-2 expression and its cellular localization. EMBO J. 1999;18:3419–3430. doi: 10.1093/emboj/18.12.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, et al. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and c-MYB-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci USA. 1997;94:3296–3301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Ensor EA, Coffin RS, Boxer LM, Latchman DS. Bcl-2 transcription from the proximal P2 promoter is activated in neuronal cells by the Brn-3a POU family transcription factor. J Biol Chem. 1998;273:16715–16722. doi: 10.1074/jbc.273.27.16715. [DOI] [PubMed] [Google Scholar]
- Tamatani M, Che YH, Matsuzaki H, Ogawa S, Okado S, Miyake S, et al. Tumor necrosis factor induces Bcl-2 and Bcl-x expression through NFκB activation in primary hippocampal neurons. J Biol Chem. 1999;274:8531–8538. doi: 10.1074/jbc.274.13.8531. [DOI] [PubMed] [Google Scholar]
- Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apopotosis. Mol Cell Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Gao L, Xia T, Tencomnao T, Yu RK. Characterization of the 5′-flanking fragment of the human GM3-synthase gene. Biochim Biophys Acta. 2003;1625:30–35. doi: 10.1016/s0167-4781(02)00573-0. [DOI] [PubMed] [Google Scholar]