Abstract
Activation of fibroblast growth factor (FGF) receptors elicits diverse cellular responses including growth, mitogenesis, migration, and differentiation. The intracellular signaling pathways that mediate these important processes are not well understood. In Caenorhabditis elegans, suppressors of clr-1 identify genes, termed soc genes, that potentially mediate or activate signaling through the EGL-15 FGF receptor. We demonstrate that three soc genes, soc-1, soc-2, and sem-5, suppress the activity of an activated form of the EGL-15 FGF receptor, consistent with the soc genes functioning downstream of EGL-15. We show that soc-2 encodes a protein composed almost entirely of leucine-rich repeats, a domain implicated in protein–protein interactions. We identified a putative human homolog, SHOC-2, which is 54% identical to SOC-2. We find that shoc-2 maps to 10q25, shoc-2 mRNA is expressed in all tissues assayed, and SHOC-2 protein is cytoplasmically localized. Within the leucine-rich repeats of both SOC-2 and SHOC-2 are two YXNX motifs that are potential tyrosine-phosphorylated docking sites for the SEM-5/GRB2 Src homology 2 domain. However, phosphorylation of these residues is not required for SOC-2 function in vivo, and SHOC-2 is not observed to be tyrosine phosphorylated in response to FGF stimulation. We conclude that this genetic system has allowed for the identification of a conserved gene implicated in mediating FGF receptor signaling in C. elegans.
Fibroblast growth factor (FGF) receptors comprise a family of transmembrane receptor tyrosine kinases (RTKs) that mediate diverse cellular responses including growth, mitogenesis, migration, and differentiation. FGF receptors are activated by the concerted action of secreted polypeptide growth factor ligands and heparin sulfate proteoglycans (reviewed in refs. 1 and 2). Like other RTKs, activation is associated with receptor dimerization and autophosphorylation on specific intracellular tyrosine residues (3). Autophosphorylation leads to activation of receptor kinase activity and generates potential binding sites for Src homology 2 or phosphotyrosine binding domain proteins (4).
Six major autophosphorylation sites (Y463, Y583, Y585, Y653, Y654, and Y730) on human FGF receptor 1 have been identified. Two of these sites, Y653 and Y654, are required for the stimulation of receptor kinase activity (5). Phospholipase C-γ has been shown to associate with human FGF receptor 1 requiring Y766 (6); however, the significance of this interaction is unclear because mutation of Y766 abolishes phosphatidylinositol hydrolysis without affecting mitogenic or differentiation responses to FGF in several cell lines (7–9). In fact, FGF receptors lacking all autophosphorylation sites except those required for kinase activity are capable of inducing proliferation and differentiation in a variety of cell lines (5), suggesting that, unlike certain other RTKs, target recruitment to autophosphorylation sites plays a limited role in FGF receptor signaling.
Members of the Ras/mitogen-activated protein kinase pathway have been implicated in FGF receptor-mediated signaling in a number of cell types (10, 11) as well as in mesoderm induction in Xenopus (12). Interestingly, there appear to be two pathways linking FGF receptor activation to the Ras/mitogen-activated protein kinase cascade. The adaptor protein Shc (13) is tyrosine phosphorylated in response to FGF and recruits a GRB2/Sos complex, presumably leading to the activation of Ras (10). Recently, a second mechanism for GRB2/Sos recruitment has been identified. FRS2, a lipid-anchored phosphotyrosine binding domain protein, is tyrosine phosphorylated in FGF-stimulated cells and also recruits the GRB2/Sos complex (14). While the Ras pathway appears to be involved in FGF receptor responses, the relative contribution of these two pathways to Ras activation remains unclear, as is the contribution of Ras-independent pathways.
The finding that egl-15 encodes an FGF receptor with multiple distinct functions in Caenorhabditis elegans development (15) offers the opportunity to use genetics to dissect FGF receptor pathways. One role of the EGL-15 FGF receptor is uncovered by a genetic interaction with clr-1, which encodes a receptor tyrosine phosphatase (16). Hypomorphic mutations in egl-15 and clr-1 display mutual suppression, suggesting that these genes act antagonistically. Genetic analysis using putative null alleles of egl-15 and clr-1 demonstrate that EGL-15 is required for the clear (Clr) phenotype of clr-1. These findings are consistent with a model in which CLR-1 acts as a negative regulator of EGL-15 signaling and the Clr phenotype results from hyperactive EGL-15 FGF receptor signaling. This model is supported by the finding that an activated egl-15 construct can confer a Clr phenotype when introduced into wild-type C. elegans (16).
Because the Clr phenotype appears to be caused by excess EGL-15 signaling, suppressors of clr-1 can identify mutations that reduce signaling through this FGF receptor pathway. Screens for suppressors of clr-1 (soc) have identified multiple alleles of four genes: egl-15, sem-5, soc-1, and soc-2 (15). As our model predicts, reduction of function mutations in the EGL-15 FGF receptor are isolated as suppressors of clr-1. Mutations affecting SEM-5, the structural and functional homolog of the GRB2 adaptor protein (17), also suppress clr-1 (15), supporting the hypothesis that suppressors of clr-1 can identify FGF receptor pathway components. The two previously uncloned genes identified in this screen may also participate in this FGF receptor pathway. We have cloned and characterized soc-2 to understand the molecular basis of soc-2 function in FGF receptor signaling.
MATERIALS AND METHODS
Strains.
C. elegans strains were maintained according to standard methods (18). The parental strain for all work was C. elegans N2 with the exception of strains used for restriction fragment length polymorphism mapping of soc-2, which contained DNA from the Bergerac strain N62.
Mapping of soc-2.
Dpy non-Unc and semi-Dpy Unc recombinants were isolated at 15°C from clr-1(e1745ts); dpy-13(e184sd) + unc-5(e53)/+ soc-2(n1774) + heterozygotes. Their progeny were shifted to 25°C and scored for the Soc phenotype. Soc progeny were segregated by 9/11 Dpy non-Unc and 2/8 Unc semi-Dpy recombinants. clr-1(e1745ts); soc-2(n1774)/mDf4 animals were found to be viable and Soc when shifted to 25°C, demonstrating that the deficiency mDf4 deletes at least a portion of soc-2.
The soc-2 gene was mapped with respect to restriction fragment length polymorphisms between the N2 Bristol and the N62 Bergerac strains. Soc non-Unc recombinants were isolated from clr-1(e1745ts); soc-2(n1774) unc-5(e53)/soc-2(+, Berg) unc-5(+, Berg) heterozygotes. Southern blots of genomic DNA isolated from strains homozygous for the recombinant chromosome were probed with ASL6 (A. Spence, personal communication) and C14D5 to detect eP14 and ayP4, respectively. 2/8 Soc-2 non-Unc-5 recombinants contain ayP4 and an additional 3/8 contain eP14.
Germ Line Transformation.
Germ line transformation of clr-1(e1745ts); soc-2(n1774) hermaphrodites was performed as described for sem-5 (19). The pRF4 plasmid, which confers a dominant Roller (Rol) phenotype, was used as a cotransformation marker. Injected hermaphrodites were propagated at 15°C. A fraction of the extrachromosomal array-bearing F1 animals were shifted to 25°C for 8–12 hr and scored for the Clr phenotype (F1 rescue). The remaining F1 Rol animals were maintained at 15°C to generate stable lines. Stable transgenic lines were temperature shifted and scored for rescue.
The activated egl-15(neu*) construct (16) was injected at 20 ng/μl with 100 ng/μl pRF4 as a cotransformation marker. F1 Rol transformants were scored for the Clr phenotype and for abnormal morphology at 20°C.
Isolation and Analysis of soc-2 cDNAs.
soc-2 cDNAs were isolated from a mixed stage cDNA library (20) using an 8.5-kb XhoI fragment from ZL158 as a probe. Sequence analysis of six positive cDNA clones indicated that all were independent clones from the same gene. One of the six clones had sequence from this gene fused to sequence derived from chromosome II of C. elegans and was not further analyzed. The remaining five clones had identical 3′ untranslated regions and polyadenylation sites, but differed in the extent of sequence at the 5′ end. An ORF extended to the very end of even the longest cDNA clone, indicating that none of the clones were full length. 5′ rapid amplification of cDNA ends (RACE) reactions were performed to obtain sequences corresponding to the 5′ end of this transcript (Marathon cDNA Amplification kit, CLONTECH). Subsequently, a soc-2 expressed sequence tag (EST) (GenBank accession no. M75931) was identified by the C. elegans genome consortium that ends two nucleotides beyond the 5′ end predicted by our rapid amplification of cDNA ends (RACE) products.
Determination of soc-2 Genomic Organization.
To determine the intron/exon boundaries of the soc-2 locus, the soc-2 cDNA sequence was compared with the genomic sequence of soc-2 determined by the C. elegans genome consortium (GenBank accession no. U61957). This genomic sequence was initially reported to be from chromosome III (21) but was subsequently linked to the region of chromosome IV to which we mapped soc-2 (A. Coulson, C. elegans Genome Consortium, personal communication).
Allele Sequencing.
The soc-2 coding regions and intron/exon boundaries were PCR amplified from genomic DNA prepared from clr-1(e1745ts); soc-2 strains. The products were purified (Qiagen, Chatsworth, CA) and sequenced. Mutations were confirmed by sequencing an independent PCR product.
shoc-2 Cloning.
The predicted SOC-2 protein sequence was compared with those in the database of ESTs (dBEST) by using the blast algorithm (22). A PCR product corresponding to an EST (Genbank accession no. F11387) with the potential to encode a protein fragment with high homology to a portion of SOC-2 was amplified from human genomic DNA and used to isolate cDNAs from a human fetal brain cDNA library (CLONTECH). Six clones were isolated and analyzed. One 2.3-kb clone was sequenced and found to contain a single ORF of 582 amino acids flanked by regions of 5′ and 3′ untranslated regions. We confirmed that the remaining library clones shared portions of the ORF by PCR amplification using primers derived from the 2.3-kb clone. The accession number of the 2.3-kb SHOC-2 cDNA clone is AF054828. We assembled a contig representing a full-length cDNA of 4.0 kb by determining the sequence of the ends of two additional clones and performing 5′ and 3′ rapid amplification of cDNA ends (RACE) (Marathon cDNA Amplification kit, CLONTECH).
shoc-2 Genomic Clones and Fluorescent in Situ Hybridization Analyses.
The shoc-2 cDNA was used to screen a gridded human genomic P1-derived artificial chromosonal (PAC) library (Roswell Park Cancer Institute, Buffalo, NY). Three positive PACs shared a common restriction digest banding pattern. One of the PACs, 80H10, was used for fluorescent in situ hybridization. Both cytogenetic mapping and fractional length analysis (23) localize shoc-2 to 10q25.
Northern Blot Analysis.
Northern blots of poly(A)+ RNA prepared from mixed stage wild-type N2 C. elegans hermaphrodites were prepared as described (15). Hybridization with a soc-2 cDNA probe was performed in Expresshyb (CLONTECH) following the manufacturer’s instructions. Radiolabeled shoc-2 cDNA was used to probe a human multiple tissue northern blot (CLONTECH) in Expresshyb.
soc-2 Constructs.
The soc-2 cDNA was used as a template for site-directed mutagenesis (CLONTECH Transformer kit). Mutagenized fragments were cloned into the 9.0-kb genomic rescuing construct.
Antibody Production and Purification.
A 560-bp shoc-2 fragment (HpaI to NotI, amino acid 392 to stop codon) was cloned into pGex4T-1 to generate a glutathione S-transferase fusion protein. Recombinant protein was expressed in bacteria, purified on a glutathione column and injected into rabbits to generate polyclonal anti-SHOC-2 antibodies.
The SHOC-2 antisera was affinity purified by incubating 1 ml of sera with 1 mg of nitrocellulose-bound antigen for 12 hr at 4°C. The nitrocellulose was washed several times in PBS. Antibody was eluted with 100 mM glycine pH 2.5 and brought to pH 7.0 with 100 μl Tris, pH 8.8.
Immunofluorescence.
COS 7 (American type Culture Collection, Manassas, VA) cells were transiently transfected with a DNA construct consisting of the SHOC-2 cDNA inserted in the pCB7 expression vector. Transfections were done by using LipofectAMINE transfection reagent (GIBCO/BRL) following the manufacturer’s recommended protocol. Twenty-four hours after the start of transfection, the cells were trypsinized, seeded onto glass coverslips, and allowed to grow for 24 hr.
The SHOC-2 transfected COS 7 cells were fixed in methanol/acetone and blocked for 1 hr with PBS containing 2% normal goat serum (GIBCO/BRL). Coverslips were incubated for 2 hr in affinity purified SHOC-2 antibodies, preimmune sera, or affinity purified SHOC-2 antibodies preincubated for 10 min with 50 μg purified antigen (antigen block). The cells were washed several times with blocking buffer and incubated in FITC-conjugated goat anti-rabbit antibody (Sigma) for 1 hr. Coverslips were washed several times and mounted in Vectastain (Vector Laboratories).
FGF Stimulation of NIH 3T3 Cells.
NIH 3T3 fibroblasts (ATCC) were maintained in DMEM (Yale Cell Biology Tissue Culture Facility, New Haven, CT) supplemented with 10% fetal bovine serum (GIBCO/BRL) until cells reached 80–90% confluence. Cells were starved for 2 hr in DMEM lacking fetal bovine serum and then treated for 5 min with aFGF (100 ng/ml, R & D Systems) plus heparin (10 μg/ml, Sigma) in DMEM with 10% fetal bovine serum. Unstimulated controls were treated similarly except aFGF and heparin were omitted. Cells were rinsed with ice-cold PBS and lysed in 1 ml of lysis buffer [20 mM Tris, pH 7.6/150 mM NaCl/50 mM NaF/1 mM Na3V04/5 mM benzamidine/1 mM EDTA/1% Nonidet P-40/10 μg/ml leupeptin/10 μg/ml aprotinin/1 mM Pefabloc (Boehringer Mannheim)].
Immunoprecipitation and Western Blot Analysis.
Lysates containing 500 μg of total protein were incubated overnight at 4°C with appropriate antisera. Immunocomplexes were precipitated for 1 hr using 50 μl of protein A agarose (Boehringer Mannheim) at 4°C. The complexes were washed three times with cold lysis buffer. Proteins were transferred to polyvinylidene difluoride membrane (Millipore) after SDS/PAGE.
For anti-phosphotyrosine blotting, membranes were blocked for 1 hr in 1% BSA in PBS and 0.1% Tween-20. For all other primary antibodies, membranes were blocked in 5% nonfat dry milk in PBS and 0.1% Tween-20. Membranes were incubated with primary antibody prepared in blocking buffer for 1 hr at room temperature, washed for 10 min in PBS, and incubated with peroxidase-linked secondary antibodies (Amersham). Blots were washed for 40 min in PBS and 0.1% Tween-20 with several changes of buffers. Results were detected by enhanced chemiluminescence (Amersham).
RESULTS
soc-2 Encodes a Leucine-Rich Repeat (LRR) Protein.
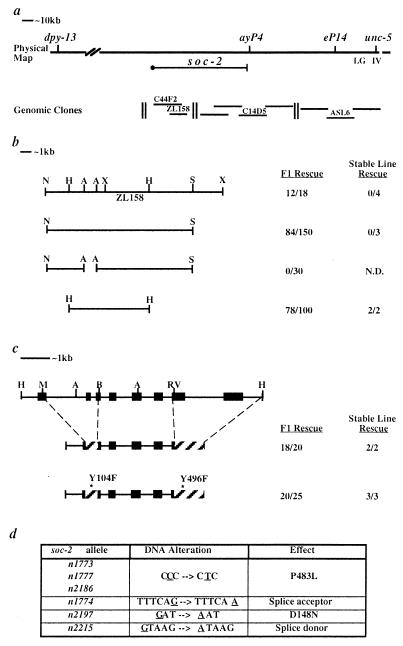
A contig of overlapping yeast artificial chromosomes, cosmids and λ phage assembled by the C. elegans genome consortium was used to clone soc-2. Three-factor mapping positioned soc-2 between two cloned genes, dpy-13 and unc-5, and close to unc-5. The soc-2 position was refined by mapping with respect to restriction fragment length polymorphisms, which placed soc-2 to the left of eP14 and ayP4 (Fig. 1a). Yeast artificial chromosomes, cosmids, and λ phage clones from this region were tested for rescue of the Soc phenotype of clr-1(e1745ts); soc-2(n1774). Phage clone ZL158 was found to confer rescuing activity in germ line transformation experiments. A 9.0-kb HpaI fragment of ZL158 was the smallest subclone to retain this rescuing activity (Fig. 1b). A single transcript was identified from this region, and a corresponding 2.3-kb cDNA was assembled from 5′ rapid amplification of cDNA ends (RACE) products and cDNA clones (see Materials and Methods). Northern blot analysis revealed a single band corresponding to the 2.3-kb transcript size predicted from this cDNA (data not shown). The genomic organization of this locus (Fig. 1c) was determined by comparing the cDNA sequence to the genomic sequence determined by the C. elegans genome consortium. The cDNA contains a single long ORF with the potential to encode a 559-amino acid protein. Molecular lesions associated with six soc-2 alleles were found to affect this ORF, indicating that this transcript corresponds to the soc-2 gene (Fig. 1d and 2a). Comparison of the SOC-2 protein sequence to proteins in the GenBank databases, indicated a high level of similarity to proteins containing LRRs (24). SOC-2 is composed almost exclusively of 18–19 reiterations of LRRs (Fig. 2a).
Figure 1.
Cloning of soc-2. (a) The position of soc-2 is shown with respect to nearby genes and Bristol/Bergerac restriction fragment length polymorphisms. The clones named were tested for soc-2 rescuing activity. Of the clones shown, only phage ZL158 rescued the Soc phenotype of soc-2(n1774). (b) Germline transformation rescue results for subclones of ZL158. A break in the bar designates a deletion. (c) The genomic organization of soc-2 is shown at the top. The positions of exons, ▪; cDNA replacements, ▨. Relevant restriction sites are indicated: A, AgeI; B, BspEI; RV, EcoRV; H, HpaI; M, MluI; N, NotI; S, SacII; X, XhoI. (d) Sequence analysis of soc-2 mutants. Sequence alterations found in soc-2 mutants are underlined. The resulting amino acid substitutions or splice site mutations are shown in the third column.
Figure 2.
(a) Protein alignment of SOC-2 and SHOC-2. Amino acid positions are numbered at the right. LRRs are underlined and assigned a repeat number. The YXNX motifs are double underlined. Sequence identities and similarities are shaded and boxed, respectively. The position of mutations in soc-2 mutants are designated by +. Splice site mutations are designated by a vertical line. (b) Alignment of SHOC-2 LRRs and a representative LRR from S. cerevisiae adenylyl cyclase. Residues identical in the majority of LRRs are shaded in black. Allowable substitutions for leucines within the LRRs are boxed (24).
The existing soc-2 alleles were isolated as viable ethyl methanesulfonate (EMS)-induced mutants that suppress all of the phenotypes associated with clr-1(e1745ts) at 25°C. Three lines of evidence suggest that this suppression is due to mutations that compromise, but do not eliminate, SOC-2 function. (i) These soc-2 mutations arise at a frequency (3.2 × 10−5) that is far lower than the rate at which mutations that eliminate gene function are normally isolated for this type of mutagenesis in C. elegans (25). Thus, null alleles of soc-2 are likely to confer a phenotype that precludes their isolation as viable, fertile suppressors of clr-1(e1745ts). (ii) None of the alterations identified in soc-2 mutants (Figs. 1d and 2a) strongly suggest that they eliminate SOC-2 function. Two alleles, n1774 and n2215, have distinct mutations predicted to affect splicing of the soc-2 message (26), consistent with these mutations reducing the level of the wild-type soc-2 product. The n2197 mutation changes an aspartic acid to asparagine within LRR12. Interestingly, three independently isolated alleles have the same lesion, resulting in a proline-to-leucine substitution within LRR17. (iii) While these mutations do not appear to be null alleles, gene dosage experiments suggest that they are not gain-of-function mutations. soc-2 mutations are complemented by soc-2(+) genes both in soc-2/+ heterozygotes and in soc-2(n1774); Ex[soc-2(+)] germ line transformants. Furthermore, the deficiency mDf4 fails to complement soc-2(n1774) for the Soc phenotype. Taken together, these results suggest that the Soc phenotype of these mutants results from mutations that reduce the function of SOC-2.
SOC-2 May Function Downstream of the EGL-15 FGF Receptor.
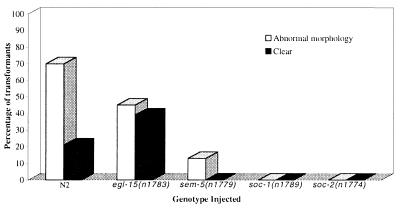
While genetic evidence suggests that suppressors of clr-1 are FGF receptor pathway components, it does not distinguish those genes that modulate receptor activity from those that mediate signaling downstream of the FGF receptor. An EGL-15(neu*) FGF receptor chimera that behaves as a constitutively activated receptor (16) was used to test where the soc genes might function in FGF receptor signaling. This chimera, which can confer a Clr phenotype when introduced into wild-type animals (16), was introduced into soc mutants to test whether the soc genes are required for the activity of EGL-15(neu*). Mutations in soc-1, soc-2, and sem-5 suppress the activity of EGL-15(neu*) (Fig. 3), indicating that these genes do not function to activate EGL-15. In contrast, a soc allele of egl-15 does not suppress this effect. These data are consistent with soc-1, soc-2, and sem-5 functioning as downstream mediators of FGF receptor signaling.
Figure 3.
Suppression of the effects of egl-15 (neu*) by mutations in sem-5, soc-1, and soc-2. An activated egl-15 construct, was introduced into N2 (wild-type) and the representative soc mutants egl-15(n1783), sem-5(n1779), soc-1(n1789) and soc-2(n1774). Injection of egl-15(neu*) into N2 (wild-type) and egl-15(n1783) results in many transformants displaying a severely abnormal morphology and/or a Clr phenotype. Mutations in sem-5, soc-1, and soc-2 suppress this effect. The abnormal morphology found in some transformants is characterized by variably dumpy animals with bulging extrusions. Clr animals have all the characteristics of clr-1 mutants including a fluid-filled pseudocoelom and enhanced cell boundaries. The number of transformants scored for each genotype: N2, n = 284; egl-15(n1783), n = 76; sem-5(n1779), n = 84; soc-1(n1789), n = 92; soc-2(n1774), n = 159.
shoc-2 Is a Putative Human Homolog of soc-2.
Blast searches with the predicted SOC-2 protein sequence identified a human EST with the potential to encode a protein fragment of 119 amino acids that is 63% identical to SOC-2. Six cDNAs corresponding to this EST were isolated (see Materials and Methods). The cDNAs represent a single transcript with the potential to encode a 582-amino acid protein, termed SHOC-2, that is 54% identical and 67% similar to SOC-2 (Fig. 2a). Both the number and organization of LRRs are conserved, suggesting that this gene is the human homolog of SOC-2. Both SOC-2 and SHOC-2 contain 18, and possibly 19, 22–23-amino acid stretches with leucines or other aliphatic residues at positions 2, 5, 7, 12, 15, and 22 (Fig. 2b). Like many of the LRRs found in other proteins, most of the SOC-2 and SHOC-2 LRRs have asparagine at position 10, proline at position 16 and an aliphatic residue at position 19 (24). Interestingly, both residues that are altered in SOC-2 mutants are conserved in SHOC-2. Together, these observations indicate that SOC-2 and SHOC-2 are closely related proteins that may have similar cellular functions.
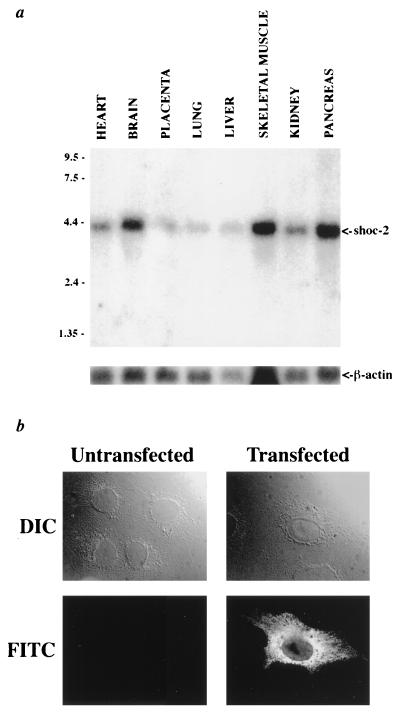
Northern blot analysis demonstrates that shoc-2 mRNA is detectable at varying levels in all tissues tested (Fig. 4). The shoc-2 cDNA hybridizes to a single band of 4.2 kb, consistent with the size predicted from shoc-2 cDNA analysis. A genomic P1-derived artificial chromosome clone containing the shoc-2 gene was isolated and used to determine the chromosomal localization of shoc-2 by fluorescent in situ hybridization. Both cytogenetic mapping and fractional length analysis (FLpter value 0.80–0.83) localize shoc-2 to 10q25. Immunofluorescent detection of SHOC-2 in COS 7 cells transfected with a SHOC-2 expression construct demonstrate that SHOC-2 is localized to the cytoplasm (Fig. 4). This punctate cytoplasmic staining is not seen in preimmune or antigen block controls or untransfected cells (data not shown), demonstrating that this cytoplasmic staining represents the localization of transfected SHOC-2. Attempts to detect endogenous SHOC-2 in NIH 3T3 cells have not generated any specific staining, perhaps due to levels of expression that are too low for visualization using this protocol.
Figure 4.
(a) A multiple tissue Northern blot was probed with the shoc-2 cDNA. A single 4.2-kb band is present in all tissues. Lower, A loading control in which the blot was stripped and reprobed for β-actin. (b) Subcellular localization of SHOC-2. Fluorescent [fluorescein isothiocyanate (FITC)] and corresponding Nomarski [DIC (differetial interference contrast)] images of transiently transfected and untransfected COS 7 cells stained with anti-SHOC-2 antisera are shown. Cytoplasmic staining is evident in cells transiently transfected with a SHOC-2 expression construct, but not in the untransfected control.
Tyrosine Phosphorylation at YXNX Is Not Required for soc-2 Function.
Within the LRRs of SOC-2 are two YXNX motifs. These motifs, when tyrosine phosphorylated, are the preferred recognition sequence of the Src homology 2 domain of the product of another soc gene, SEM-5 (27). Two lines of evidence suggest that these motifs might be important for SOC-2 function. (i) The mutation in soc-2(n1774) affects a splice site within the first YXNX motif. (ii) These motifs are conserved in SHOC-2; the YXNX motifs are at similar positions within each protein and one of the YXNX motifs, YLND, is 100% conserved (Fig. 2a). The functional significance of these motifs was tested by substituting phenylalanine for tyrosine within both YXNX motifs and assaying soc-2 rescuing activity. The mutant construct was as active as its wild-type control (Fig. 1c), suggesting that tyrosine phosphorylation of these residues is not required for SOC-2 function in this aspect of EGL-15 FGF receptor signaling.
SHOC-2 Is Not Tyrosine Phosphorylated in Response to FGF.
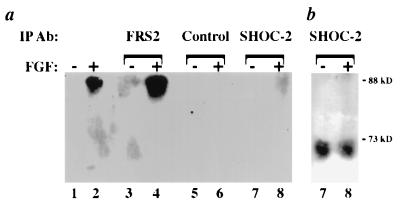
As a putative homolog of SOC-2, SHOC-2 is implicated in functioning downstream of an FGF receptor. Tyrosine phosphorylation plays a key role in the activation of FGF signaling pathways; therefore, SHOC-2 may be a target for tyrosine phosphorylation. To determine whether SHOC-2 is tyrosine phosphorylated, endogenous SHOC-2 was immunoprecipitated from lysates of FGF-treated NIH 3T3 cells and subjected to anti-phosphotyrosine blotting. Immunoprecipitated SHOC-2 does not react with anti-phosphotyrosine antibodies, whereas the tyrosine-phosphorylated protein FRS2 does when subjected to the same analysis (Fig. 5). Similar results were obtained in SHOC-2 transfected COS 7 cells (data not shown), providing evidence that SHOC-2 is not tyrosine phosphorylated in response to FGF under these conditions.
Figure 5.
(a) Anti-phosphotyrosine immunoblot. NIH 3T3 cells were treated with FGF (lanes 2, 4, 6, and 8) or left untreated (lanes 1, 3, 5, and 7) and subjected to immunoprecipitation as indicated. Lanes 1 and 2 were loaded with 25 μg total lysate. SHOC-2 immunoprecipitates from FGF-stimulated NIH 3T3 cell lysates are not detected by the anti-phosphotyrosine antibody, PY20 (lane 8). (b) SHOC-2 immunoprecipitates (lanes 7 and 8 from a) stripped and reprobed with anti-SHOC-2 antibodies.
DISCUSSION
The phenotypic and genetic similarities between egl-15, a gene encoding a C. elegans FGF receptor, and the soc genes suggest that their products could function together as components of an FGF signaling pathway. We have tested how the soc genes might function in FGF receptor signaling by assessing their requirement for phenotypes caused by extrachromosomal arrays of egl-15(neu*). We found that the activity of EGL-15(neu*), which behaves as a constitutively active EGL-15 FGF receptor in C. elegans (16), requires the function of three soc genes: sem-5, soc-1, and soc-2. Our data indicate that these genes do not function in the activation of the EGL-15 FGF receptor, but rather are likely to mediate intracellular signaling downstream of EGL-15.
We have cloned soc-2 and a putative human homolog, shoc-2, and found that they encode LRR proteins (24). The high degree of sequence identity between SOC-2 and SHOC-2 indicates that these two proteins may share important cellular functions. We have shown that the SHOC-2 protein is localized in the cytoplasm of transiently transfected cells. In addition, we mapped shoc-2 to 10q25, a region to which certain skeletal disorders (28) and cancers (29, 30) have been linked. Given that aberrant FGF receptor signaling is implicated in skeletal disorders (31) and cancer progression (32), SHOC-2 is a reasonable candidate for being affected in some of these disorders.
The LRRs are likely to be essential for defining the function of SOC-2/SHOC-2. LRR proteins define a diverse group of molecules with differing functions and cellular localizations. Proteins containing these repeats are thought to be involved in protein-protein interactions (24). As a cytoplasmic LRR-containing protein, SHOC-2 may associate with an FGF receptor signaling pathway component. One possibility is suggested by the close similarity to the LRRs found in adenylyl cyclase of Saccharomyces cerevisiae. The LRRs of adenylyl cyclase are required for the regulation of its activity by Ras (33, 34) and are sufficient for its association with Ras (35). Because FGF signaling pathways are thought to involve the activation of Ras, the function of SOC-2/SHOC-2 in FGF receptor signaling may also involve an interaction with Ras. Alternatively, the SOC-2/SHOC-2 LRRs may mediate an interaction with other FGF signaling pathway components.
In addition to LRRs, SOC-2 contains two YXNX motifs that are also conserved in SHOC-2. The Src homology 2 domain of SEM-5/GRB2 has been shown to selectively bind these motifs when tyrosine phosphorylated (27). Due to the phenotypic similarity between sem-5 and soc-2 mutants and their similar placement with respect to EGL-15, we tested whether SOC-2 might function by associating with SEM-5 utilizing these motifs. Altering the tyrosine residues in these motifs to phenylalanine did not compromise soc-2 function, indicating that tyrosine phosphorylation of these motifs is not essential for SOC-2 function in vivo. Although these data indicate that SOC-2 and SEM-5 do not associate in this manner, it does not preclude their interaction by other mechanisms.
Tyrosine phosphorylation is a key mechanism for early events in RTK signaling. We have tested whether SHOC-2 is tyrosine phosphorylated in response to FGF and found that SHOC-2 is not detectably tyrosine phosphorylated in a number of FGF-stimulated cell lines. Many components have been implicated in FGF signaling based initially on either their tyrosine phosphorylation or previously established roles in other RTK signaling pathways. In contrast, the genetic identification of the soc genes has allowed for the characterization of two nontyrosine phosphorylated components, SEM-5 and SOC-2/SHOC-2. The analysis of SEM-5/GRB-2 was pivotal in linking RTK signaling to the activation of Ras (19). Elucidating the function of SOC-2/SHOC-2 may be equally critical to our understanding of FGF receptor signal transduction.
Acknowledgments
We thank P. Ward and D. Ward for fluorescent in situ hybridization analysis of shoc-2, J. Schlessinger for FRS2 antibodies, R. Lifton for PAC filters, A. Spence for the eP14 probe, A. Coulson and the C. elegans Genome Consortium for genomic clones, D. Stern for advice on immunofluorescence studies, and members of the M. Stern and D. Stern laboratories for discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Lucille P. Markey Charitable Trust.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: FGF, fibroblast growth factor; Clr, clear; RTK, receptor tyrosine kinase; Soc, suppression of clr-1; EST, expressed sequence tag; LRR, leucine-rich repeat.
References
- 1.Basilico C, Moscatelli D. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 2.Burgess W H, Maciag T. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]
- 3.Ullrich A, Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 4.van der Geer P, Pawson T. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- 5.Mohammadi M, Dikic I, Sorokin A, Burgess W H, Jaye M, Schlessinger J. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammadi M, Honegger A M, Rotin D, Fischer R, Bellot F, Li W, Dionne C A, Jaye M, Rubinstein M, Schlessinger J. Mol Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadi M, Dionne C A, Li W, Li N, Spivak T, Honegger A M, Jaye M, Schlessinger J. Nature (London) 1992;358:681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- 8.Peters K G, Marie J, Wilson E, Ives H E, Escobedo J, Del Rosario M, Mirda D, Williams L T. Nature (London) 1992;358:678–681. doi: 10.1038/358678a0. [DOI] [PubMed] [Google Scholar]
- 9.Spivak-Kroizman T, Mohammadi M, Hu P, Jaye M, Schlessinger J, Lax I. J Biol Chem. 1994;269:14419–14423. [PubMed] [Google Scholar]
- 10.Wang J-K, Gao G, Goldfarb M. Mol Cell Biol. 1994;14:181–188. doi: 10.1128/mcb.14.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer N E, D’Arcangelo G, Thomas S M, DeMarco M, Brugge J S, Halegoua S. J Cell Biol. 1991;115:809–819. doi: 10.1083/jcb.115.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umbhauer M, Marshall C J, Mason C S, Old R W, Smith J C. Nature (London) 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- 13.Pelicci G, Giordano S, Zhen Z, Salcini A E, Lanfrancone L, Bardelli A, Panayotou G, Waterfield M D, Ponzetto C, Pelicci P G, et al. Oncogene. 1995;10:1631–1638. [PubMed] [Google Scholar]
- 14.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 15.DeVore D L, Horvitz H R, Stern M J. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 16.Kokel M, Borland C Z, DeLong L, Horvitz H R, Stern M J. Genes Dev. 1998;12:1425–1437. doi: 10.1101/gad.12.10.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern M J, Marengere L E M, Daly R J, Lowenstein E J, Kokel M, Batzer A, Olivier P, Pawson T, Schlessinger J. Mol Biol Cell. 1993;4:1175–1188. doi: 10.1091/mbc.4.11.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark S G, Stern M J, Horvitz H R. Nature (London) 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 20.Barstead R J, Waterston R H. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- 21.Wilson R, Ainscough R, Anderson K, Baynes C, Berks C, Berks M, Bonfield M, Burton J, Connell M, Copsey T, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Lipman D J. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bray-Ward P, Menninger J, Lieman J, Desai T, Mokady N, Banks A, Ward D C. Genomics. 1996;32:1–14. doi: 10.1006/geno.1996.0070. [DOI] [PubMed] [Google Scholar]
- 24.Kobe B, Deisenhofer J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 25.Herman R K. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, New York: Cold Spring Harbor Lab. Press; 1988. pp. 17–45. [Google Scholar]
- 26.Krause M. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. Vol. 48. San Diego: Academic; 1995. pp. 483–512. [Google Scholar]
- 27.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 28.Raas-Rothschild A, Manouvrier S, Gonzales M, Farriaux J P, Lyonnet S, Munnich A. J Med Genet. 1996;33:996–1001. doi: 10.1136/jmg.33.12.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier D, Comparone D, Taylor E, Zhang Z, Gratzl O, Van Meir E G, Scott R J, Merlo A. Oncogene. 1997;15:997–1000. doi: 10.1038/sj.onc.1201209. [DOI] [PubMed] [Google Scholar]
- 30.Nagase S, Yamakawa H, Sato S, Yajima A, Horii A. Cancer Res. 1997;57:1630–1633. [PubMed] [Google Scholar]
- 31.Muenke M, Schell U. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 32.Penault-Llorca F, Bertucci F, Adelaide J, Parc P, Coulier F, Jacquemier J, Birnbaum D, deLapeyriere O. Int J Cancer. 1995;61:170–176. doi: 10.1002/ijc.2910610205. [DOI] [PubMed] [Google Scholar]
- 33.Colicelli J, Field J, Ballester R, Chester N, Young D, Wigler M. Mol Cell Biol. 1990;10:2539–2543. doi: 10.1128/mcb.10.6.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Field J, Xu H P, Michaeli T, Ballester R, Sass P, Wigler M, Colicelli J. Science. 1990;247:464–467. doi: 10.1126/science.2405488. [DOI] [PubMed] [Google Scholar]
- 35.Minato T, Wang J, Akasaka K, Okada T, Suzuki N, Kataoka T. J Biol Chem. 1994;269:20845–20851. [PubMed] [Google Scholar]