Summary
Mitochondrial fusion and division play important roles in the regulation of apoptosis. Mitochondrial fusion proteins attenuate apoptosis by inhibiting release of cytochrome c from mitochondria, in part by controlling cristae structures. Mitochondrial division promotes apoptosis by an unknown mechanism. We addressed how division proteins regulate apoptosis using inhibitors of mitochondrial division identified in a chemical screen. The most efficacious inhibitor, mdivi-1 (for mitochondrial division inhibitor) attenuates mitochondrial division in yeast and mammalian cells by selectively inhibiting the mitochondrial division dynamin. In cells, mdivi-1 retards apoptosis by inhibiting mitochondrial outer membrane permeabilization. In vitro, mdivi-1 potently blocks Bid-activated Bax/Bak-dependent cytochrome c release from mitochondria. These data indicate the mitochondrial division dynamin directly regulates mitochondrial outer membrane permeabilization independent of Drp1-mediated division. Our findings raise the interesting possibility that mdivi-1 represents a novel class of therapeutics for stroke, myocardial infarction, and neurodegenerative diseases.
Keywords: mitochondrial dynamics, mitochondrial outer membrane permeabilization, apoptosis, mitochondrial division dynamin, chemical screen, small molecule inhibitor
Introduction
Mitochondrial fusion and division proteins have been identified in model systems such as flies and yeast (Hoppins et al., 2007). Amongst these proteins are three highly conserved dynamin-related GTPases (DRPs), which function via self-assembly to regulate membrane dynamics in a variety of cellular events. DRPs are relatively large proteins that contain, in addition to a canonical GTPase domain, several regions that facilitate self-assembly via both intra- and intermolecular interactions (Danino and Hinshaw, 2001). Self-assembly of dynamin-1, which functions in endocytosis and Dnm1, which functions in mitochondrial division, greatly stimulates the hydrolysis of GTP (Ingerman et al., 2005; Warnock et al., 1996). Both of these DRP activities, self-assembly and self-assembly stimulated GTP hydrolysis, are critical for the cellular functions of these proteins.
Two distinct DRPs are required for mitochondrial fusion, the transmembrane proteins, Fzo1 (yeast)/Mfn1/2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), which drive outer and inner mitochondrial membrane fusion, respectively (Meeusen et al., 2006; Meeusen et al., 2004). A single DRP, Dnm1 (yeast)/Drp1 (mammals), is required for mitochondrial division. Current models suggest that self-assembly of mitochondrial fusion DRPs functions to tether membranes together, at least in part, by mediating intermolecular trans interactions (Griffin and Chan, 2006; Ishihara et al., 2004; Koshiba et al., 2004; Meeusen et al., 2006; Meeusen et al., 2004). In contrast, biochemical and structural analyses indicate that the self-assembly of the mitochondrial division DRP into ring-like structures around mitochondria directly drives membrane constriction and fission during division (Ingerman et al., 2005; Naylor et al., 2006).
In addition to the role of mitochondrial dynamics in the regulation of mitochondrial distribution and mitochondrial DNA maintenance, detailed analysis of mitochondrial behavior during apoptosis revealed that mitochondrial division and fusion regulate mitochondrial dependent or intrinsic apoptosis (Youle, 2005). Intrinsic apoptosis is critically dependent on mitochondrial outer membrane permeabilization (MOMP), which results in the release of mitochondrial intermembrane space proteins, such as cytochrome c, that are mediators of cell death (Antignani and Youle, 2006; Chipuk et al., 2006; Newmeyer and Ferguson-Miller, 2003). Although the exact mechanism for MOMP is unknown, it is regulated by interactions amongst the pro- and anti-apoptotic Bcl-2 proteins, a protein family defined by the presence of up to four BH domains (Adams and Cory, 1998). Current models suggest that BH3-dependent activation of the pro-apoptotic Bcl-2 members, such as the multidomain proteins Bax and Bak, causes them to oligomerize and insert into the outer mitochondrial membrane, where they promote the release intermembrane space proteins.
Recent data indicate that mitochondrial fusion protects cells from apoptosis (Neuspiel et al., 2005; Olichon et al., 2003; Sugioka et al., 2004). While the exact molecular mechanism is not completely understood, the mitochondrial inner membrane fusion protein, Opa1, may, through its role in cristae maintenance, exert an anti-apoptotic effect by attenuating the MOMP-induced release of cytochrome c (Frezza et al., 2006; Scorrano et al., 2002). This model has been recently substantiated by work showing that Mgm1, the yeast ortholog, also plays a role in cristae maintenance (Meeusen et al., 2006). Recent studies have also shown that the pro-apoptotic Bcl-2 family members, Bax and Bak, play a reciprocal, housekeeping role in mitochondrial fusion in non-apoptotic cells. Overexpression of the C. elegans Bcl-2 protein, Ced-9, induces mitochondrial fusion in mammalian cells and interacts with the outer membrane fusion protein, Mfn2 (Delivani et al., 2006). In addition, in apoptotic cells, Mfn2 co-localizes with mitochondrial Bax and Bak containing clusters (Karbowski and Youle, 2003). In non-apoptotic cells, Bax and Bak influence the mitochondrial distribution and motility of Mfn2, suggesting that these proteins regulate Mfn2 activity (Karbowski et al., 2006). These data imply that interactions amongst Bcl-2 proteins and Mfn2 reciprocally regulate and integrate mitochondrial fusion and the apoptotic activity of Bcl-2 family members.
In contrast to fusion, inhibition of Drp1-dependent mitochondrial division delays and partially inhibits intrinsic apoptosis (Frank et al., 2001; Jagasia et al., 2005; Lee et al., 2004). Concomitant with MOMP during apoptosis, Drp1 self-assembly and its recruitment to mitochondria are increased, resulting in an enhanced rate of Drp1-dependent mitochondrial division and mitochondrial fragmentation. Evidence suggests that mitochondrial division and fragmentation per se may not contribute to apoptosis, raising the possibility that mitochondrial division proteins directly regulate this event (Martinou and Youle, 2006; Rolland and Conradt, 2006) (Breckenridge et al., 2003; Frank et al., 2001; Germain et al., 2005). Consistent with this, assembled Drp1 co-localizes with activated Bax clusters on mitochondria, some of which are at presumptive division sites. Here, we used a small molecule inhibitor of the mitochondrial division dynamin to probe the mechanistic role that mitochondrial division plays in apoptosis.
Results
A chemical screen for inhibitors of mitochondrial division
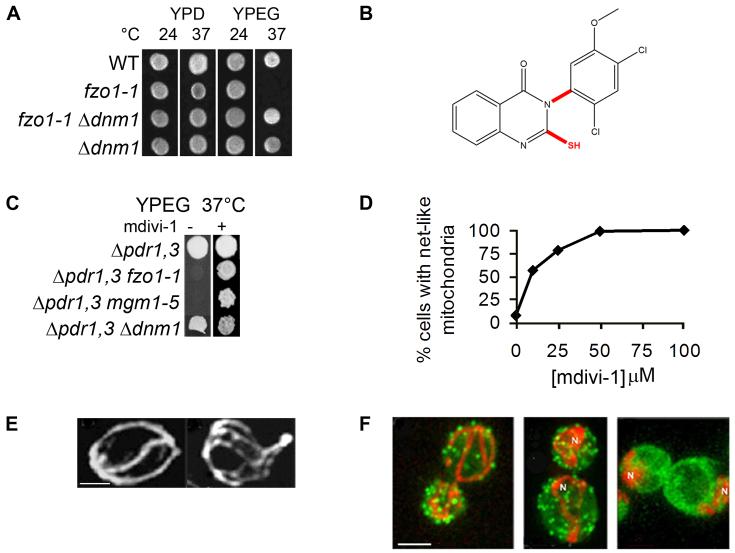
The growth phenotypes of mitochondrial division- and mitochondrial fusion-defective mutants formed the basis of our screen for small molecules that inhibit mitochondrial division. Specifically, exposure of yeast cells harboring the temperature-sensitive fzo1-1 allele to the non-permissive temperature causes mitochondrial membranes to fragment and as a consequence, cells quantitatively lose mtDNA and are unable to grow on the non-fermentable carbon source glycerol (Fig. 1A, fzo1-1, YPEG, 37°C). These cells can still be propagated if grown using a fermentable carbon source, such as glucose (Fig. 1A, fzo1-1, YPD, 37°C). Mutations in components required for division, such as DNM1, which encodes the mitochondrial division dynamin, suppress division-mediated mitochondrial fragmentation and also mitochondrial DNA loss in fzo1-1 cells and thus also suppress the glycerol growth defect at the non-permissive temperature (Fig. 1A, fzo1-1 Δdnm1). In yeast, loss of mitochondrial division has virtually no associated growth phenotype under laboratory conditions (Fig. 1A, Δdnm1, YPD and YPEG).
Figure 1. Chemical screen for mitochondrial division inhibitors.
A. Growth phenotypes of mitochondrial fusion (fzo1-1) and division (Δdnm1) mutants. B. Chemical structure of the quinazolinone, mdivi-1. Important structural features are highlighted in red and were determined by comparing the efficacies of mdivi-1 like compounds shown in Figure 2. C. mdivi-1 suppresses the growth defect of mitochondrial fusion mutants, fzo1-1 and mgm1-5 at the restrictive temperature . D and E. mdivi-1 causes the formation of mitochondrial net-like structures (E, right panel, mdivi-1; left panel, DMSO) in Δprd1 Δprd3 yeast cells in a dose-dependent manner (D, representative experiment shown, n ≥ 100). In E, left panel is the DMSO control, right panel is mdivi-1. F. mdivi-1 has no effect on the F-actin cytoskeleton. Mitochondria are in red, and Phalloidin is in green. Left panel: DMSO control cells. Center panel: mdivi-1-treated cells. Right panel: Latrunculin-A and mdivi-1-treated cells. N=mitochondrial nets. Scale bar = 2μ.
Thus, to identify mitochondrial division inhibitors, we performed a straightforward growth-based screen to identify small molecules that suppress the glycerol growth defect of fzo1-1 cells. To enhance the steady state intracellular concentration of the drugs in yeast cells, null mutations in the PDR1 and PDR3 genes, which encode for transcriptional regulatory proteins that positively control the expression of multi-drug resistance ABC transporters, were created in the strains used in the screen and in the characterization of the small molecules (Rogers et al., 2001). These additional mutations had no effect on mitochondrial division and fusion in cells (not shown). Initially, small molecules were screened at single concentrations between 10-100 μM in primary and secondary assays due to the limited amount of the compounds obtained. When tested alone, DMSO, the solvent used to solubilize the small molecules, had no significant effects in any of the assays described.
We screened approximately 23,000 compounds, representative of several commercially available libraries, using the primary growth assay-based screen (Table 1, 1° screen). All compounds identified were further tested in a secondary analysis for their effects on steady state mitochondrial morphology in yeast (Table 1, 2° screen). The steady state structure of mitochondria in yeast and mammalian cells is an indicator of the relative rates of mitochondrial division and fusion in cells (Bleazard et al., 1999; Hermann et al., 1998; Nunnari et al., 1997; Sesaki and Jensen, 1999). Specifically, the presence of fragmented mitochondrial structures indicates that mitochondrial fusion is selectively attenuated. In contrast, the presence of net-like mitochondrial structures indicates that mitochondrial division is selectively attenuated. We assayed for these morphological phenotypes using a mitochondrially targeted GFP that is efficiently localized to both wild type and respiratory deficient mitochondria. In this secondary assay, small molecules were judged to be positive if they produced a mutant phenotype in greater than 20% of the cell population. As summarized in Table 1, the overall frequency of division inhibitor hits (total of 3) identified using our primary and secondary assays was extremely low, indicating our screening strategy was selective.
Table 1.
Mitochondrial Division Inhibitor Screen
| Library | Library Size | Hits 1° Screen | Hits 2° Screen |
|---|---|---|---|
| Bionet | 4,800 | 16 | 1 (mdivi-1) |
| Cerep | 4,800 | 3 | 0 |
| Maybridge | 8,800 | 42 | 2 |
| NCI Diversity | 1,900 | 6 | 0 |
| Peakdale | 2,800 | 0 | 0 |
| Total | 23,100 | 67 (0.3%) | 3 (0.013%) |
Characterization of the mitochondrial division inhibitor, mdivi-1
We identified three potential mitochondrial division inhibitors and pursued the most efficacious, which is a derivative of quinazolinone, termed mdivi-1 (for mitochondrial division inhibitor, Fig. 1B). As expected, we observed that mdivi-1 suppresses the glycerol growth defects in fzo1-1 cells (Fig. 1C). Significantly, mdivi-1 also suppressed the glycerol growth defects observed in other mutants defective in the mitochondrial fusion pathway, such as mgm1-5 cells, which contain a mutated copy of the gene encoding the mitochondrial inner membrane fusion dynamin, Mgm1 (mgm1-5, Fig. 1C). In addition, we observed that mdivi-1 causes the rapid (≤ 5 min), reversible and dose-dependent formation of net-like mitochondria in wild type cells, with an IC50 of approximately 10 μM (Fig. 1D and E). These observations indicate that mdivi-1 acts as a general suppressor of mitochondrial fusion defects by selectively inhibiting mitochondrial division.
We also directly measured the rates of division and fusion events in yeast by time-lapse fluorescence microscopy after the addition mdivi-1. Time-lapse analysis of mitochondria in mdivi-1 treated cells indicates that no detectable division events occurred, but that fusion events were observed (not shown). In addition, mdivi-1 did not change the net-like morphology of mitochondria in Δdnm1 cells, further suggesting that it blocks division by acting in the Dnm1-dependent division pathway (not shown). Taken together, our results indicate that mdivi-1 is a selective inhibitor of mitochondrial division.
To address the specificity of mdivi-1 effects on mitochondrial division, we examined its effect on two cellular structures that, when perturbed, can cause indirect changes in mitochondrial morphology: the actin cytoskeleton and the peripheral ER network. These structures are routinely examined in yeast mitochondrial morphology mutants as a test for the specificity of the mitochondrial phenotype (McConnell et al., 1990). Treatment of cells with 100 μM mdivi-1 caused the formation of mitochondrial net-like structures, but did not result in significant changes in either the actin cytoskeleton (Fig. 1F, 100%, n=100, left panel) or the peripheral ER network (not shown, 100%, n=50), as compared to control DMSO-treated cells. In contrast, addition of the F-actin depolymerizing compound Latrunculin-A after mdivi-1 treatment caused disassembly of actin cables and patches and caused mitochondrial nets to collapse and aggregate, consistent with published observations (Bleazard et al., 1999)(Fig, 1F, right panel). These observations indicate that the effect of mdivi-1 on mitochondrial morphology is not the result of secondary changes in either the actin cytoskeleton or ER network and are consistent with our data indicating that mdivi-1 produces net-like structures by directly attenuating mitochondrial division.
Structure-activity analysis of mdivi-1
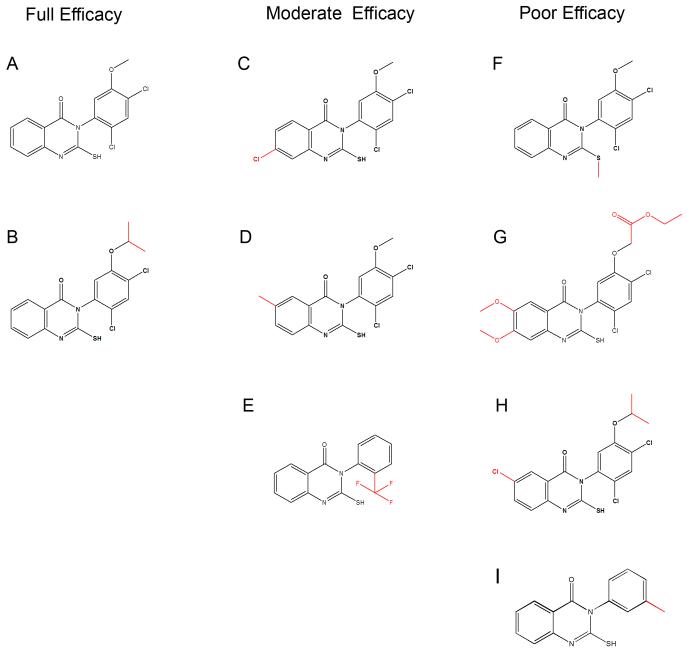
To determine which structural features are important for the effects of mdivi-1 on mitochondrial division, we used ChemNavigator to search available compound databases for small molecules that uniquely represented key structural features of mdivi-1. We tested a total of over 30 mdivi-1 like molecules for their effects on mitochondrial morphology in yeast. A summary of representative compounds, termed A-H, and their efficacy is shown in Figure 2 and Table 2. In no case did we identify a compound that was more efficacious than mdivi-1 (compound A); rather most compounds had the same (Fig. 2, compound B), moderate (Fig. 2, compounds C-E), or poor/no efficacy (Fig. 2, compounds F-H) when examined in our assay for mitochondrial morphology (Table 2). We utilized these derivatives as tools to help determine the target and the specificity of mdivi-1.
Figure 2. Compounds related to mdivi-1 have different efficacies.
Compounds (A-H) are grouped by their relative efficacy to form mitochondrial net-like structures in yeast. The structural differences between each molecule and mdivi-1 (A) are highlighted in red.
Table 2.
mdivi-1 structure-activity analysis
As indicated in Figure 2. All compounds at 50 μM.
Represents n ≥ 100 cells.
Analysis of our structure-function results indicates that at least two structural features are important for the efficacy of mdivi-1 (Fig. 1B, shown in red): an unblocked sulfhydryl moiety on the 2-position of the quinazolinone and limited rotation about the 3-position nitrogen-phenyl bond. Indeed, the bulky ortho chloro substituent of the phenyl ring attached at the N-3 of mdivi-1 predicts that mdivi-1 is a mixture of two atropisomers: isomers that in this case are distinct because rotation about the nitrogen-phenyl bond is prevented or greatly slowed by the rotational energy barrier created by the bulky ortho chloro substituent. Consistent with this, mdivi-1 can be resolved into two distinct species by chiral chromatography (not shown). Assuming that one mdivi-1 isomer is selectively active in attenuating mitochondrial division, the efficacy of mdivi-1 would be two fold greater than our experimental data indicate. Taken together, our preliminary structure-activity analysis indicates that the ability of mdivi-1 to inhibit mitochondrial division is dependent upon stringent structural requirements, consistent with it being a selective inhibitor.
mdivi-1 is a selective inhibitor of the mitochondrial division dynamin
Using a coupled assay for GTPase activity and EM analysis, we previously characterized the kinetic and structural properties of recombinant Dnm1 (Ingerman et al., 2005)). Our analysis indicates that Dnm1 self-assembly greatly stimulates GTP hydrolysis and in the presence of non-hydrolyzable GTP analogs, Dnm1 forms spiral structures, whose diameters correspond to those of mitochondrial constriction sites in vivo. These data strongly suggest a model where the self-assembly of Dnm1 drives mitochondrial constriction during division in vivo.
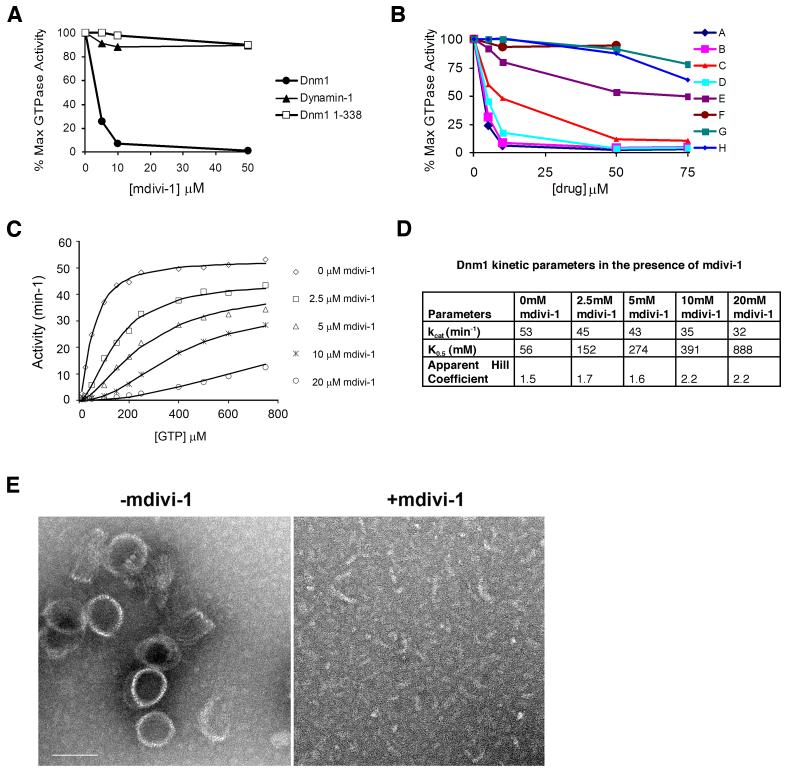
To test whether mdivi-1 targets Dnm1, we tested its effects on self-assembly stimulated Dnm1 GTPase activity. As shown in Figure 3A, mdivi-1 inhibited Dnm1 GTPase activity in a dose dependent manner, with an estimated IC50 of 1-10 μM, which is lower, but consistent with the IC50 observed for the effects of mdivi-1 on the formation of mitochondrial net-like structure in vivo. These observations suggest that mdivi-1 attenuates mitochondrial division in vivo by inhibiting Dnm1.
Figure 3. The target of mdivi-1 is the mitochondrial division dynamin, Dnm1.
A. The dose-dependent effect of mdivi-1 on the GTPase activity of Dnm1, Dnm1 1-388 (Dnm1 GTPase domain) and dynamin-1. 100% activity: Dnm1, 50 min-1; Dnm1 1-388 0.9 min-1; dynamin-1 3.3 min-1 B. The effects of mdivi-1 analogs (see Fig. 2) on Dnm1 GTPase activity. C. Kinetic analysis of the effects of mdivi-1 on Dnm1 GTPase activity. Representative experiments shown in parts A-C. D. Analysis of Dnm1 self-assembly by negative stain electron microscopy. Representative images of Dnm1 incubated in the presence of GMPPCP (left panel) and Dnm1 pre-incubated with mdivi-1 prior to the addition of GMPPCP (right panel). Scale bar = 100 nm.
Our observations raise the question of whether mdivi-1 is simply a general inhibitor of GTPase super family members and/or DRPs. Thus, we examined the effects of mdivi-1 on Dnm1 1-338, the monomeric GTPase domain of Dnm1, which lacks other DRP-specific regions (Ingerman et al., 2005). As shown in Figure 3A, mdivi-1 had no effect on Dnm1 1-338 GTP hydrolysis, indicating that it is not a general inhibitor of GTPases and suggesting that mdivi-1 inhibits Dnm1 by binding to other DRP regions or, more interestingly, to a region dependent on multiple domains. We also tested whether mdivi-1 targets other DRP family members by testing its effects on dynamin-1, which functions during endocytosis in the scission of clathrin coated pits from the plasma membrane. Significantly, mdivi-1 had no effect on either basal (not shown) or assembly-stimulated rates of GTP hydrolysis for dynamin-1 (Fig. 3A). Together these results suggest that mdivi-1 is a selective inhibitor of the mitochondrial division DRP.
To further test the hypothesis that mdivi-1 blocks mitochondrial division in vivo by inhibiting Dnm1 GTPase activity, we tested mdivi-1 like molecules of variable efficacy for their ability to inhibit division in vivo (Fig. 2 and Table 2). The results from multiple independent double-blinded experiments demonstrated that there is a tight correlation between the efficacy of a given derivative to block mitochondrial division in vivo and Dnm1 GTPase activity in vitro (Fig. 3B and Table 2). This observation further supports the conclusion that the mitochondrial division DRP, Dnm1, is the target of mdivi-1 in vivo.
To gain insight into the mechanism of mdivi-1 inhibition of Dnm1, we performed a detailed kinetic analysis of the effects of mdivi-1 on Dnm1 GTPase activity (Fig. 3C and D). From our analysis, we estimate that the Ki of mdivi-1 for Dnm1 is 1-50 μM, which is in the range of the IC50 for mdivi-1in vivo. Our kinetic data fit well to the concerted transition model of Monod, Wyman, and Changeux, which describes the behavior of allosteric proteins that can form oligomers of identical subunits (Fig. S1). Thus, based on this model, the Dnm1 dimer, which we have previously shown to be the building block for assembled Dnm1, is predicted to exist in either one of two states: R (relaxed and assembled) or T (taut and unassembled), that are in equilibrium with one another, where the R state has a relatively high affinity for GTP and T has a relatively low affinity for GTP. This model predicts that mdivi-1 is an allosteric inhibitor with relatively high affinity for the T (unassembled) state and relatively low affinity for the R (assembled) state of Dnm1 and thus implies that mdivi-1 inhibits GTP hydrolysis by blocking the self-assembly of Dnm1. Indeed, the kinetic effects that mdivi-1 has on Dnm1 mimic those observed under high ionic conditions, which antagonize assembly (Ingerman et al., 2005). Specifically, we observed that mdivi-1 increases the apparent K0.5 for GTP, lowers the apparent Vmax for GTP hydrolysis, and causes an increase in the Hill coefficient observed for GTP in the Dnm1 GTP hydrolysis reaction (Fig. 3C and E).
We directly tested the hypothesis that mdivi-1 inhibits Dnm1 self-assembly by examining its effects by EM on Dnm1 spirals formed in the presence of the non-hydrolyzable GTP analog, GMPPCP. As shown in Figure 3E, when present at the start of the self-assembly reaction, mdivi-1 quantitatively blocked GMPPCP-dependent Dnm1 self-assembly in a concentration range similar to its effects on both Dnm1 GTPase activity in vitro and mitochondrial division in vivo. Interestingly, when mdivi-1 was added after the formation of GMPPCP-Dnm1 spirals, the compound had no discernable effect, i.e. failed to promote their disassembly (not shown). Given that Dnm1 spirals formed in the presence of non-hydrolyzable GMPPCP are likely stable, not dynamic structures, our observations suggest that mdivi-1 blocks self-assembly by inhibiting Dnm1 polymerization and not by promoting disassembly.
Consistent with this, we observed that mdivi-1 inhibits GTP hydrolysis of the dimeric middle domain mutant, Dnm1G385D, which is defective for self-assembly (Ingerman et al) (Fig. S2A and B). Analysis of the kinetic data for Dnm1G385D indicated that, in contrast to the concerted transition mechanism for Dnm1, mdivi-1 likely functions as a mixed type inhibitor of Dnm1G385D with a Ki of 1-4 μM, which significantly lowers the affinity of Dnm1 for GTP. These observations indicate that mdivi-1 targets the fundamental building block of the Dnm1 spiral structure to block polymerization. This mechanism is similar to the action of latrunculin A, which alters the actin monomer subunit interface, alters nucleotide binding, and prevents polymerization of F-actin filaments (Morton et al., 2000). Taken together, our analysis of the mechanism of mdivi-1 effects on Dnm1 activity suggests that it inhibits division by binding to an allosteric site that blocks or retards a conformational change required for Dnm1 self-assembly and GTP hydrolysis.
mdivi-1 attenuates mammalian mitochondrial division
Drp1, the mammalian mitochondrial division DRP, has a high degree of identity to its yeast ortholog, Dnm1 (Labrousse et al., 1999). This encouraged us to exploit the chemical genetic approach and examine the effects of mdivi-1 on mitochondrial morphology in mammalian cells. In mammalian cells, when mitochondrial division is retarded by expression of dominant-negative Drp1 or by RNAi of mitochondrial division proteins, tubular mitochondria become progressively more interconnected to form net-like structures, and also collapse into degenerate perinuclear structures (Smirnova et al., 2001; Smirnova et al., 1998).
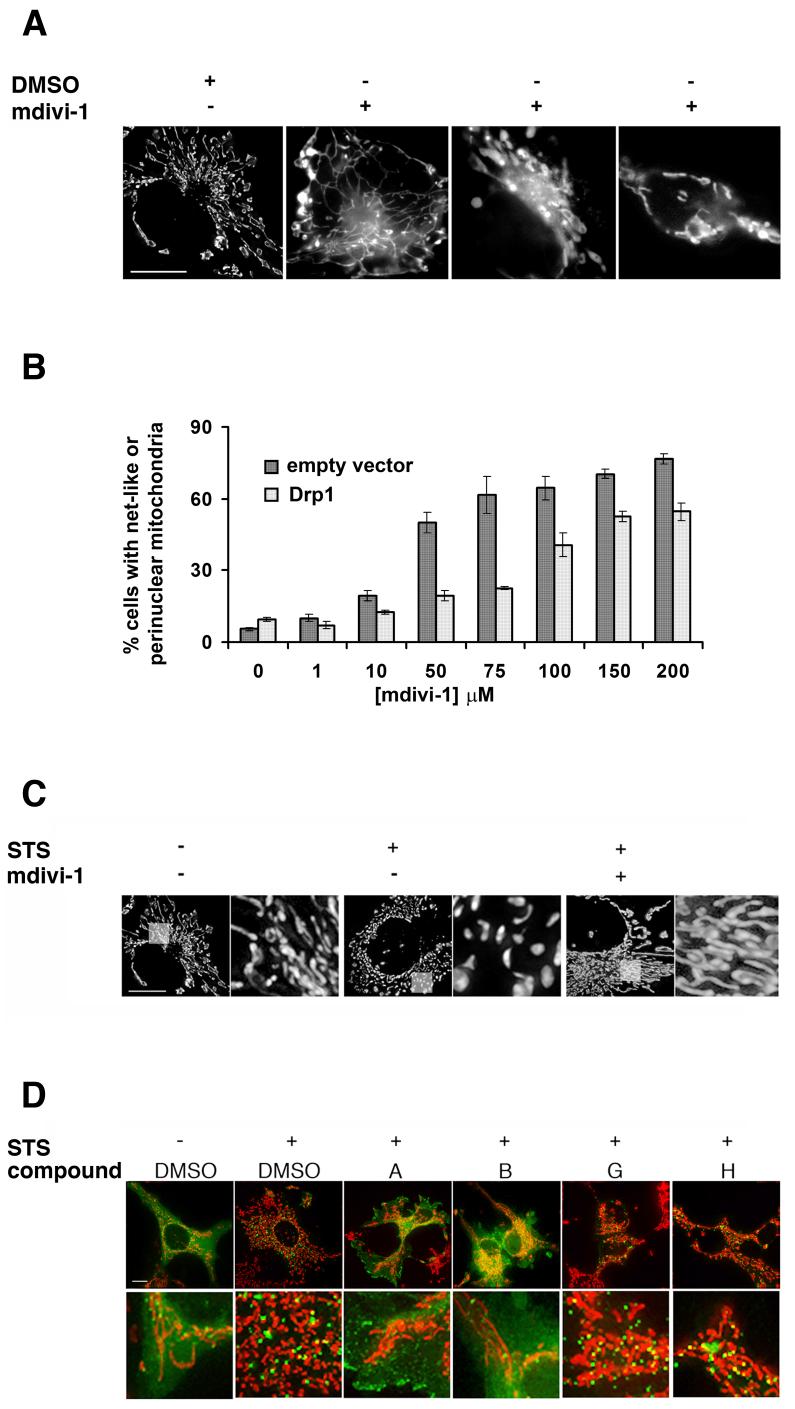
The addition of mdivi-1 to mammalian cells (COS) in culture caused a rapid and reversible formation of mitochondrial net-like and degenerate perinuclear structures, consistent with an attenuation in mitochondrial division (Fig. 4A and Supplemental Table 1). The IC50 of mdivi-1 for its effects on mitochondrial morphology in mammalian cells (IC50 ≈ 50 μM) is comparable to that observed for the effect of mdivi-1 on mitochondrial morphology in yeast (IC50 ≈ 10 μM). In addition, we observed that the mdivi-1 structural derivatives that do not affect mitochondrial morphology in yeast and do not inhibit Dnm1 GTPase activity, also do not affect mitochondrial morphology in COS cells (Supplemental Table 4). Thus, the characteristics of mdivi-1’s effect on mitochondria in mammalian cells are similar to those observed in yeast cells and, by extension, suggest that mdivi-1 inhibits mitochondrial division in mammalian cells by inhibiting Drp1 activity.
Figure 4. mdivi-1 inhibits mitochondrial division in mammalian cells by attenuating Drp1 self-assembly.
A. Mitochondrial morphology in COS cells in the absence (left panel, DMSO control) and presence (right panels, 50 μM) of mdivi-1. B. The concentration of mdivi-1 required to produce mitochondrial net-like and perinuclear structures in COS cells increases in cells overexpressing Drp1 (light grey bars) as compared to cells transfected with a control empty vector (dark grey bars). C. The reticular morphology of mitochondria (left panel) in COS cells becomes fragmented upon addition of staurosporine (STS, center panel). mdivi-1 attenuates STS-induced mitochondrial fragmentation (50 μM, right panel). D. mdivi-1 (50 μM, compound A) and the active derivative, but not the inactive derivatives G and H (each at 50 μM), inhibit self-assembly of GFP-Drp1 stimulated by STS treatment (in green) in COS cells. The bottom panels are a representative region of each cell shown in the top panels and are magnified seven fold. Mitochondria are labeled with MitoTracker Red CMXRos and shown in red. All images were obtained using identical exposure and gain settings. Size bars are 10 μm.
To test whether mdivi-1 targets Drp1, we examined its effects on recombinant Drp1 GTPase activity in vitro. In contrast to its effects on Dnm1, mdivi-1 had no effect on Drp1 GTP hydrolysis. However, the maximal GTP hydrolysis rate of Drp1 was relatively low (2.1 min-1) and we failed to detect the formation of GMPPCP-dependent Drp1 spirals in vitro by EM, even under molecular crowding conditions (not shown), indicating that recombinant Drp1 is not capable of self-assembly and thus is not fully functional.
Thus, we asked whether over expression of Drp1 in mammalian cells could rescue the effects mdivi-1 on mitochondrial morphology. As shown in Figure 4B, the concentration of mdivi-1 required to observe either net-like or collapsed/degenerate perinuclear mitochondrial structures in cells was significantly higher in the population of cells over expressing Drp1 (IC50 between 75-100 μM) as compared to those transfected with a control empty vector (IC50 between 10-50 μM). In addition, in agreement with published work, depletion of Drp1 by RNAi also caused the formation of net-like or collapsed perinuclear mitochondrial structures in cells and treatment of these cells with mdivi-1 did not produce any additional changes to mitochondrial morphology (not shown). These observations substantiate our conclusion that the mitochondrial division dynamin is the target of mdivi-1 in both yeast and mammalian cells.
mdivi-1 attenuates mammalian mitochondrial division and Drp1 self-assembly during apoptosis
Drp1-mediated mitochondrial division in mammalian cells is stimulated by apoptotic signals, such as staurosporine (STS), which promote intrinsic apoptotic cell death via Bcl-2 proteins (Frank et al., 2001). We examined the effects of mdivi-1 on mitochondrial fragmentation caused by STS in mammalian COS cells (Fig. 4C and Table 5). As shown previously, STS stimulation caused a significant increase in mitochondrial fragmentation in cells ((Frank et al., 2001), Fig. 4C and Supplemental Table 2). In comparison, mitochondrial fragmentation was significantly reduced in cells treated with both STS and mdivi-1, (Fig. 4C and Table 5). As a control, we observed that expression of dominant-negative Drp1 or RNAi mediated depletion of Drp1 also inhibited STS-induced mitochondrial fragmentation to a similar degree as mdivi-1, which is in agreement with published observations (not shown) (Frank et al., 2001). These observations indicate that mdivi-1 inhibits apoptosis-stimulated Drp1-dependent mitochondrial division.
To gain insight into the mechanistic basis of mdivi-1 inhibition of Drp1-mediated mitochondrial division, we exploited the fact that during apoptosis, Drp1 self-assembly and recruitment to mitochondria are significantly increased and these events can easily be assayed by monitoring the behavior of GFP-Drp1 in cells (Frank et al. 2001). As shown in Figure 4D, addition of the apoptotic stimulant STS caused a decrease in diffusely localized GFP-Drp1 and a concomitant increase in the number of GFP-Drp1 clusters and GFP-Drp1 clusters associated with mitochondria (compare green fluorescence in - STS/DMSO with +STS/DMSO cells). In contrast, in cells treated with STS and mdivi-1 (compound A) or the active compound B, the majority of GFP-Drp1 was diffusely distributed in the cytoplasm, in a manner similar to that observed for GFP-Drp1 in cells that were not treated with STS (Fig. 4D, compare -STS/DMSO and +STS/DMSO with +STS/A and +STS/B). Consistent with our previous observation (Fig. 4C) and as expected, mitochondria in cells treated with STS and mdivi-1 (compound A) or the active compound B were tubular as compared to fragmented mitochondria present in cells treated with STS only (Fig.4D, in red). In addition, in cells treated with STS and either of the inactive mdivi-1 compounds, G and H, the localization pattern of GFP-Drp1 and the mitochondrial morphology was similar to cells that were treated only with STS (Fig. 4D, compare +STS/DMSO with +STS/G and +STS/H). These observations indicate that mdivi-1 attenuates mitochondrial division during apoptosis by blocking Drp1 self-assembly and the recruitment of Drp1 assembled structures to mitochondria and are consistent with our biochemical data indicating that mdivi-1 inhibits the yeast ortholog, Dnm1, by blocking polymerization. Together, these observations indicate that mdivi-1 targets the mitochondrial division dynamin and acts in a mechanistically conserved manner in yeast and mammalian cells
mdivi-1 attenuates apoptosis by inhibiting mitochondrial outer membrane permeabilization
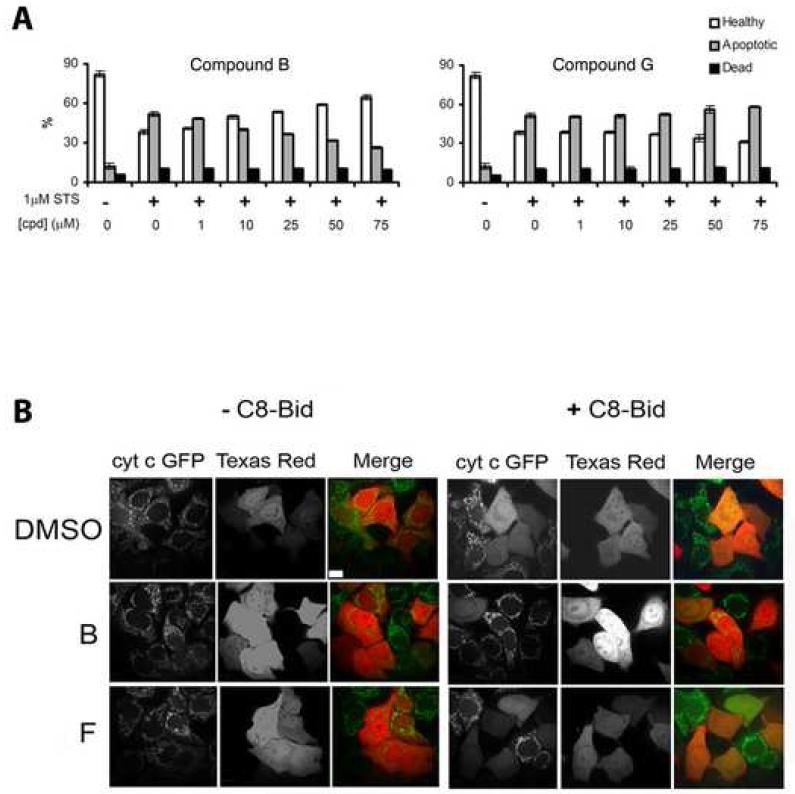
Given the inhibitory effect of mdivi-1 on STS-induced mitochondrial division, we tested whether mdivi-1 also retards apoptosis in mammalian HeLa cells. We initially examined the effects of mdivi-1 on the externalization of plasma membrane phosphatidylserine (PS), which is a relatively late event in apoptotic cell death (Fadok and Henson, 2003). We quantified this event using fluorescently labeled (FITC)-annexin V, which is a Ca2+-dependent phospholipid binding protein with a high affinity for PS, in conjunction with established fluorescent-activated cell sorter (FACS) methodology. As shown in Figure 5A, we observed that mdivi-1, but not the inactive mdivi-1 derivative, G, significantly inhibits STS-induced annexin V staining of non-necrotic cells as assessed by FACS analysis, indicating that mdivi-1 inhibits apoptosis. Significantly, although the mdivi-1 inhibition of STS-induced apoptosis was fractional, the extent of inhibition was comparable to that observed when the dominant negative Drp1K38A mutant is over expressed in HeLa cells and is consistent with mdivi-1 targeting Drp1 in vivo (Frank et al. 2001 and STS-induced apoptotic cells in Drp1: Drp1 K38A = 0.8 by annexin V analysis).
Figure 5. mdivi-1 and its active analogs attenuate apoptosis.
A. FACS analysis of staurosporine-treated Hela cells in the presence and absence of the active compound B (left panel) and inactive compound G (left panel). B. Analysis of the effects of mdivi-1 derivatives (B and F) on the release of cytochrome c stimulated by C8-Bid injection of HeLa cells. The injection marker is Texas Red (in red) and cytochrome c release is monitored with cytochrome c-GFP (in green). Size bar is 10 μm.
To resolve the point at which mdivi-1 inhibits apoptosis, we examined its effect on the early MOMP event, measured by cytochrome c release. To directly stimulate MOMP and cytochrome c release, HeLa cells were microinjected with caspase-8 cleaved recombinant Bid (C8-Bid). C8-Bid directly (Kuwana et al., 2002; Walensky et al., 2006) or indirectly (Willis et al., 2007) activates Bax/Bak, causing MOMP and cytochrome c release. As expected, cytochrome c release, as monitored by a cytochrome c-GFP fusion, was not stimulated in control cells injected with the marker Texas Red with or without the active mdivi-1 derivative, B (Fig. 5B and Supplemental Table 3). In contrast, but also as expected, cytochrome c-GFP release was greatly stimulated in cells injected with C8-Bid, (Fig 5B, DMSO, cells stained with Texas red and Supplemental Table 3). Significantly, the active mdivi-1 derivative, B, dramatically inhibited C8-Bid stimulated cytochrome c-GFP release, whereas the inactive mdivi-1 derivative, F had virtually no effect on C8-Bid stimulated cytochrome c-GFP release (Fig. 5B and Supplemental Table 3). Similar effects of mdivi-1 on STS-induced cytochrome c release were also observed (Fig. S3). Together our results indicate that mdivi-1 inhibits the activity of the mitochondrial division dynamin Drp1 and as a result impedes apoptosis early in the intrinsic pathway by blocking Bax/Bak dependent MOMP.
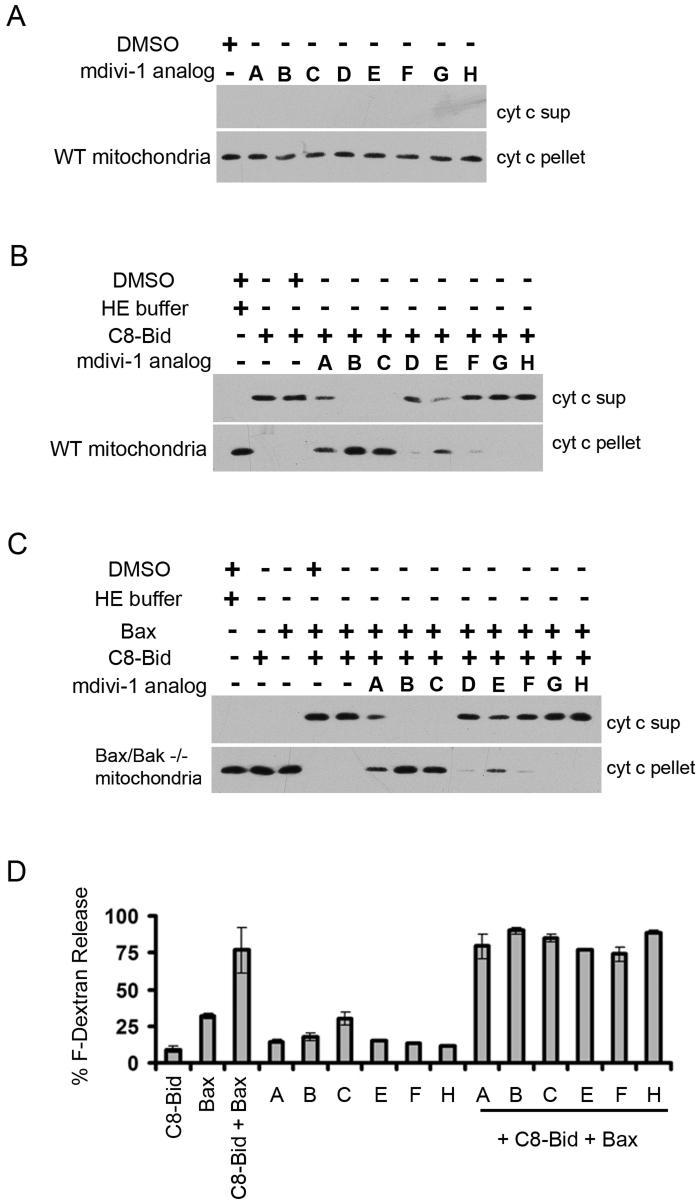
To further test this conclusion, we examined the effect of mdivi-1 on C8-Bid induced, BAK dependent cytochrome c release from isolated wild-type murine liver mitochondria (Fig. 6). As assessed by western blot analysis of mitochondrial derived supernatant and pellet fractions with anti-cytochrome c, mdivi-1 derivatives had no effect on cytochrome c release in vitro when tested alone (Fig. 6A). In contrast and as expected, cytochrome c release was greatly stimulated by the addition of recombinant active C8-Bid (Fig. 6B). Significantly, the addition of mdivi-1, as well as the active mdivi-1 like compounds B and C, appreciably blocked C8-Bid induced cytochrome c release, whereas mdivi-1 derivatives lacking efficacy, either failed or had relatively minor effects on cytochrome c release (Fig 6B, compare compounds A-C to D-H). In addition, the ability of mdivi-1 and active mdivi-1 compounds to inhibit C8-BID induced, BAX dependent cytochrome c release was examined in bak/bax double knockout mitochondria from MxCre bak-/-bax-/-livers (Fig. 6C) supplemented with monomeric full-length BAX.
Figure 6. mdivi-1 and its active analogs inhibit activated Bax/Bak-dependent MOMP in vitro.
A. Analysis of the effects of mdivi-1 and its analogs on MOMP by monitoring cytochrome c release in vitro from murine liver mitochondria. B. Analysis of the effects of mdivi-1 and its analogs on MOMP in vitro as in A. with murine liver mitochondria stimulated by C8-Bid. C. The effects of mdivi-1 and its analogs on C8-Bid stimulated MOMP depend on the presence of Bax. The assays were as described in A and B using mitochondria isolated from the livers of polydIdC-treated MxCre bak-/- baxf/- mice. D. mdivi-1 and its analogs do not directly inhibit Bid-activated Bax permeabilization of large unilamellar vesicles. Permeabilization is monitored by the release of encapsulated fluorescein-dextran by filtration analysis as described in methods.
These findings indicated that mdivi-1 and active derivatives inhibit cytochrome c release by preventing the full Bid-dependent activation of Bax and Bak. Finally, as a control, we examined whether mdivi-1 derivatives directly affect Bid-dependent Bax induced membrane permeabilization using an established large unilamellar vesicle (LUV) release assay in which LUVs are preloaded with fluorescein dextran (Kuwana et al., 2002). As shown in Figure 6D, the amount of fluorescein dextran released from LUVs by C8-Bid/Bax induced permeabilization was not affected by the addition of mdivi-1 and mdivi-1 derivatives, nor did the mdivi-1 compounds cause significant permeabilization on their own. These observations indicate that mdivi-1 does not block MOMP by directly inhibiting C-8 Bid activated Bax. Thus, together our data suggest that mdivi-1 attenuates MOMP by inhibiting the self-assembly of the mitochondrial division dynamin, which functions upstream, together with Bcl-2 proteins to directly stimulate Bcl-2 mediated outer membrane permeabilization. Consistent with this conclusion, we observed that a significant level of Drp1 is present and cofractionates with mitochondrial membranes in vitro (not shown).
Discussion
The mechanism of DRP self-assembly
In this study, we identify mdivi-1 as the first selective inhibitor of mitochondrial division dynamins. The mechanism of mdivi-1 inhibition is distinct from the more general DRP inhibitor, Dynasore, which was recently discovered in a chemical screen for inhibitors of dynamin-1 GTPase activity (Macia et al., 2006). Dynasore inhibits the GTP hydrolysis of dynamin-1, dynamin-2, and Drp1, in a non-competitive manner by binding to the GTPase domain in both assembled and unassembled states. Our data demonstrate that mdivi-1 selectively inhibits the activity of mitochondrial division DRPs by binding to an allosteric site that does not exclusively act through the GTPase domain. Upon binding, mdivi-1 creates or stabilizes a conformational form of unassembled, likely dimeric Dnm1 that can bind GTP, but at a significantly lower affinity. This mdivi-1 dependent conformational state is not able to assemble into a Dnm1 filament/spiral, indicating that mdivi-1 inhibits division DRPs by blocking their polymerization, not by causing disassembly. The behavior of the Dnm1/mdivi-1 complex has both similar and interesting differences as compared to the Dnm1 mutant, Dnm1K41A. Similar to Dnm1/mdivi-1, Dnm1K41A is able to bind, but not hydrolyze GTP, however, unlike Dnm1/mdivi-1, Dnm1K41A, is able to polymerize into GMPPCP -dependent spiral-like structures (Naylor et al., 2006). This raises the possibility mdivi-1 binding specifically blocks GTP-induced conformational changes in Dnm1 that are necessary to promote self-assembly. Interestingly, mdivi-1 also inhibits GTP hydrolysis by the assembly deficient dimer, Dnm1G385D. This finding indicates that Dnm1G385D is not a zero order assembly intermediate, which is consistent with our data that, at higher concentrations, Dnm1G385D can undergo conformational changes required for self-assembly (not shown). The mechanism of action of mdivi-1 underscores the conformational plasticity of DRPs and the fact that regulated conformational changes are critical for the self-assembly of these proteins.
The role of Drp1 in MOMP
The molecular role of Drp1 in apoptosis in mammalian cells has been elusive because, during apoptosis, Drp1-mediated mitochondrial division and MOMP are both early apoptotic events that have not been temporally resolved (Martinou and Youle, 2006). This temporal link raises the possibility that Drp1 acts to permit or facilitate MOMP and apoptosis as a result of mitochondrial fragmentation. Our data suggests that Drp1 functions during MOMP in a manner independent of mitochondrial division and fragmentation per se and thus suggests that Drp1 possesses multiple functions in mammalian cells. Remarkably, we found that mdivi-1 inhibits Bak/Bax-dependent MOMP induced by C8-Bid in isolated mitochondria in vitro, where EM analysis of mitochondria before and after MOMP indicate that mitochondrial division does not occur (Scorrano et al., 2002). In addition, our data indicate that mdivi-1 does not inhibit C8-Bid/Bax dependent permeabilization of liposomes, which lack Drp1, consistent with mdivi-1 inhibiting MOMP via Drp1. Thus, our data suggests that Drp1 acts upstream or together with Bcl-2 proteins to directly modulate MOMP.
Our analysis of mdivi-1 mechanism in mammalian cells indicates that it blocks Drp1 self-assembly during apoptosis and indicates that Drp1 self-assembly is critical for its role in MOMP. One possibility is that Drp1 directly interacts and co-assembles with Bid-activated Bax/Bak, creating a complex that is more active for both MOMP and mitochondrial division. Consistent with this, by both EM and light microscopy, Drp1 co-localizes with Bax in clusters on the mitochondrial outer membrane associated with mitochondrial constriction sites (Karbowski et al., 2002b). In addition, the dynamic behavior exhibited by Drp1 clusters on mitochondria is dramatically lost after Bax recruitment to mitochondria during apoptosis, indicating that the biochemical properties of Drp1 are altered in a Bax dependent manner (Wasiak et al., 2007). Thus, the MOMP and division activities of a Drp1/Bax/Bak complex could be altered by conformational changes or more indirectly via the recruitment of lipids, such as cardiolipin, that have been shown to be required in vitro for Bax-dependent membrane permeabilization.
Another possible mechanism for Drp1 during apoptosis is that it exerts its effects on MOMP by regulating the mitochondrial fusion machinery. Recent studies have shown that in healthy cells, Bax and Bak are required to maintain normal levels of mitochondrial fusion activity and alter the behavior of the mitochondrial outer membrane fusion protein, Mfn2 (Karbowski et al., 2006). This is in contrast to the role of Bak/Bax during apoptosis, where, when activated, Bax and Mfn2, like Drp1, are co-localized in clusters on the outer membrane and mitochondrial fusion is attenuated (Karbowski et al., 2004; Karbowski et al., 2002a). Thus, it is possible that Drp1 recruitment to an activated Bax/Bak complex alters it, causing inhibition of Mfn2-dependent fusion activity, which in turn facilitates MOMP, perhaps via the control of cristae structure (Germain et al., 2005). Indeed, it has been reported that RNAi depletion of Drp1 in HeLa cells does not affect the release of SMAC/Diablo from mitochondria during apoptosis, but attenuates cytochrome c release, which is sequestered in cristae (Estaquier and Arnoult, 2007; Parone et al., 2006). In contrast, we observed that in vitro mdivi-1 blocked C8-Bid Bax/Bak dependent release of SMAC/Diablo from mitochondria, indicating that mdivi-1 has a general influence of MOMP and likely does not exert its effects through changes in cristae structure (not shown). This discrepancy is likely the result of the vastly different approaches used to inhibit Drp1 function in these studies i.e. RNAi depletion of Drp1 versus a drug-mediated block in Drp1 function.
Regardless of the mechanism, it seems likely that Bcl-2 and mitochondrial division and fusion proteins have evolved to form a regulatory network that functions to sense the health status of cells. Recent studies in the invertebrate worm and fly models have demonstrated a regulatory role for Drp1 in mitochondrial fragmentation and cell death, indicating that this network is conserved. However, it appears that mitochondrial division and fusion proteins are not essential components of the apoptotic pathway (Abdelwahid et al., 2007; Estaquier and Arnoult, 2007; Frank et al., 2001; Goyal et al., 2007; Parone et al., 2006; Wasiak et al., 2007). Consistent with this observation, we find that mdivi-1 only partially blocks apoptosis by acting early in the pathway to inhibit MOMP. Thus, it is possible that post-translational modifications of Drp1 are regulated by stress in cells, and modulate Drp1 self-assembly to coordinately and positively activate apoptosis. Indeed, evidence suggests that Drp1 is covalently modified by phosphorylation, ubiquitination and sumoylation and that these modifications regulate Drp1 activity (Harder et al., 2004; Nakamura et al., 2006; Taguchi et al., 2007).
The key positive regulatory role that Drp1 plays in apoptosis makes it an appealing pharmaceutical target for neurodegenerative diseases, stroke and myocardial infarction, where the inhibition of apoptosis may be therapeutically beneficial. In addition, Drp1 is a logical target for the neurodegenerative diseases, Charcot-Marie Tooth type 2A and autosomal dominant optic atrophy, which are caused by mutations in mitochondrial fusion proteins Mfn2 and OPA1, respectively. One of the primary phenotypes associated with loss of fusion in mammalian cells is loss of mitochondrial respiratory function, caused in part by loss of mtDNA within a subset of mitochondria due to excessive mitochondria division, which is likely an important factor contributing to the etiology of these neurodegenerative diseases (Chan, 2006a, b; Chen and Chan, 2006; Chen et al., 2005; Olichon et al., 2006; Parone et al., 2006). Indeed, one of the fundamental roles of mitochondrial fusion is to provide an exchange mechanism that allows all mitochondria within a cell access to mtDNA and its products (Hoppins et al., 2007). Attenuating mitochondrial division and increasing mitochondrial connectivity could benefit some of the many heteroplasmic mtDNA-linked diseases in humans. Thus, our results showing that mdivi-1 is a selective inhibitor of mitochondrial division and apoptosis in mammalian cells are of potentially great clinical significance because they have revealed a promising novel target and treatment for many diseases.
Experimental Procedures
Chemical genetic screen for mitochondrial division inhibitors
Strains: RDY84 (W303 background: ade2-1 his3 -11,-15 trp1-1 can1-100 pdr1::KanR pdr3::His5+, a gift of Russell Dorer, Department of Pathology, Brigham and Women’s Hospital) was crossed to JNY539 (Hermann et al. 1998, W303 background: ade2-1 his3-11,-15 trp1-1 can1-100 fzo1::fzo1-1), diploids were sporulated and tetrads were dissected to obtain ACY201 (ade2-1 his3-11,-15 trp1-1 can1-100 pdr1::KanR pdr3::His5+ fzo1::fzo1-1). RDY84 was crossed to JNY905 (ade2-1 his3-11,-15 trp1-1 can1-100 dnm1::His3), diploids sporulated and tetrads dissected to obtain ACY208 (ade2-1 his3 -11,-15 trp1-1 can1-100 pdr1::KanR pdr3::His5+ dnm1::His3).
The screen was conducted at the ICCB-Longwood Screening Facility (Boston, MA). ACY201 cells at a density of 1.2×104 cells per well were aliquoted into 384-well plates using a Bio-Tek Precision 2000 Robot. 100 nl of each compound (each at 5 mg/ml or ∼15 mM in DMSO) was added to each well using a Seiko pin-transfer robot. Each plate was created in quadruplicate. Two copies were incubated at 24°C in a moist chamber and two were incubated at 37°C in a moist chamber. After 3 days, cells in individual wells were scored for growth by assessing turbidity. Representative small molecules from the following commercial libraries were screened: Bionet, Cerep, Maybridge, NCI Diversity and Peakdale.
Commercially available structural analogs of mdivi-1 were identified using the web-based ChemNavigator program. Small molecules tested in all assays were resuspended in DMSO at 10 mg/ml and stored desiccated at -20°C. All commercial compounds used in this study were verified by mass spectrometry.
Assays
All of the experiments described in this study using the small molecules were double blinded. Representative images were chosen and quantification was performed without knowledge of the identity of the compounds being tested. The compounds were analyzed using a battery of established yeast and mammalian cell-based and in vitro assays. These are described in detail in the Supplememtal Data Section of the manuscript.
Supplementary Material
Acknowledgements
The chemical screen was performed at the former ICCB Facility at Harvard (currently ICCB-Longwood) and we would like to thank Drs. Caroline Shamu, Tim Mitchison, and John A. Tallarico for their excellent advice and support. We are also grateful to Dr. Irwin Segel for his insightful comments and generous help in interpreting and modeling our kinetic data. We would like to thank Joseph Opferman, Department of Biochemistry, St. Jude Children’s Research Hospital for providing the MxCre bak-/-baxf/- livers. We thank Dr. Shelly Meeusen and Mr. James Partridge for help with experiments and the Nunnari lab for their comments on the manuscript. This work was supported by NIH grants 1 R01 EY015924 to JN and CA69301, AI40646, and GM52735 to DG. This research was supported in part by the Intramural Research Program of NIH, NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPases, Dnm1, regulates mitochondrial fission in yeast. Nature Cell Biology. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006a;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondrial Fusion and Fission in Mammals. Annu Rev Cell Dev Biol. 2006b doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006;18:453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr Opin Cell Biol. 2001;13:454–460. doi: 10.1016/s0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. Mol Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Estaquier J, Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- Fadok VA, Henson PM. Apoptosis: giving phosphatidylserine recognition an assist--with a twist. Curr Biol. 2003;13:R655–657. doi: 10.1016/s0960-9822(03)00575-x. [DOI] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner W, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Developmental Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, et al. OPA1 Controls Apoptotic Cristae Remodeling Independently from Mitochondrial Fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. Embo J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal G, Fell B, Sarin A, Youle RJ, Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Chan DC. Domain interactions within Fzo1 oligomers are essential for mitochondrial fusion. J Biol Chem. 2006;281:16599–16606. doi: 10.1074/jbc.M601847200. [DOI] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. The Journal of Cell Biology. 1998;143:359–374. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The Machines that Divide and Fuse Mitochondria. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. Epub 2004 Feb 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002a;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002b;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris K, Cleland M, Jeong S, Youle R. Role of Bax and Bak in Mitochondrial Morphogenesis. Nature. 2006 doi: 10.1038/nature05111. in press. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Detmer S, Kaiser J, Chen H, McCaffery J, Chan D. Structural basis of mitochondrial tethering by mitofusin complexes acting in trans. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1controls severing of the mitochondrial outer membrane. Molecular Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006;13:1291–1295. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive mutants defective in mitochondrial inheritance. The Journal of Cell Biology. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, Devay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial Inner-Membrane Fusion and Crista Maintenance Requires the Dynamin-Related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Marshall W, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in S. cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mtDNA. Molecular Biology of the Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Olichon A, Guillou E, Delettre C, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Daloyau M, Hamel C, Amati-Bonneau P, et al. Mitochondrial dynamics and disease, OPA1. Biochim Biophys Acta. 2006;1763:500–509. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Parone PA, James DI, Da Cruz S, Mattenberger Y, Donze O, Barja F, Martinou JC. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol Cell Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B, Decottignies A, Kolaczkowski M, Carvajal E, Balzi E, Goffeau A. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. Journal of molecular microbiology and biotechnology. 2001;3:207–214. [PubMed] [Google Scholar]
- Rolland S, Conradt B. The role of mitochondria in apoptosis induction in Caenorhabditis elegans: more than just innocent bystanders? Cell Death Differ. 2006;13:1281–1286. doi: 10.1038/sj.cdd.4401980. [DOI] [PubMed] [Google Scholar]
- Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. Journal of Cell Biology. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van Der Bliek AM. Dynamin-related protein drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland D, Ryazantsev S, Van Der Bliek A. A human dynamin-related protein controls the distribution of mitochondria. Journal of Cell Biology. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka R, Shimizu S, Tsujimoto Y. Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J Biol Chem. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. Journal of Biological Chemistry. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Youle RJ. Morphology of mitochondria during apoptosis: worms-to-beetles in worms. Dev Cell. 2005;8:298–299. doi: 10.1016/j.devcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.