Abstract
The asexual blood stage of Plasmodium falciparum in the human host is comprised of morphologically distinct ring, trophozoite and schizont stages, each of which possesses a distinct pattern of gene expression. Episomal promoter recombination has been recently reported in malaria parasites. We aim to investigate the nature of this process, and its relationship with promoter activity, by employing a series of nested deletions of the ring specific hrp3 promoter. Our results showed a discrete promoter region that is preferentially used for recombination. The P. falciparum hrp3 RNA gene is only seen in ring-stage parasites but deletion of the recombination region was associated with decreased ring-stage expression and concurrent detection of transcripts in trophozoite-stage parasites. Our results describe a ring-stage specific regulatory region possibly involved in episomal promoter recombination, suggesting that common sequences might mediate both processes.
1. Introduction
Plasmodium falciparum infection kills 2–3 million people every year, mostly children under 5 years of age (Greenwood and Mutabingwa, 2002). The blood stage of the parasite’s life cycle is comprised of four distinct morphological forms (rings, trophozoites, schizonts and merozoites) and development through these stages is linked to regulation of gene expression.
Comprehensive evidence for stage specific gene expression comes from microarray expression analysis of distinct global patterns of RNA synthesis at different phases of parasite growth (Ben Mamoun, et al., 2001, Bozdech, et al., 2003, Hayward, et al., 2000, Munasinghe, et al., 2001, Patankar, et al., 2001). The role of Plasmodium promoters on regulation of gene expression has been assessed using a small number of asexual and sexual gene promoters. Thus, the structure of basal and stage specific promoters are poorly described. The advent of transfection to the blood stage parasite allowed the use of 5’ end regions from asexual genes such as hsp86, cam, hrp3, PfEMP1 (var) as well sexual genes such as pfs16 and pfs25 to drive reporter gene expression suggesting that they function as promoters (Crabb and Cowman, 1996, Crabb, et al., 1997, Dechering, et al., 1999, Deitsch, et al., 2001, Horrocks and Lanzer, 1999, Voss, et al., 2000, Wu, et al., 1995).
Transfection efficiency of blood stage malaria parasite remains very low despite recent progress in transfection methodology by several laboratories (Balu, et al., 2005, Nkrumah, et al., 2006). A possible explanation for the low transfection efficiency is the formation of large concatameric extrachromosomal plasmid forms (O'Donnell, et al., 2001). Plasmids appear to integrate into the malaria parasite genome in 1–3 tandem copies, suggesting that they may be present as concatamers prior to integration (Menard, et al., 1997).Episomal plasmid recombination in malaria has been reported to generate large stably replicating forms (O'Donnell, et al., 2001). This recombination appears to occur preferentially at the promoter region suggesting a correlation between promoter activity and promoter recombination (Kadekoppala, et al., 2001). Recently, plasmid recombination at non-promoter region has been reported in P. falciparum (Frank, et al., 2006). Plasmid chimeras occurred by intramolecular uneven recombination at the 3’ end region of HRP 2 (3’hrp2) used as termination region for expression of recombinant protein (Frank, et al., 2006). Together these results suggest the presence of different mechanisms of homologous recombination in the parasite.
The role of malaria parasite promoter and non-promoter sequences on plasmid homologous recombination has not been assessed in detail. Here we investigated promoter-mediated recombination by cotransfecting malaria parasites with two plasmids, one carrying non-selectable marker GFP gene driven by the hrp3 promoter, the other carrying a selectable marker (TgDHFR) driven by the same promoter. Successful recombination of these plasmids will generate pyrimethamine-resistant cell lines expressing GFP. Nested deletions on the 1.7-kb ring-specific hrp3 promoter decreased promoter-mediated recombination and switched promoter stage specificity from ring to trophozoite parasites. Deletion events that switched promoter specificity were associated with low recombination efficiency suggesting that timing of transcription might be important for efficient promoter recombination. In addition, all plasmid chimeras “rescued” from the transfected parasite cell lines conform to at least one of the three chimera types reported earlier (Kadekoppala, et al., 2001). Replacement of hrp3 for the trophozoite-specific msp1 generated a parasite subpopulation expressing GFP; these two promoters displayed a distinct efficiency of recombination despite their similar length. Together, our results suggest that recombination efficiency might correlate with the promoter’s strength and/or stage specificity.
2. Material and Methods
2.1. Plasmid constructs
The hrp3 promoter deletions were made using Erase-a-base® (Promega) on pHRPCAT (Wu, et al., 1995) containing the hrp3 promoter driving the CAT gene. Kpn I-Dra III-digested pHRPCAT was incubated with exonuclease III (exoIII) at predetermined times to obtain specific-sized promoters of 1.1-kb, 0.6-kb and 0.5-kb. The CAT gene in exoIII-digested pHRPCAT was substituted with the GFP gene to generate pHRP1.7G (Kadekoppala, et al., 2000), pH1.1G, pH0.6G, pH0.5G. The CAT gene in exoIII-digested pHRPCAT was also substituted with the GFP/hDHFR-fusion gene (HDGFP, (Kadekoppala, et al., 2000)) to generate pH1.7HG (Kadekoppala, et al., 2000), pH1.1HG and pH0.6HG.
2.2. In vivo recombination assay and plasmid rescue
Episomal plasmid recombination was assessed by the method described previously (Kadekoppala, et al., 2001). Plasmids containing hrp3 1.7-kb and truncated promoters driving GFP expression (pH1.7G, pH1.1G, pH0.6G, pH0.5G), were individually mixed with the pDT.Tg23 plasmid (Wu, et al., 1996) carrying the 1.7-kb hrp3 promoter driving Toxoplasma gondii’s bifunctional enzyme gene dhfr-ts (fig 1A). Plasmids, at 1:1 ratio (50 µg per plasmid) were cotransfected into infected erythrocytes (Fidock and Wellems, 1997). Forty-eight hpi, 100 ng/ml of pyrimethamine was added to the cultures for two days. Subsequently parasite cultures were incubated and maintained at 20 ng/ml of pyrimethamine. The pmspTgDHFR plasmid was generated by digesting the TgDHFR gene from pDT.Tg23 with Nsi I and Hind III. The Nsi I-Hind III TgDHFR fragment used to replace the GFP gene in the pMSPGFP (Kadekoppala, et al., 2001). Live parasite cells expressing GFP (gfp+) and/or pyrimethamine-resistant (pyrr) were stained with 5 µg/ml of DAPI and used to determine the total parasite number using a epifluorescent Nikon eclipse 400. gfp+-parasites were scored and reported as a percentage of total pyrimethamine-resistant (pyrr) cells stained with DAPI.
Figure 1. Construction of plasmids used to transfect P. falciparum parasites.
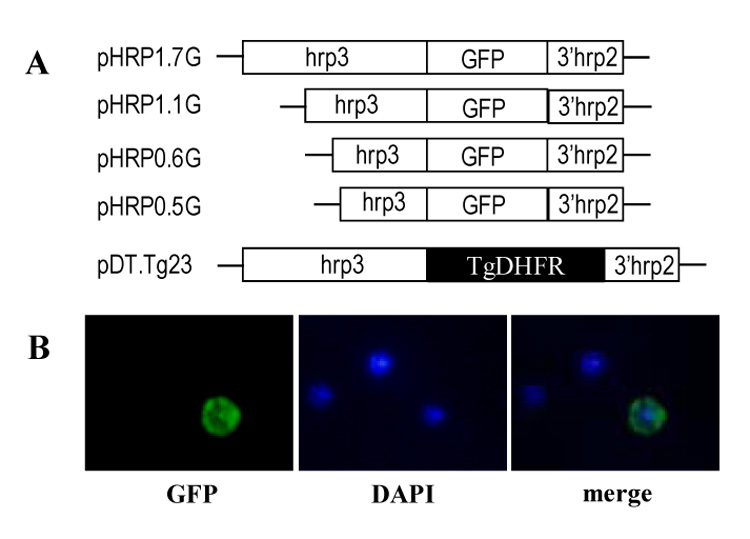
(A) Schematic representation of plasmids containing 1.7-kb promoter (pHRP1.7G), 1.1-kb (pHRP1.1G), 0.6-kb (pHRP10.6G) and 0.5-kb (pHRP0.5G) hrp3 truncated promoters driving the GFP gene. The plasmid pDT.Tg23 plasmid is also shown containing the 1.7-kb hrp3 promoter driving the T. gondii dhfr-ts gene (Kadekoppala, et al., 2001). (B) Stable pyr-resistant parasite cultures cotransfected with pHRP1.7G and pDT.Tg23 plasmids show green fluorescent GFP (left panel) in early trophozoite. Nucleic acid staining is shown in blue in the nucleus (Center panel) and the merge image (right panel).
Total DNA, for plasmid rescue experiments, was isolated from saponin-released gfp+:pyrr parasite stable cell lines. Released parasites were lysed with SDS extraction buffer (10mM Tris-HCl pH 8.0, 0.1 M EDTA pH 8.0, 0.5% SDS) and DNA extracted with buffered phenol:chloroform:isoamyl alcohol (25:24:1) at room temperature. Plasmids from transfected cell lines were recovered by transforming Escherichia coli competent cells with purified parasite DNA. E. coli recovered plasmid from two independent transfections were digested with Xho I and/or Hpa I and separated on agarose gel.
2.3. Parasite transfection and Northern blot Analysis
P. falciparum stable cell lines were generated by transfecting parasite-infected erythrocytes (3D7 strain) with 100 µg of experimental plasmids (Fidock and Wellems, 1997). Stable cells were selected with methotrexate (2.2 µM, Sigma) 48 hours post-transfection and maintained until the cell lines were established (3–4 weeks). Parasites were percoll/sorbitol synchronized and monitored by light microscopy of a giemsa stained infected red cells smear. Synchronization was repeated until 99.9% of the parasites were in a specific stage. Total RNA from stable cell lines carrying pH1.7HG, pH1.1HG or pH0.6HG were isolated using TRIzol® LS reagent (Gibco BRL). Ten micrograms of total RNA was electrophoresed in agarose gel and transferred to Hybond N+ membranes (Amersham) and subsequently hybridized with a 32P-antisense GFP probe generated by asymmetric PCR (Bird, 2005).
3. Results
Recombination of episomal plasmid carrying the hrp3 promoter has been previously described (Kadekoppala, et al., 2001). To investigate which promoter region is responsible for in vivo recombination, we co-transfected two separate plasmids, one plasmid carrying a non-selectable reporter gene (GFP in pHRPGFP) and the other a drug selection marker from Toxoplasma gondii (dhfr-ts in pDT.Tg23) (Kadekoppala, et al., 2001). Long-term maintenance of the GFP-containing plasmid (pHRPGFP) will depend upon successful recombination with the selectable plasmid pDT.Tg23. GFP-encoding plasmids were mixed at a 1:1 ratio with the pDT.Tg23 (Fig. 1A). In order to avoid differences in parasite growth due to drug selection, the 1.7-kb hrp3 promoter was used to drive the selectable marker dhfr-ts gene and truncations were made on the hrp3 promoter driving the non-selectable maker GFP. The percentage of in vivo recombination at a given time was calculated by counting the number of GFP-expressing parasites and the total parasites were determined by scoring the number of intraerythrocytic DAPI-stained parasite’s nuclei in the sample. After pyrimethamine (pyrr) selection, a subpopulation of the parasite showed gfp+ expression (Fig. 1B, parasites transfected with pHRP1.7G/pDT.Tg23 plasmids). A sizable number of parasite were pyrimethamine resistant but did not express parasite gfp+ as evidenced by the DAPI-stained nuclei in fig. 1B. Parasites cotransfected with the hrp3 1.7-kb promoter in both GFP and TgDHFR-containing plasmids, showed 8% of green parasites at 12 days post-transfection (Fig. 2, pHRP1.7G). At 24 days post-transfection the gfp+-expressing parasite reached a peak around 45% and stabilized at 30% after 30 days post-transfection, which is consistent with previous results (Kadekoppala, et al., 2001). The deletion of 600 bp from the 5’ end on the hrp3 to give rise to the 1.1-kb truncated promoter in the pH1.1G plasmid, reduced green parasites to approximately 15% after 30 days post-transfection (Fig. 2, pHRP1.1G). This reduction in parasites carrying chimeric plasmids, suggests the presence of a recombinogenic element between −1.7-kb and −1.1 kb (Fig. S1 supplementary data, underlined nucleotides). Further deletion of the 1.1-kb hrp3 promoter to 0.6-kb and 0.5-kb in pH0.6G and pH0.5G respectively, reduced green parasites below 5% (Fig. 2, pHRP0.6G and pHRP0.5G) suggesting a second recombinogenic region located between −1.1-kb and −0.6-kb region (Fig. S1 supplementary data, bold faced nucleotides). Parasites were also transfected with pDT.Tg23 and pMSPGFP in order to evaluate whether recombination was specific to the promoter region. Pyrimethamine-resistant parasites but not gfp+-expressing parasites were recovered; this result indicates that effective recombination requires homologous promoter as previously shown (Kadekoppala, et al., 2001).
Figure 2. In vivo episomal recombination in P. falciparum parasites.
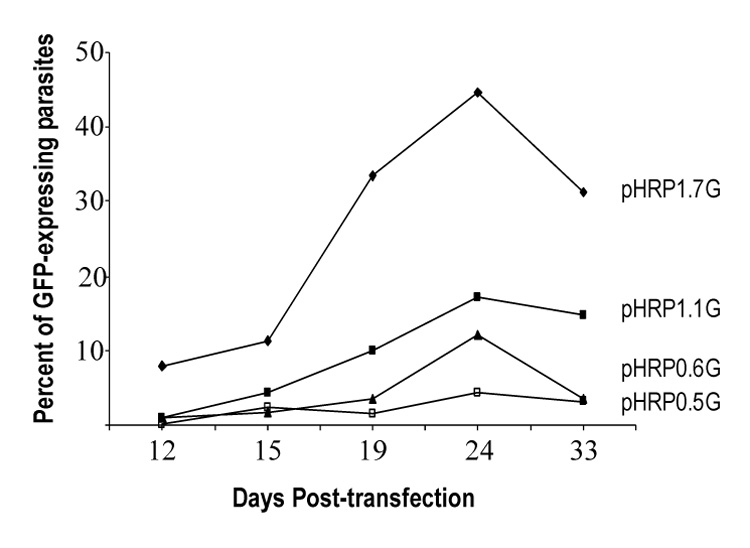
Parasite cultures were cotransfected with pHRP1.7G, pHRP1.1G, pHRP10.6G or pHRP0.5G hrp3 truncated promoters driving the GFP gene and pDT.Tg23 as shown in figure 1. Parasites were collected at 12, 15, 19, 24 and 33 days post transfection. Green fluorescent parasites were scored and expressed as percent of total.
To assess whether the gfp+ and pyrr containing plasmids recombined and remained as an episomal chimera, we performed plasmid rescue experiments. This method has been previously used to identify plasmid chimeras in the parasite (Frank, et al., 2006, Kadekoppala, et al., 2001). There are three different types of chimeras depending on the number of concatamers according to (Kadekoppala, et al., 2001); the simplest chimera comprised of two plasmid units, one pDT.Tg23 (1 pyrr) and one pH1.7G (1 gfp+) namely type 1. The second class of chimeras contained three concatamers from which two additional types were identified. Type 2 comprised of 1 pyrr and 2 gfp+, whereas type 3 comprised 2 pyrr and 1 gfp+ (Kadekoppala, et al., 2001). Total DNA purified from gfp+-pyrr parasite cell lines, grown in continuous culture for more than a year, was used to transform E. coli competent cells. Purified plasmids isolated from individual bacteria colonies (10 colonies for each parasite cell line) were digested with Xho I and/or Hpa I. These restriction enzymes cut only once in the dhfr-ts and gfp genes respectively, but not in the plasmid backbone (Kadekoppala, et al., 2001), which allows for the identification of chimera types in the parasites. Plasmid rescued from pDT.Tg23/pH1.7G-transfected parasites digested with Xho I or Hpa I yielded a single band of 13-kb (Fig. 3, lanes 2 and 3). This result is consistent with a chimera containing one copy of each original plasmid in the parasite. Double digestion with Xho I and Hpa I restriction enzymes generated bands of 7-kb and 6-kb, consistent with the presence of pDT.Tg23 and pH1.7G plasmids respectively (Fig. 3 lane 4). These results indicate the presence of stable episomal chimeras in Plasmodium and demonstrate that chimeras are stably maintained even after long-term culturing of transfected parasites (Kadekoppala, et al., 2001). Episomal chimeras were also rescued from parasite cell lines transfected with pDT.Tg23/pH1.1G, pDT.Tg23/pH0.6G or pDT.Tg23/pH0.5G plasmids. Digestion of these chimeras with Xho I generated two bands of 13-kb and 7-kb respectively (Fig. 3, lanes 6, 10 and 14 respectively); a finding that is diagnostic of episomal chimeras comprised of 2 pyrr and 1 gfp+ plasmids. Conversely, Hpa I digestion generated only one band of approximately 20-kb, confirming the presence of only one gfp+-containing plasmid in the chimera (Fig. 3, lanes 7, 11 and 15 respectively). Double digestion with Xho I/Hpa I confirmed the presence of the two plasmid types (pyrr and gfp+) in the chimera (Fig. 3, lanes 8, 12 and 16 respectively). In addition, episomal chimeras containing 1 pyrr-2 gfp+ (data not shown) were also detected in pyrr-gfp+ Plasmodium cell lines transfected with pDT.Tg23/pH1.1G, pDT.Tg23/pH0.6G or pDT.Tg23/pH0.5G. The rescue of chimeras indicates the presence of long-term plasmids generated by homologous recombination of the pDT.Tg23 with the pH1.1G, pH0.6G or pH0.5G.
Figure 3. Heteromultimeric plasmids recovered in E. coli.
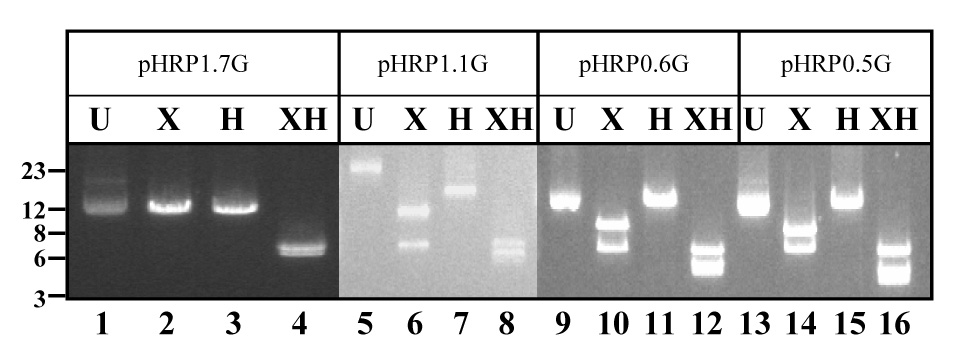
(A) Schematic representation of heteromultimeric plasmids containing 1pyrr:1gfp copy of pH1.7G and pDT.Tg23 (type 1); 1pyrr:2gfp (type 2) or 2pyrr:1gfp (type 3) (Kadekoppala, et al., 2001). (B). E. coli competent cells were transformed with total DNA from parasite cell lines. Plasmid recovered in E. coli transformed with pDT.Tg23/pH1.7HG (pH1.7HG), pDT.Tg23/pH1.1HG (pH1.1HG), pDT.Tg23/pH0.6HG (pH0.6HG) or pDT.Tg23/pH0.5HG (pH0.5HG) plasmid mixtures were digested with Hpa I (H) and/or Xho I (X) and separated on agarose gel with undigested (U) plasmid as control. DNA molecular weight marker (in kb) is presented. Detected bands did not change size after long-term culture (more than a year). Plasmid backbone is pBluescript (not shown).
To test whether promoter recombination is a common feature of other malaria promoters, we cotransfected two new plasmids containing the 1.6-kb trophozoite specific msp1 promoter. The first plasmid carries the TgDHFR-ts and the second plasmid carries the gfp gene (pMSPGFP, (Kadekoppala, et al., 2001). Fig. S2, supplemental data). Drug selection of transfected parasites gave rise to a pyrimethamine-resistant population, consistent with the presence of the selectable marker plasmid pmspTgDHFR. Approximately 1% of the parasites were gfp+ one week after the start of selection. Thirty days post-transfection the number of gfp+ parasites remained constant (approximately 1%). No further increase in gfp+- expressing parasites was observed. These results suggest that homologous recombination also occurs in the trophozoite-specific msp1 promoter, suggesting that homologous recombination is a general feature associated with Plasmodium promoters. Although the msp1 and the hrp3 promoters are similar in size, the latter recombined more efficiently suggesting that promoter size might not play a major role on the overall recombination rates.
Because of the putative correlation between promoter activity and recombination, we evaluated the role of hrp3 promoter nested deletions on gene expression. The full length (1.7-kb) and deleted (1.1-kb and 0.6-kb) promoters were placed upstream of the HDGFP fusion gene (Kadekoppala, et al., 2000) (Fig. 4A). Methotrexate-resistant stable cell lines were percoll/sorbitol synchronized and used to purify RNA at 12–14 hpi (ring-stage) and subjected to northern blot analysis using a 32P-antisense GFP probe. The 1.7-kb promoter (Fig 4A, pH1.7HG) had the maximum steady state mRNA accumulation (Fig. 4Bi and 4Biii) in the ring-stage of the parasite. Deletions of the hrp3 5’ end to generate the 1.1-kb and 0.6-kb truncated promoters (Fig 4A, pH1.1HG and pH0.6HG) decreased mRNA accumulation in ring-stage parasites (Fig. 4Bi and 4Biii) with little mRNA detectable in the 0.6-kb promoter truncation. We then investigated the effect of the same 5’ deletions on trophozoite-stage promoter activity at 32–34 hpi. Steady state accumulation of HDGFP mRNA from the 1.7-kb promoter cell line pH1.7HG showed a significant reduction of mRNA compared to ring parasites (Fig. 4Bii), consistent with the ring-stage specificity of the 1.7-kb promoter (Cheresh, et al., 2002). HDGFP mRNA from 1.1-kb truncated promoter was also detected in trophozoite-stage parasites (Fig 4Bii, 4Biv). Detection of HDGFP mRNA from the 1.1-kb promoter in both ring and trophozoite stage parasites indicates that the deletion of the −1.7-kb to −1.1-kb region correlates with loss of ring specificity. This truncation may affect the regulation of promoter activity by increasing transcription or HDGFP mRNA stability from trophozoite stage. Unexpectedly, deletion of hrp3 to a 0.6-kb in pH0.6HG, showed an increase level of HDGFP mRNA accumulation in the trophozoite stage parasites (Fig. 4Bii and 4Biv), but with little transcript in ring stage parasites. Comparison of HDGFP mRNA accumulation in the ring and trophozoite stage parasites clearly shows a switch of promoter stage specificity. One interesting observation was that the 0.6-kb hrp3 promoter showed similar recombination efficiency to the msp1 promoter despite the size difference (0.6-kb and 1.6-kb respectively). Alignment analysis of these two promoters failed to show a significant homology. However both 0.6-kb hrp3 and 1.0-kb msp1 promoters have a peak of mRNA accumulation at trophozoite-stage. These data suggest the possibility that homologous recombination is less favored at the trophozoite stage of the parasites. Our results strongly suggest that the region from −1.7-kb to −1.1-kb of hrp3 promoter restricts promoter activity to ring stage parasites.
Figure 4. Deletion of the hrp3 promoter switched stage specific expression from ring to trophozoite stage.
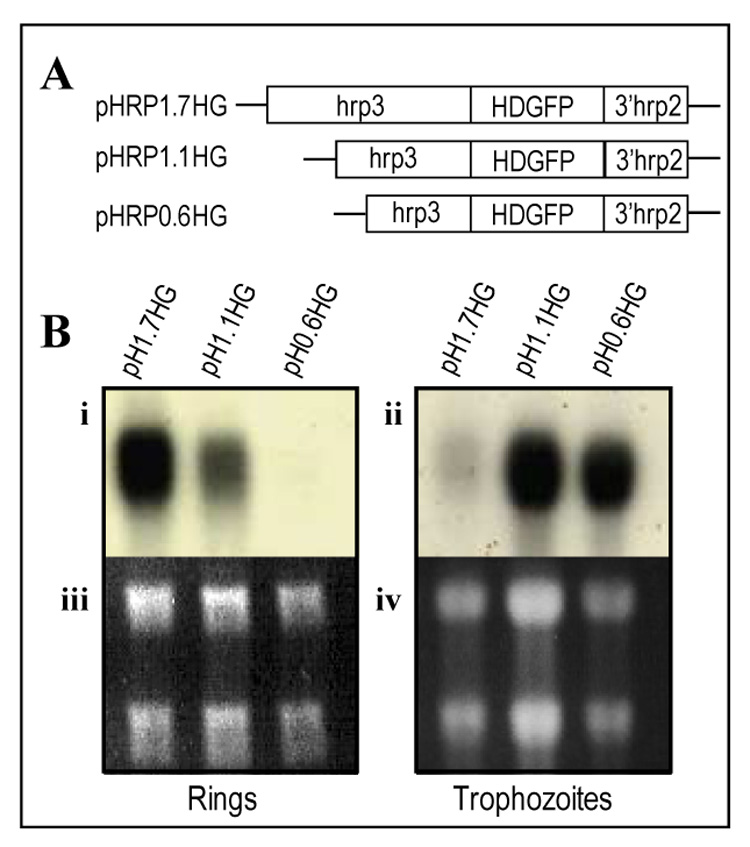
(A) Schematic representation of hrp3 promoter truncations (diagonal lines) driving human DHFR fused to GFP (HDGFP, filled box) in plasmid pH1.7HG containing the 1.7-kb full length promoter as well as the plasmids pH1.1HG, pH0.6HG carrying the truncated promoters of 1.1-kb and 0.6-kb respectively. (B) Total RNA from stable cell lines expressing HDGFP was purified from ring (i, iii) and trophozoite (ii, iv) stages, separated on agarose gels stained with ethidium bromide (iii, iv) and analyzed by northern blot (i, ii) using 32P-GFP antisense probe.
Discussion
In this study we report a correlation between promoter recombination and promoter activity in the human malaria parasite P. falciparum. Truncation of a hrp3 promoter decreased the number of gfp+-expressing parasites, suggesting a reduced recombination efficiency of the truncated promoters. Two region of approximate size were associated with the overall recombination efficiency. Truncation of the region between −1.7-kb to −1.1-kb decreased recombination from 30% to 15%. Further truncation of the region between −1.1-kb and −0.6-kb further reduced the number of gfp+-expressing cells and thus recombination from 15% to 5%. The formation of episomal chimeras due to plasmid recombination was assessed by plasmid rescue experiments on E. coli cells. All rescued chimeras conformed the 3 basic structures previously described (Kadekoppala, et al., 2001), indicating that recombination most likely occurred at the promoter sequences. It was rather unexpected that only “type” 1 chimeras were obtained from the cell line transfected with pDT.Tg23 and pH1.7HG. In contrast, “types” 2 and 3, but not “type” 1, chimeras were obtained from cell lines transfected with pDT.Tg23 and pH1.1HG, pH0.6HG or pH0.5HG. It is possible that long-term cell culture may result in the 1.7-kb promoter forming large self-replicative chimera (O'Donnell, et al., 2001) leaving only “type” 1 chimera available for transforming competent bacterial cells.
Homologous recombination at regions known as hotspots have been described in yeast and human genomes (Chakravarti, et al., 1984, Jeffreys, et al., 2001, McVean, et al., 2004, Petes, 2001) as well as the Plasmodium parasite (Mu, et al., 2005). Recombination events in the parasite’s genome appear to be clustered in the middle and near the chromosome end (telomeric regions). Although recombination rates are highly variable, hotspot locations seems to be conserved among distinct population of malaria parasites, suggesting a common mechanism (Mu, et al., 2005). Telomeric regions are packed with genes involve in antigenic variation and cell surface interaction genes such as var, rifin and stevor (Bowman, et al., 1999, Gardner, et al., 2002). Interestingly, hrp3 is also located at the end of chromosome 13 (Wellems, et al., 1987), suggesting the possibility that promoter-mediated recombination is a feature of telomeric-located genes. However, whether promoter region and recombination hotspot share the same distribution along Plasmodium’s chromosomes has yet to be determined. Homologous recombination on non-promoter regions has been recently reported in episomal plasmid used to study variant var gene silencing regulation (Frank, et al., 2006). Specific uneven recombination at the 3’ end of the hrp2 region (3hrp2) was generated after transfecting the parasite with a double cassette plasmid carrying two copies of 3hrp2. All recombination events detected on the plasmid and into the chromosome has at least one of the two 3hrp2 copies immediately adjacent to the var intron bidirectional promoter (Calderwood, et al., 2003) used to drive a downstream-located selectable gene (hDHFR) (Frank, et al., 2006). Interestingly, recombination between 3hrp2 regions distant from promoter sequences was not detected and thus, it is possible that intron promoter activity might play a role in recombination mediated by 3hrp2 in this plasmid. gfp+-expressing parasites were not detected when parasites were cotransfected with plasmids carrying non homologous promoters (Kadekoppala, et al., 2001), suggesting that chimeric gfp+pyrr plasmids were not present in the recovered pyrr-resistant parasite population. Our results strongly indicate that 3hrp2 regions were not involved in chimera formation. However, we cannot rule out the possibility that very low levels of 3hrp2-mediated recombination was just not detected in our assays. Recombination efficiency does not appear to be determined exclusively by promoter length. For instance, the msp1 promoter displayed lower recombination efficiency than its hrp3 counterpart albeit the similar size of these two promoters. In this context, promoter activity might account for the difference in recombination efficiency.
The change in promoter recombination was associated with loss of hrp3 promoter stage specificity. The reporter HDGFP mRNA was present in both ring and trophozoite parasite in the promoter lacking the −1.7-kb to −1.1- kb region. This truncation turned the hrp3 promoter’s activity constitutive. Further truncation of the hrp3 promoter region located between −1.1-kb to −0.6-kb showed mRNA accumulation in trophozoite parasites but not in ring parasites. The pattern of mRNA accumulation was therefore switch from ring to trophozoite after transition from 1.7-kb to 0.6-kb truncated promoter. Interestingly, the hrp3 0.6-kb and msp1 behave as trophozoite specific promoters with relatively low recombination efficiency compared to the 1.7-kb hrp3 promoter, suggesting that recombination in trophozoite parasite might be lees efficient than in ring parasites. Together, these results suggest the presence of common sequences that might regulate both promoter recombination and stage specificity. However, the role of transcription in recombination has to be further evaluated.
Genome wide analysis has been impaired due to the propensity of episomal plasmids to form large chimeras in the parasite, precluding random integration into the genome. The understanding of homologous episomal recombination will facilitate the design of improved vectors for reverse genetic studies in the parasite. To this goal, a more comprehensive analysis of the cis and trans factors mediating episomal recombination is needed. This is stressed by the fact that uneven plasmid recombination has been recently reported (Frank, et al., 2006). This may be undesirable when it results in the loss of plasmid regions required for the proper analysis of biological processes. Questions such as the nature of the DNA sequences involved in these processes, the role of transcription as well as the role of stage specific promoter activity are currently been addressed in our laboratory.
Supplementary Material
Acknowledgments
We thank Dr. Steve D. Schwartzbach, Dr. Charles Lessman and Dr. Stephan Schoech, University of Memphis for critical reading of the manuscript. This work was funded by NIH grants 1R03AI054529 (to KH and CLE), R01HL69630 and R01AI39071 (to KH). JPS was supported by a fellowship from INSERM (France).
Index descriptors and abbreviations
- Plasmodium
transcription; regulation; histidine-rich protein 3; stage specificity; expression.
- dhfr-ts
dihydrofolate reductase-thymidylate synthase
- cam
calmodulin
- msp1
merozoite surface protein 1
- hrp3
Histidine-rich protein 3
- hrp2
Histidine-rich protein 2
- GFP
Aequorea victoria green fluorescent protein
- PfEMP1
Plasmodium falciparum erythrocyte membrane protein 1
- hsp86
Heat shock protein 86
- pfs16
Plasmodium falciparum sexual-stage antigen 16
- pfs25
Plasmodium falciparum sexual-stage antigen 25
- RNA
Ribonucleic acid
- rRNA
ribosomal ribonucleic acid
- CAT
chloramphenicol acetyl transferase
- PCR
Polymerase chain reaction
- DAPI
4′-6-Diamidino-2-phenylindole
- hpi
hours post-invasion
- mRNA
messenger ribonucleic acid
- rifin
Repetitively Interspersed Family
- stevor
subtelomeric variable open reading frame
- HD
Human dihydrofolate reductase (hDHFR)
- HDGFP
Human dihydrofolate reductase fused to Aequorea victoria Green fluorescent protein
- SDS
Sodium dodecyl sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balu B, Shoue DA, Fraser MJ, Jr, Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proceedings of the National Academy of Sciences. U S A. 2005;102:16391–16396. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Mamoun C, Gluzman IY, Hott C, MacMillan SK, Amarakone AS, Anderson DL, Carlton JM, Dame JB, Chakrabarti D, Martin RK, Brownstein BH, Goldberg DE. Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Molecular Microbiology. 2001;39:26–36. doi: 10.1046/j.1365-2958.2001.02222.x. [DOI] [PubMed] [Google Scholar]
- Bird IM. Generation of high-sensitivity antisense cDNA probes by asymmetric PCR. Methods in Molecular Medicine. 2005;108:199–213. doi: 10.1385/1-59259-850-1:199. [DOI] [PubMed] [Google Scholar]
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, Gentles S, Gwilliam R, Hamlin N, Harris D, Holroyd S, Hornsby T, Horrocks P, Jagels K, Jassal B, Kyes S, McLean J, Moule S, Mungall K, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutter S, Skelton J, Squares R, Squares S, Sulston JE, Whitehead S, Woodward JR, Newbold C, Barrell BG. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The Transcriptome of the Intraerythrocytic Developmental Cycle of Plasmodium falciparum. Public Library of Science. 2003;Biol 1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. Journal of Biological Chemistry. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Buetow KH, Antonarakis SE, Waber PG, Boehm CD, Kazazian HH. Nonuniform recombination within the human beta-globin gene cluster. The American Journal of Human Genetics. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- Cheresh P, Harrison T, Fujioka H, Haldar K. Targeting the Malarial Plastid via the Parasitophorous Vacuole. Journal of Biological Chemistry. 2002;277:16265–16277. doi: 10.1074/jbc.M109331200. [DOI] [PubMed] [Google Scholar]
- Crabb BS, Cowman AF. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proceedings of the National Academy of Sciences. U S A. 1996;93:7289–7294. doi: 10.1073/pnas.93.14.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, Triglia T, Waterkeyn JG, Cowman AF. Stable transgene expression in Plasmodium falciparum. Molecular and Biochemical Parasitology. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- Dechering KJ, Kaan AM, Mbacham W, Wirth DF, Eling W, Konings RN, Stunnenberg HG. Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite Plasmodium falciparum. Molecular and Cellular Biology. 1999;19:967–978. doi: 10.1128/mcb.19.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proceedings of the National Academy of Sciences. U S A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Costantini D, Amulic B, Berdougo E, Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. Journal of Biological Chemistry. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood B, Mutabingwa T. Malaria in 2002. Nature. 2002;415:670–672. doi: 10.1038/415670a. [DOI] [PubMed] [Google Scholar]
- Hayward RE, Derisi JL, Alfadhli S, Kaslow DC, Brown PO, Rathod PK. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Molecular Microbiology. 2000;35:6–14. doi: 10.1046/j.1365-2958.2000.01730.x. [DOI] [PubMed] [Google Scholar]
- Horrocks P, Lanzer M. Differences in nucleosome organization over episomally located plasmids coincides with aberrant promoter activity in P. falciparum. Parasitology International. 1999;48:55–61. doi: 10.1016/s1383-5769(99)00002-1. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genetics. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Kadekoppala M, Cheresh P, Catron D, Ji D, Deitsch K, Wellems TE, Seifert HS, Haldar K. Rapid recombination among transfected plasmids, chimeric episome formation and trans gene expression in Plasmodium falciparum. Molecular and Biochemical Parasitology. 2001;112:211–218. doi: 10.1016/s0166-6851(00)00368-6. [DOI] [PubMed] [Google Scholar]
- Kadekoppala M, Kline K, Akompong T, Haldar K. Stable expression of a new chimeric fluorescent reporter in the human malaria parasite Plasmodium falciparum. Infection and Immunity. 2000;68:2328–2332. doi: 10.1128/iai.68.4.2328-2332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Menard R, Sultan AA, Cortes CC, Altszuler R, van Dijk MR, Janse CJ, Waters AP, Nussenzweig RS, Nussenzweig V. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. 1997;385:336–340. doi: 10.1038/385336a0. [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ. Recombination hotspots and population structure in Plasmodium falciparum. Public Library of Science. 2005;Biol 3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munasinghe A, Patankar S, Cook BP, Madden SL, Martin RK, Kyle DE, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression (SAGE) in Plasmodium falciparum: application of the technique to A–T rich genomes. Molecular and Biochemical Parasitology. 2001;113:23–34. doi: 10.1016/s0166-6851(00)00378-9. [DOI] [PubMed] [Google Scholar]
- Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nature Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell R, Preiser PR, Williamson DH, Moore PW, Cowman AF, Crabb BS. An alteration in concatameric structure is associated with efficient segregation of plasmids in transfected Plasmodium falciparum parasites. Nucleic Acids Research. 2001;29:716–724. doi: 10.1093/nar/29.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Molecular Biology of the Cell. 2001;12:3114–3125. doi: 10.1091/mbc.12.10.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD. Meiotic recombination hot spots and cold spots. Nature Reviews Genetics. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- Voss TS, Thompson JK, Waterkeyn J, Felger I, Weiss N, Cowman AF, Beck HP. Genomic distribution and functional characterisation of two distinct and conserved Plasmodium falciparum var gene 5' flanking sequences. Molecular and Biochemical Parasitology. 2000;107:103–115. doi: 10.1016/s0166-6851(00)00176-6. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Walliker D, Smith CL, do Rosario VE, Maloy WL, Howard RJ, Carter R, McCutchan TF. A histidine-rich protein gene marks a linkage group favored strongly in a genetic cross of Plasmodium falciparum. Cell. 1987;49:633–642. doi: 10.1016/0092-8674(87)90539-3. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kirkman LA, Wellems TE. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proceedings of the National Academy of Sciences. U S A. 1996;93:1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proceedings of the National Academy of Sciences. U S A. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


