Abstract
A classic problem in analytical chemistry has been determination of individual components in a mixture without availability of the pure individual components. Measurement of the distribution of isotopomers in a labeled compound or mixture of labeled compounds is an example of this problem that is commonly encountered when stable isotopically labeled metabolites are used to determine in vivo kinetics and metabolism. We present a method that uses the measured mass spectral data of the unlabeled material to represent any and all combinations of isotopomer variations of that material and to determine abundances of these isotopomers. Although examples of the method are presented for gas chromatography-mass spectrometry, the method is applicable to any type of mass spectrometry data. The method also accounts for errors induced by mass spectrometer ionization and resolution effects. To demonstrate this method, we determined the isotopomer distributions of samples of 13C-labeled leucine and glucose for both highly enriched isotopomers and labeled isotopomers present in low abundance against a natural isotopic abundance background. The method accurately and precisely determined isotopomer identity and abundance in the labeled materials without adding noise or error that was not inherent in the original mass spectral data. In examples shown here isotopomer uncertainties were calculated with relative standard errors of <1% from good quality mass spectral data.
INTRODUCTION
A challenge in biology is to measure the production, disposal, transformation and flows of metabolites in living cells, tissues and whole animals, including humans. To measure these metabolic kinetic rates, isotopically labeled molecules are commonly used to follow movement of metabolites in vivo against a background of existing endogenous metabolites in cells, tissues and/or the body. Stable isotopically labeled molecules are identical in structure to the naturally occurring species, allowing the body to process the tracers similarly to existing endogenous metabolites. The difference between the tracer and the naturally occurring species is the increase in molecular weight with isotopic substitution, e.g. from substitution of 13C or 2H for 12C or 1H, and this difference can be measured by mass spectrometry.
The challenge for those who use stable isotopically labeled tracers has been determining the specific enrichment or pattern of labeling in the labeled tracer compound prior to and after administration into cells, tissues, or whole animals. The second challenge has been determining the isotopic pattern and enrichment of the labeled tracer after it has been metabolically transformed into other metabolites in vivo. The third challenge is that multiple combinations of isotopes of same compound are often administered or generated in vivo requiring the need to distinguish different isotopomers (isotopic combinations such as 13C1-, 13C2-, … 13C6-glucose). Because accurate and precise tracer enrichment measurements are needed to calculate kinetic rates of metabolic processes, approaches have been developed for measuring tracer isotopomer distributions, but there is little consensus or agreement about how the measurements should be made.
Biemann1 first proposed a simple calculation scheme to determine isotopic enrichments in compounds from the measured mass spectrum based upon a stepwise calculation of the differences of the measured isotope abundances of a fragment ion against the expected abundances based upon the natural isotopic abundances of the different elements in the fragment. Although conceptually simple, this stepwise approach propagated errors from the calculation of the first isotopic abundance into calculation of subsequent isotopic ions. An alternative and simpler approach specific for metabolic studies using tracers was to develop a standard curve for the measurement of the observed mass spectrometric isotope ratio against the expected tracer to unlabeled material for the tracer being used.2 This approach requires a calibration curve be developed for each isotopically labeled compound to be measured and does not allow for measurement of tracers that are metabolically created in vivo rather than are administered exogenously.
Other methods that have been developed require only measurement of the natural material and the labeled material to calculate the isotopic content of the isotopomers in a sample. These approaches are general in that a priori assumptions about a specific labeling pattern are not required before the calculations are begun. Rosenblatt et al. developed a method using a system of linear equations to transform raw gas chromatography-mass spectrometry (GCMS) data, consisting of relative abundance ratios of the natural species and the labeled sample, into the molar ratio of the tracer versus unlabeled species.3 Although this method does not require standards of the measured material, many of the correction factors that take into account isotopic impurity and ionization effects in the mass spectrometer are calculated theoretically and are not measured empirically.
Matrix mathematics have been used to solve a series of linear equations where different terms in the matrix correspond to different isotopomers to be determined. Lee developed a matrix approach for solving simultaneous equations of measured isotopic ratios by mass spectrometry for different possible isotopomer combinations.4–6 A similar approach was used by Lin et al.7 except that the natural abundance isotopic background was calculated based upon elemental abundances, rather than measured experimentally for the compound being studied. Unfortunately, theoretical elemental isotopic abundances do not take into account instrumental noise or differential ionization effects that occur in the production of the actual measured ions and their abundances.
Fernandez et al.8 used measured natural abundance ion intensity measurements to adjust for instrumental offsets in the measured isotope ratios. The derived spectrum of the unlabeled material was compared to the measured spectrum of the unlabeled compound. Once the derived spectrum closely matched the measured spectrum by adjusting different parameters in the method, the derived isotopic distributions of the unlabeled and labeled species were used to determine the isotopic distribution of the sample. Finally, Vogt et al.9 defined a series of equations for different isotopomer combinations where the natural abundance background had already been subtracted from the measured species, reducing the need for accounting for natural abundances.
We have reflected on the various methods described above that have been published for determination of isotopomers in labeled compounds and have found them lacking due to either specificity of the method to a particular tracer application in physiology or metabolism, limited ability to account for instrumental and chemical noise that accompanies all measurements, or complicated sets of assumptions and algorithms requiring proprietary computer programs to implement the methods. We have developed a general model applicable to all types of mass spectrometry from GCMS to electrospray ionization liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization time of flight mass spectrometry. We also wanted the method to account for instrumental and chemical noise and to accommodate any isotopic pattern of natural abundance isotopes. Our development of such a method has centered around the paper published by Brauman10 in 1966 that provided a simple matrix method using least squares reduction of error to determine isotopic distributions above natural abundance. The method is simple in that it uses the measured spectrum of the natural abundance material from which isotopomer distributions are calculated in labeled materials. The method does not rely on theoretically derived spectra. We have developed this method to measure isotopic and isotopomer distributions above natural abundance in a range of applications. The method accommodates ionization, instrumental, and chemical biases. Further the method allows the ability to measure more isotopic peaks than number of isotopomers determined (i.e. have more equations than unknowns) and to account for complicated isotopomer distributions involving multiple substitutions of different isotopes.
EXPERIMENTAL SECTION
Definitions of terms and key equations of the method
The term isotopomer refers to molecules that have the same structure but differ by the presence of a combination of one or more isotopically labeled atoms. For example, the different leucine isotopomers, unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine, [1,2-13C2, 15N]leucine and [1-13C, 5,5,5-2H3]leucine differ by mass and are discernable by mass spectrometry. The problem is that there will be natural abundance isotopes associated with each isotopomer that will produce overlapping ions in the mass spectrum any mixture of leucine isotopomers.
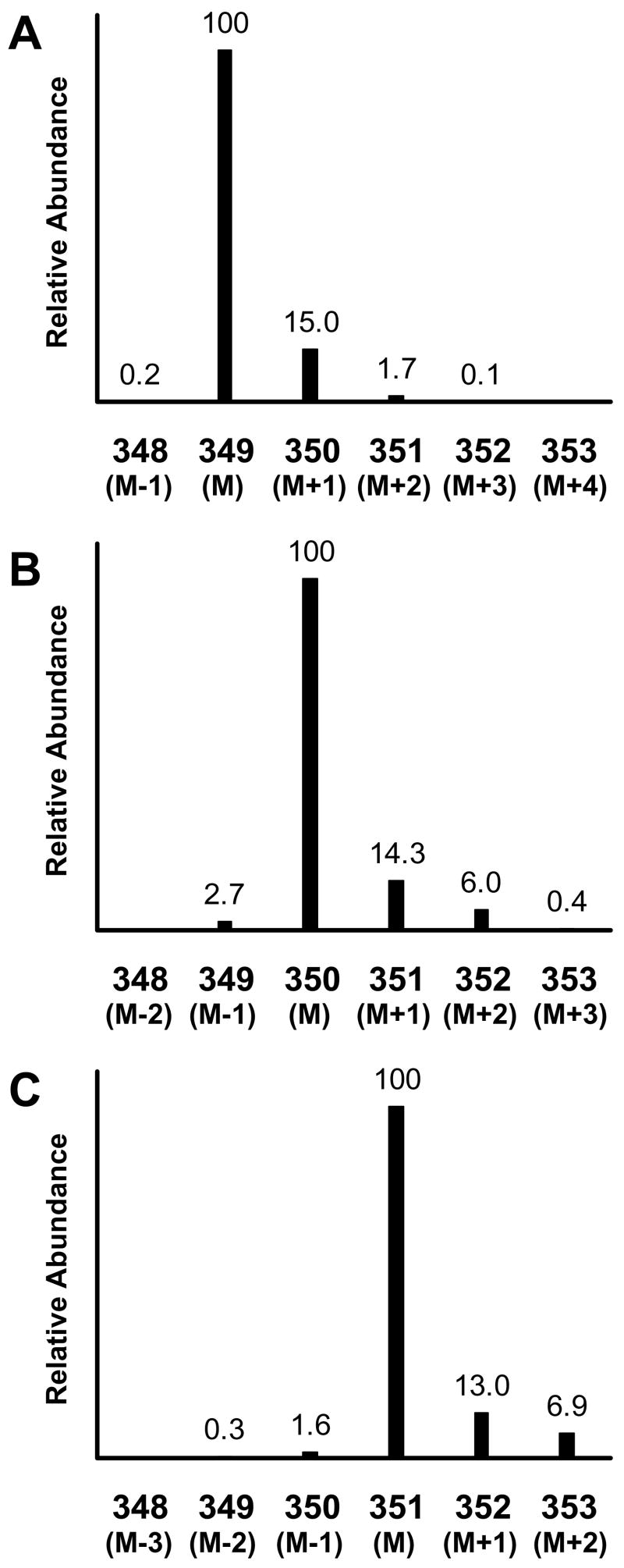
The mass spectrum around the molecular ion is shown for the N-heptafluorobutyryl, n-propyl derivative (HFBP) of unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine in Figure 1. These leucine isotopomers have isotopic distributions of ions of increasing mass from their most abundant mass, the base mass M. The lesser abundant higher mass ions exist due to the natural occurrence of the less abundant stable isotopes, e.g. 13C, 15N, 2H, and 18O. The natural abundance contributions at higher mass also occur in the labeled isotopomers. For example, unlabeled HFBP-leucine produces a strong [M-HF]− ion at m/z = 349 and has minor isotopes at m/z = 350 and 351.11 The base mass of HFBP [1-13C]leucine is at m/z = 350 with minor isotopes at m/z = 351 and 352, and the base mass of [1,2-13C2]leucine is at m/z = 351 with minor isotopes at m/z = 352 and 353. The overlap of natural abundance isotopes between the different labeled isotopomers in Figure 1 complicates determining the amount of each isotopomer in a mixture.
Figure 1.
Mass spectra of the N-heptafluorobutyryl, n-propyl derivative of unlabeled leucine (panel A), [1-13C]leucine (panel B), and [1,2-13C2]leucine (panel C) obtained by negative chemical ionization GCMS. The presented spectra are focused on the [M-HF]− ion and its naturally occurring isotope ions for each compound. Intensities are shown as relative abundance for each ion relative to the base mass M of each isotopomer. The numerical values for the x-axis are the measured m/z values. The figure represents data obtained from Experiment 1.
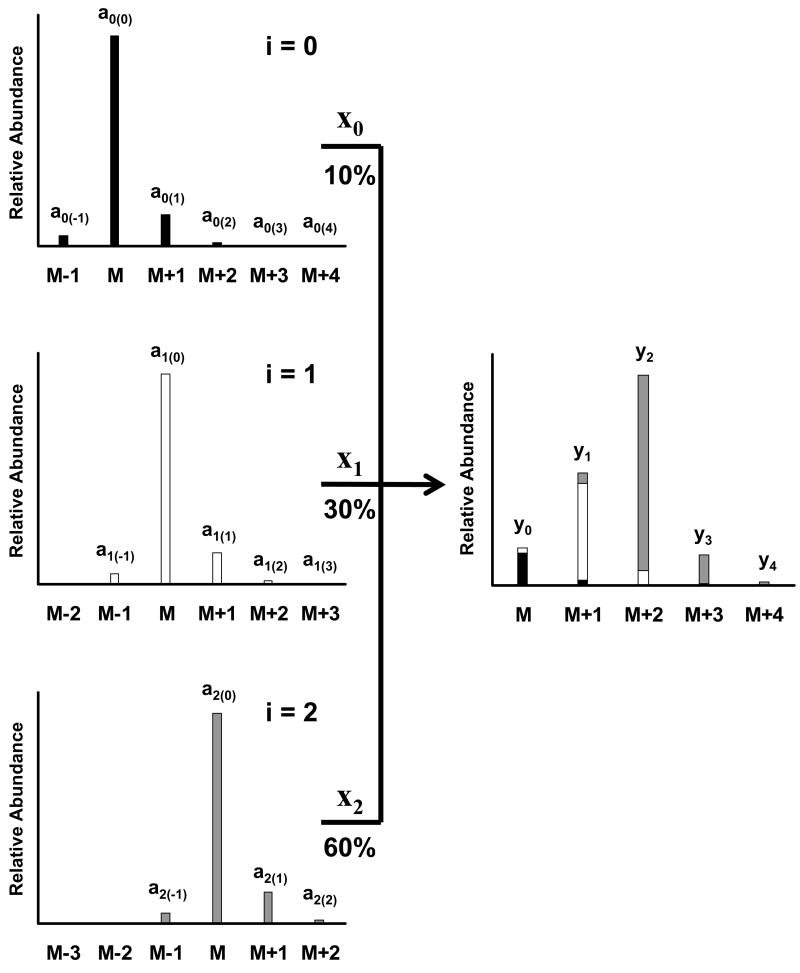
All mixtures are the sum of their components. Figure 2 shows how a mass spectrum of a mixture of unlabeled, singly, and doubly labeled isotopomers is the sum of the individual isotopic distributions for each isotopomer. We represent each isotopomer in Figure 2 by i, where the unlabeled isotopomer is i = 0. Isotopomers that increase the base mass of the compound by one will have an i = 1, and isotopomers that increase the base mass of the compound by two will have an i = 2. Each isotopomer, i, contributes to the mixture by its fractional abundance, xi.
Figure 2.
Theoretical representation of a mixture of three isotopomers of a single compound. The left side spectra show the individual spectra for the pure labeled isotopomers: unlabeled (top panel, solid bars), singly (middle panel, open bars), and doubly labeled (bottom panel, gray bars) isotopomers. Intensities are shown as relative abundance for each ion relative to the base mass M of each isotopomer’s spectrum. The right side panel shows the mixture of all three isotopomers. The contributing components of each isotopomer are shown by the different shading of the bars. The mixture is composed of 10% unlabeled material, 30% singly labeled isotopomer, and 60% doubly labeled isotopomer. Each isotopomer is designated by a value of i, where i = 0, 1, and 2 for the unlabeled, singly, and doubly labeled isotopomers, respectively. The relative abundance of each ion in each individual isotopomer’s spectrum is designated by ai(j), where j is the mass difference from the base mass, M, for each isotopomer, i. The value xi defines the fractional abundance for each isotopomer in the mixture on the right. The relative abundance of each ion in the mixture is represented by values of yj. For the mixture, M is indexed to the base mass of the unlabeled isotopomer.
Next, we need to define reference points in the mass spectrum. We will use the subscript j to represent the relationship of masses relative to the base mass (M) in a given isotopomer’s mass spectrum. The mass above M (i.e. M+1) has a value of j = 1, and the mass below M (i.e. M−1) has a value of j = −1. In the spectrum of the singly labeled isotopomer, the base mass, M, is one mass greater than the base mass for the unlabeled isotopomer, indicating i = 1. However, j is still defined as equal to 0 for the base mass of the singly labeled isotopomer.
We differentiate between the measured ions of the individual isotopomers and the ions of the mixture in Figure 2. We have defined the intensity of any ion of mass M+j for any isotopomer, i, as ai(j) where a refers to the relative intensity at mass M+j. The measured relative intensities of the ions in the mixture in Figure 2 (right side), y0, y1, y2, … yj correspond to the masses M, M+1, M+2, … M+j, respectively, where M is equal to the base mass of the unlabeled isotopomer. The intensity of any one yj ion is the sum of the intensities of the individual components (mass spectra on the left side of Figure 2) times each component’s fractional abundance represented by xi. The ai(j), yj, and xi terms are used in the following equations that define the method.
What is illustrated in Figure 2 can be defined as a set of linear equations relating the individual isotopomer mass spectral intensities (a terms) and the fractional abundances of the isotopomers (x terms) to the observed mass spectral intensities of the mixture (y terms). Equations 1–3 show this relationship for the first three ions of the mixture:
| (1-3) |
Equations 1–3 can be simplified into a matrix form:
| (4) |
where A is the matrix of ion intensities of the different isotopomers, x is the vector of the fractional abundances of each isotopomer in the mixture, and y is the vector of the mass spectral intensities of the mixture. The A matrix will have m rows and n columns, where m represents the number of measured masses and n represents the number of different isotopomers and is graphically demonstrated in Figure 2.
Note that we using a nonstandard presentation of array element subscripts. The order of the array subscripts are reversed (the first element, i, is the column indicator, not the row indicator), and we are using parentheses to denote the second element, j, to indicate the hierarchy of isotopomer, i, and its measured ions, j, in the form of ai(j) for individual elements of the A matrix.
We are interested in finding the fractional abundances, x. We can solve eq 4 for x because A is a square matrix:
| (5) |
However, the mass spectrometer can measure more masses than there are isotopomers in the mixture, for example, the y3 and y4 ions in Figure 2.
Let us rewrite eq 4 taking into account the y3 and y4 ions in Figure 2:
| (6) |
Because the A matrix above is not a square matrix, we cannot find the inverse of A directly. However, as Brauman pointed out in his method10, we can make a square matrix by multiplying by the transpose of A, denoted by AT:
| (7) |
The matrix ATA is a square matrix, and its inverse, (ATA)−1, can be found. Multiplying both sides by (ATA)−1 gives the solution for x:
| (8) |
Note that the above discussion assumes we can measure the mass spectra of the labeled isotopomers shown in the left-side spectra of Figure 2. This assumption poses a problem: we never have a pure labeled isotopomer available to measure. We may buy nearly labeled materials, but one application of this method is determining the extent of labeling of any labeled material, including highly labeled materials. Instead the method uses the mass spectrum measured for the unlabeled material (shown in the top left spectrum of Figure 2) as a substitute for the mass spectrum of a pure isotopomer’s mass spectrum. We make the assumption that the mass spectrum for a pure, labeled isotopomer will be identical to the mass spectrum of the unlabeled isotopomer, only increased in mass by a value of i. We can therefore substitute the measured abundances of the unlabeled isotopomer in eq 6 for the relative abundances of the mass spectral peaks of the labeled isotopomers. Doing so changes eq 6. For example, we can see from Figure 2 that a0(0) = a1(0) = a2(0) = … ai(0). The same is true for other values of j, such that ai(j) = a0(j). Thus, we can simplify eq 6 to:
| (9) |
Note that the negative values of j correspond to M−1 and M−2 in the unlabeled spectrum shifted upward by one and two masses in eq 9 at a0(−1) and a0(−2), respectively. Theoretically, the intensities for these masses should be zero. Practically, the ion source of the mass spectrometer may produce ions at M−1 or M−2, requiring our method to account for their existence. Correcting for the presence of an M−j species is easily carried out by measuring the M−j of the natural species and including the abundance of the M−j species in the A matrix for the isotopomers. It is unlikely for M−3 or lower fragments to occur, and these species can usually be neglected.
Another correction that needs to be considered deals with the addition of 13C labels to a molecule. When 13C labels are added to a molecule, the amount of natural abundance 13C decreases in the molecule, and the natural abundance contribution to the M+1 and M+2 isotopes decreases in the 13C labeled isotopomers. In general, substitution of any element with its corresponding stable isotope will cause a reduction in the natural abundance isotope peaks in the mass spectrum, but only 13C makes a significant enough contribution to the M+1 and M+2 ions to require a correction. We can limit our corrections to changes in the abundance of the M+1 and M+2 isotopes because changes in the higher mass isotopes (M+3, M+4 …) are not normally significant.
We can provide a simple example of the effect on the natural abundance isotope peaks when 13C’s are substituted into a molecule. Propane (C3H8) has a base mass at M = 44 Da. The M+1 mass is 45 and has a relative abundance of 3.4% with respect to M. The existence of the M+1 ion is primarily due to any one of the three carbon atoms in propane being a 13C of natural abundance 1.1%. If one of the carbons of propane were replaced by a 13C label, making the base mass of 13C1-propane 45, the relative abundance for M+1 at mass 46 is reduced to 2.3% because only two natural carbons can exist as a 13C. The (M+1)/M of the molecule will decrease to 1.2% as a second 13C is substituted, and (M+1)/M of the molecule will drop to nearly an immeasurable amount for 13C3-propane. The decreasing abundance of (M+1)/M as 13C labels are incorporated into a molecule can also be seen in Figure 1 for the different HFBP leucine isotopomers.
The effect on the M+1 ion by the addition of 13C requires a simple subtraction of the natural 13C contribution that is lost by the number of added 13C labels. The corrected (M+1)/M isotope ratio with the addition of nl 13C labels is:
| (10) |
where corr stands for the corrected (M+1)/M ratio of the 13C-containing isotopomer, obs stands for the observed or measured (M+1)/M ratio, and R13C is the ratio of 13C to 12C in nature (1.10%). Equation 10 is incorporated into eq 9 by adjusting the a0(1) value for each isotopomer having 13C labels:
| (11) |
The change in relative abundance of the (M+2)/M isotope with the addition of 13C labels is more complicated, but the change is also more minor. The abundance of the (M+2)/M isotope based on carbon can be calculated from the binomial distribution using the natural abundance of 13C:
| (12) |
where NC is the total number of carbon atoms in the molecule.12 The abundance of the (M+2)/M isotope ratio with nl 13C labels is calculated similar to eq 12:
| (13) |
The term (1-R13C)2 can be dropped because it is approximately equal to 1. The 13C natural abundance that is lost by the incorporation of 13C labels into the molecule will be the difference between eq 12 and 13. We can subtract this difference between eq 12 and 13 from the observed (M+2)/M to get the corrected (M+2)/M:
| (14) |
Equation 14 can be simplified to give the useful form of the (M+2)/M 13C correction:
| (15) |
The 13C correction for the (M+2)/M isotope can be carried out in our matrix equation by adjusting the a0(2) values for each isotopomer containing 13C labels:
| (16) |
The above 13C correction is carried out on the measured isotopic distribution of the unlabeled isotopomer’s M+1 and M+2 abundances using eq 11 and 16, respectively. The unlabeled isotopomer’s isotopic distribution with the 13C correction is used in the A matrix in eq 9:
| (17) |
where a′0(1) and a′0(2) are the 13C corrected isotopic abundances. This correction can be applied iteratively to multiple ions. For example, bromine produces two isotope contributions (79Br and 81Br) of nearly equal abundances that will produce two “M” ions, and adjustments need to be made to the M+1 and M+2 ions of each of these bromine isotope ions. After these adjustments have been made, eq 17 can now be used to find the fractional abundance, x, of the multiple 13C isotopomers. The solution for x is found as per eq 8.
For the experiments below, at least triplicates of each sample were made and analyzed by GCMS. The variance of the relative intensity for each mass in the sample was calculated and used to determine the standard error for the relative abundance of each isotopomer in the sample. The standard errors were calculated based on the statistical treatment of linear models described by Lewis et al. and Mandel.13,14
Materials
The reagents heptafluorobutyric anhydride and n-methyl-n-(t-butyldimethylsilyl)trifluoroacetamide were obtained from Regis (Morton Grove, IL). Unlabeled leucine, glucose, and butylboronic acid were purchased from Sigma-Aldrich (St. Louis, MO). [1-13C]Leucine and [1,2-13C2]leucine were purchased from Mass Trace (Woburn, MA) and Tracer Technologies (Somerville, MA), respectively. Uniformly labeled [U-13C6]glucose was obtained from MSD Isotopes (formerly from Montreal, Canada).
Overview of Experiments
Three experiments were carried out demonstrating the method. All samples were analyzed using either a Hewlett Packard 5973A GCMS (Palo Alto, CA) or a Hewlett Packard 5971A GCMS. The samples were injected into the GC with the injector at 260 °C in split mode. A 30-m, 1% phenyl methyl siloxane, 0.25-mm i.d., and 0.25-μm film thickness column (Phenomenex, Torrance, CA) was used. The analyses were run isothermally with a helium gas flow of 1 mL/min through the column. Different ionization techniques and chemical derivatives were used as outlined below. Data were collected by selected ion monitoring (SIM) of a range of masses around a fragment ion, M, for each species as stated below. The signal intensities of each mass were recorded throughout the chromatographic run; the ion current time-profiles for each mass were integrated; and the background-subtracted area ratios were expressed relative to the most abundant peak area. The area ratios were used to calculate the relative fractional abundance of each isotopomer present in the samples.
Experiment 1: abundance of isotopomers in samples of labeled [1-13C]leucine and [1,2-13C2]leucine
The isotopic content of a samples of [1-13C]leucine and [1,2-13C2]leucine was measured using two different derivatives and two different instrument methods. First, the N-heptafluorobutyryl, n-propyl (HFBP) leucine derivatives were prepared, as previously described,11 and the derivatized labeled materials and unlabeled leucine were measured by a Hewlett Packard 5973A GCMS using methane gas and negative chemical ionization (NCI). The GC was kept at 150 °C. The [M-HF]−ions m/z = 347–352 for natural leucine and m/z = 349–354 for the labeled leucines were monitored. A dwell of 10 ms for each ion was used for all three leucine samples.
We then determined the isotopic content of the same [1-13C]leucine and [1,2-13C2]leucine materials using the t-butyldimethylsilyl (TBDMS) derivatives, as previously described,15 and a Hewlett Packard 5971 GCMS with electron impact ionization (EI). The GC oven temperature was kept at 205 °C. The ions around the [M-57]+ ion were monitored (m/z = 301–306 for unlabeled leucine and m/z = 302–307 for the labeled isotopomers). A dwell of 10 ms for each ion was used.
Experiment 2: abundance of unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine in two standard mixtures
Two different mixtures of the three leucine materials used in Experiment 1 were quantitatively prepared and measured by GCMS to determine the fractional abundance of each isotopomer present in the mixtures. Known amounts of unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine were mixed together, and the TBDMS derivatives were made. Unlabeled TBDMS-leucine was also prepared separately. Samples were measured as described above in Experiment 1 for the TBDMS derivatives of leucine.
Experiment 3: abundance of different glucose isotopomers in a [U-13C]glucose sample
To demonstrate our method’s ability to measure complicated isotopomer distributions, glucose samples were analyzed using the bis-butylboronate acetyl derivative. The bis-butylboronate acetyl derivatives were prepared as previously described16 for unlabeled glucose and uniformly labeled [U-13C]glucose samples and analyzed by EI-GCMS isothermally at 220 °C. The ions around [M-57]+ were monitored: m/z = 293–300 and m/z = 297–303 for natural glucose and [U-13C]glucose samples, respectively.
RESULTS AND DISCUSSION
Determination of isotopic abundances in labeled [1-13C]leucine and [1,2-13C2]leucine
The normalized measured mass spectra intensities around the measured [M-HF]− ion for unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine samples as the HFBP derivatives are shown in Figure 1. Let us first consider the [1-13C]leucine sample and its isotopomer abundances. The [1-13C]leucine data (panel B, Figure 1) will be entered into the y vector, as shown in Table 1. The A matrix in Table 1 is constructed from the unlabeled, natural abundance leucine data (panel A, Figure 1). Once the A-matrix column entries have been set up, the isotopomer abundances of the [1-13C]leucine can be calculated (x vector of eq 17). Let us assume that we will solve for isotopomers ranging from an increase in mass from 0 (unlabeled) to 3. Therefore, we need four columns in the A matrix corresponding to isotopomers of i = 0, 1, 2, and 3. The i=0 column entry is simply the unlabeled leucine data (shown as panel A in Figure 1). The i=1 column entry will be the unlabeled leucine data shifted down one row and with the (M+1)/M and (M+2)/M entries reduced for the substitution of a 13C for one natural abundance carbon. This adjustment reduces the (M+1)/M entry for i=1 by 1.1%. The effect on the (M+2)/M entry is much smaller and nearly negligible. Because we know a priori that we have a [1-13C]leucine sample, we can assume that the i=1 isotopomer must be a 13C substitution, not for example a 2H or 15N substitution that would not require an adjustment to the (M+1)/M and (M+2)/M entries.
Table 1.
Unlabeled Leucine and [1-13C]Leucine Isotopic Abundance Values Used to Calculate the Abundance of Isotopomers in [1-13C]Leucine
| m/z | A matrix values | [1-13C]Leucine Isotopic Abundances (y) | Calculated Abundances (x,%) | Isotopomer | |||
|---|---|---|---|---|---|---|---|
| i = 0 | 1 | 2 | 3 | ||||
| 349 | 100 | 0.18 | 0.11 | 0 | 2.68 ±0.05 | x0 = 2.34 ±0.13 | Unlabeled Leucine |
| 350 | 14.98 | 100 | 0.18 | 0.11 | 100.00 ±0.06 | x1 = 93.20 ±0.14 | [1-13C]leucine |
| 351 | 1.71 | 13.88 | 100 | 0.18 | 14.27 ±0.03 | x2 = 0.36 ±0.08 | [18O]leucine |
| 352 | 0.14 | 1.57 | 14.98 | 100 | 6.01 ±0.02 | x3 = 4.10 ±0.04 | [1-13C,18O]leucine |
The measured unlabeled leucine intensities shown in Fig. 1 were used to construct the A matrix. The measured ion abundances (y vector) for the [1-13C]leucine material are shown with their respective standard deviations. The fractional abundances (xi) of the different isotopomers (i = 0, 1, 2, and 3 for unlabeled leucine, [1-13C]leucine, [18O]leucine, and [1-13C,18O]leucine, respectively) were calculated. Standard errors are given for each abundance value.
Although we have no reason to suspect that any di-labeled 13C is present in the [1-13C]leucine, we have included an i=2 isotopomer entry in the A matrix anyway. By the nature of the synthetic route used to produce the [1-13C]leucine we did know that labeled 13CO2 was used to incorporate 13C into the C1 position of leucine and that the starting 13CO2 had the possibility of containing some 18O in the CO2, i.e. 13CO18O. Thus, an i=2 isotopomer corresponding to [18O]leucine could occur. Because this isotopomer does not involve a 13C substitution, no reduction of the (M+1)/M abundance was needed in the i=2 column of the A matrix in Table 1. An i=3 isotopomer could occur by incorporation of both a 13C and 18O during synthesis to form an [1-13C,18O]leucine. Thus, an i=3 isotopomer was included in the A matrix shown in Table 1 and that column entry includes correction for one 13C substitution.
With the A matrix and y vector established, as shown in Table 1, we solved the system for the fractional abundances of each isotopomer in the [1-13C]leucine material (x vector) as per eq 17. The xi entries in Table 1 show the calculated isotopomer abundances with their respective standard errors. Most of the material was [1-13C]leucine (93.20 ±0.14%), as expected. There was a small amount of unlabeled leucine (2.34 ±0.13%) and a significant amount of triply labeled [1-13C,18O]leucine (4.10 ±0.04%) isotopomer. These results confirmed the presence of 18O in the labeled CO2 used to synthesize the labeled [1-13C]leucine.
Note that the total 13C content of the labeled [1-13C]leucine will be the sum of the [1-13C]leucine and [1-13C,18O]leucine species, i.e. 97.3%. The remainder is not labeled with respect to 13C (i.e. 2.7%). Similarly the 18O content is 0.36 + 4.10 = 4.46% of the leucine.
We can also define the isotopomer composition of the [1,2-13C2]leucine sample in the similar manner as used above for the [1-13C]leucine material. Figure 1 panel C shows the measured [1,2-13C2]leucine data that appears as the y vector in Table 2. The first column in the A matrix in column 1 in Table 2 is the unlabeled leucine data (i.e. i=0). Next are column entries into the A matrix of the isotopomers of i = 1, 2, 3, and 4 corresponding to the [1-13C]leucine, [1,2-13C2]leucine, [1-13C,18O]leucine, and [1,2-13C2,18O]leucine, respectively. The unlabeled leucine data have been shifted downward appropriately for the degree of substitution of label and have been adjusted for the substitution of 13C reducing (M+1)/M and (M+2)/M abundances. The calculated isotopomer abundances in the [1,2-13C2]leucine material correspond to the xi entries in Table 2.
Table 2.
Unlabeled Leucine and [1,2-13C2]Leucine Isotopic Abundance Values Used to Calculate the Abundance of Isotopomers in [1,2-13C2]Leucine
| m/z | A matrix values | [1-13C]Leucine Isotopic Abundances (y) | Calculated Abundances (x,%) | Isotopomer | ||||
|---|---|---|---|---|---|---|---|---|
| i = 0 | 1 | 2 | 3 | 4 | ||||
| 349 | 100 | 0.18 | 0.11 | 0 | 0 | 0.28 ±0.02 | x0 = 0.16 ±0.13 | Unlabeled Leucine |
| 350 | 14.98 | 100 | 0.18 | 0.11 | 0 | 1.57 ±0.03 | x1 = 1.28 ±0.02 | [1-13C]leucine |
| 351 | 1.71 | 13.88 | 100 | 0.18 | 0.11 | 100.00 ±0.04 | x2 = 93.29 ±0.22 | [1,2-13C2]leucine |
| 352 | 0.14 | 1.57 | 12.79 | 100 | 0.18 | 12.99 ±0.01 | x3 = 0.18 ±0.05 | [1-13C,18O]leucine |
| 353 | 0 | 0.14 | 1.44 | 13.88 | 100 | 6.91 ±0.03 | x4 = 5.09 ±0.13 | [1,2-13C2,18O]leucine |
The measured unlabeled leucine intensities shown in Fig. 1 were used to construct the A matrix. The measured ion abundances (y vector) for the [1,2-13C2]leucine material are shown with their respective standard deviations. The fractional abundances (xi) of the different isotopomers (i = 0, 1, 2, 3, and 4 for unlabeled leucine, [1-13C]leucine, [1,2-13C2]leucine, [1-13C,18O]leucine, and [1,2-13C2,18O]leucine, respectively) were calculated. Standard errors are given for each abundance value.
As shown in Table 2, most of the [1,2-13C2]leucine was this isotopomer (93.3 ±0.2%). There was a measurable amount of 18O labeled material: 5.09% [1,2-13C2, 18O]leucine. The di-labeled 13C content is the sum of the [1,2-13C2]leucine and [1,2-13C2,18O]leucine isotopomer abundances or 98.4%. The 13C content of the starting material used to label these two positions should be the square root of the di-labeled abundance; i.e. 99.2%. From this starting 13C enrichment, we can predict that there should be 2(99.2%)(100-99.2%) = 1.6% of the [1-13C]leucine isotopomer present in the [1,2-13C2]leucine. The measured abundance of the [1-13C]leucine after correcting for the presences of 18O was 1.3% ([1-13C]leucine) + 0.2% ([18O]leucine) = 1.5% – very similar to the predicted value of 1.6%. The total 18O content was 0.2 + 5.1 = 5.3%.
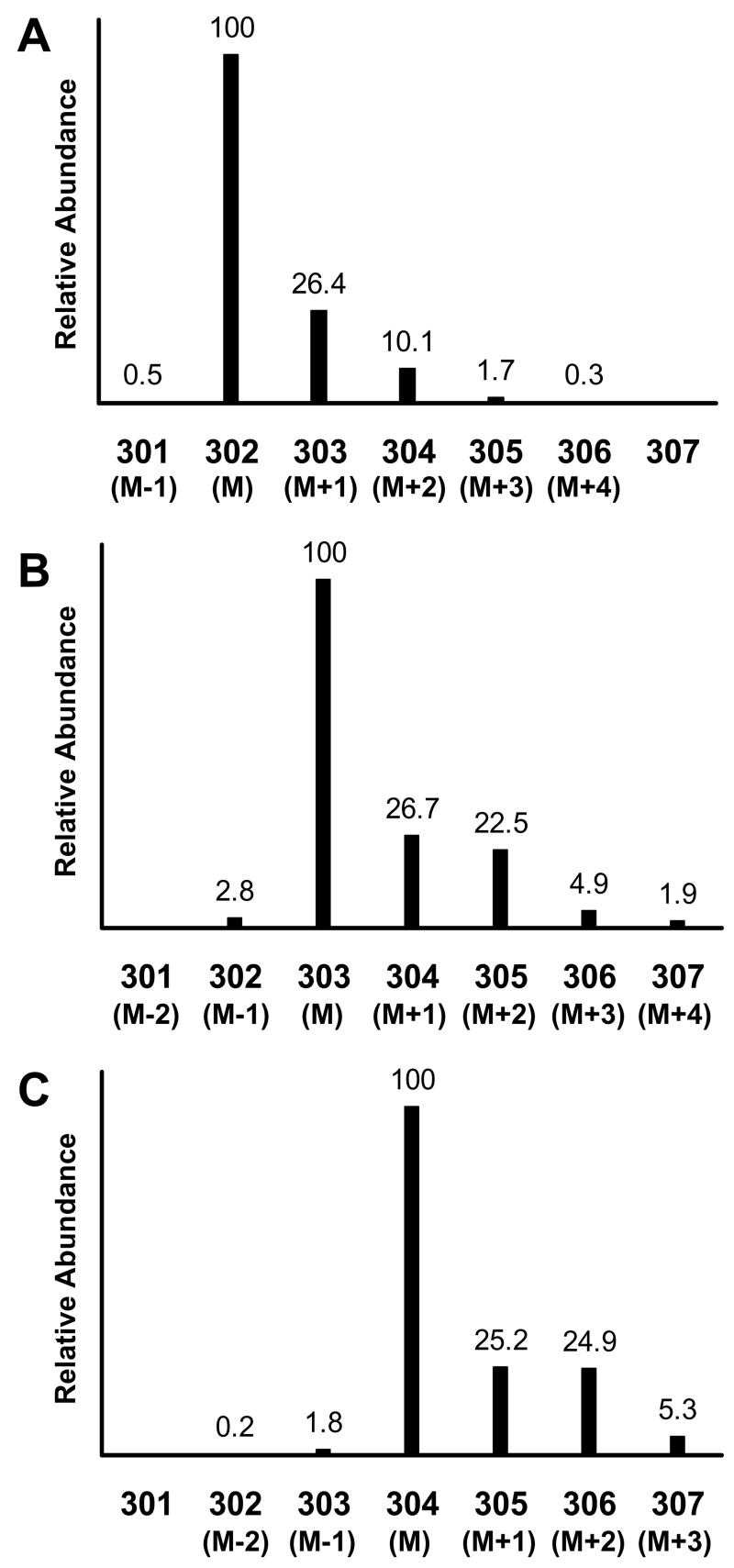
Next we measured these same two labeled leucine samples, but now using the TBDMS derivative. The measured, normalized mass spectra intensities around the [M-57]+ ion for the TBDMS derivatives of unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine materials are shown in Figure 3. The TBDMS silicon atoms contribute significantly to the M+1and M+2 abundances (28Si, 29Si, and 30Si abundances are 92.21%, 4.67%, and 3.10%, respectively). For the TBDMS-leucine molecule, two Si atoms are attached to each leucine molecule, and the contribution of silicon isotopes to the M+1 and M+2 abundances are approximately 9.3% and 6.2%, respectively, above the natural abundance contribution from the other atoms in the [M-57]+ fragment.
Figure 3.
Mass spectra of the t-butyldimethylsilyl derivative of unlabeled leucine (panel A), [1-13C]leucine (panel B), and [1,2-13C2]leucine (panel C) obtained by electron impact ionization GCMS. The presented spectra are focused on the [M-57]+ ion (defined as M for each isotopomer) and its naturally occurring isotope ions for each isotopomer. Intensities are shown as relative abundance for each ion relative to the base mass M of each isotopomer. The numerical values for the x-axis are the measured m/z values. The figure represents the data obtained from Experiment 1.
The calculated isotopomer abundances for TBDMS-[1-13C]leucine and TBDMS-[1,2-13C2]leucine samples are shown in Table 3. The total 13C content was 86.0% + 10.9% = 96.9% for TBDMS [1-13C]leucine (sum of the [1-13C]- and [1-13C,18O]leucine measured abundances) and was indistinguishable from the 97.3% 13C abundance measured for the HFBP [1-13C]leucine. Similarly, the total 13C content of the TBDMS [1,2-13C]leucine was 1.0% + 84.8% + 0.7% + 13.3% = 99.8% and was identical to the 99.8% 13C content measured using the HFBP derivative. However, the 18O content of the leucines measured as the TBDMS derivative (Table 3) was significantly higher compared to that measured as the HFBP derivative (Tables 1 and 2). The amount of 18O was 1.1% + 10.9% = 12.0% in the TBDMS [1-13C]leucine and 0.7% + 13.3% = 14.0% in the TBDMS [1,2-13C2]leucine materials.
Table 3.
Calculated Abundance Values for the Isotopomers of [1-13C]Leucine and [1,2-13C2]Leucine as the t-Butyldimethylsilyl Derivatives
| Isotopomer | [1-13C]Leucine Calculated Abundances (%) | [1,2-13C2]Leucine Calculated Abundances (%) |
|---|---|---|
| Unlabeled Leucine | 1.98 ±0.05 | 0.18 ±0.02 |
| [1-13C]Leucine | 86.03 ±0.12 | 1.04 ±0.05 |
| [18O]Leucine or [1,2-13C2]Leucine | 1.09 ±0.12 | 84.78 ±0.16 |
| [1-13C,18O]Leucine | 10.90 ±0.12 | 0.69 ±0.22 |
| [1,2-13C2,18O]Leucine | 13.31 ±0.24 |
The measured leucine intensities for unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine shown in Figure 3 were used in eq 17 to determine the isotopomer content of the labeled materials. The isotopomer content of [1-13C]leucine and [1,2-13C2]leucine materials are shown above for the TBDMS derivative. The standard error is given for each value listed.
The difference in 18O content between the TBDMS and HFBP derivatives of the [1-13C]leucine and [1,2-13C2]leucine can be explained by the derivatization chemistries used to make these different derivatives. When the TBDMS group is attached to the α-amino and hydroxyl moieties of the amino acid, no loss of oxygen occurs because the carboxyl-C is not involved in the reaction. In contrast, the first step in forming the HFBP derivative takes place by the acid catalyzed esterification of the carboxylic acid. In this reaction, there is a nucleophilic attack on the carboxyl-C atom and the hydroxyl group of leucine is lost when the propyl ester is formed. Therefore, we would expect the 18O content of the HFBP derivative to be no more than half of that observed for the TBDMS derivative. In reality we find that the 18O content is reduced in the HFBP derivative by about two thirds compared to the TBDMS derivative for both the [1-13C]leucine and [1,2-13C2]leucine materials. These results demonstrate the affect of derivative choice on the measured isotopic composition of a labeled compound. Our method was able to discern these small differences in different isotopomer combinations between the different derivatives.
Quantitative measurement of labeled leucine abundances
Two mixtures of labeled leucines added to unlabeled leucine were prepared to demonstrate our method’s ability to measure quantitative amounts of isotopomers, especially where the labeled material is in low abundance compared to the unlabeled material. The first mixture contained almost equal amounts of unlabeled leucine, [1-13C]leucine, and [1,2-13C2]leucine (referred to as the “high enrichment mixture”). The second mixture was about 96% unlabeled leucine and about 2% each of [1-13C]leucine and [1,2-13C2]leucine (the “low enrichment mixture”). The exact abundances of the expected isotopomers in the “low” and “high” enriched materials are shown in the two right-most columns of Table 4. The expected abundances of the different isotopomers in the mixtures were calculated based on the amounts of material taken and the isotopomer content found in Table 3 for the TBDMS derivatives of the [1-13C]leucine and [1,2-13C2]leucine materials.
Table 4.
Measurement of Two Different Isotopic Abundances from Two Mixtures of Leucine Isotopomers
| Isotopic Abundances (y) Enrichment Mixture | Calculated Abundances (x, %) Enrichment Mixture | Expected Abundances (%)Enrichment Mixture | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | A matrix values | Low | High | Low | High | Low | High | ||||
| i = 0 | 1 | 2 | 3 | 4 | |||||||
| 302 | 100 | 0.3 | 0 | 0 | 0 | 100.0 ±0.1 | 74.3 ±0.5 | x0 = 95.7 ±0.2 | 31.3 ±0.2 | 95.8 | 31.0 |
| 303 | 26.6 | 100 | 0.3 | 0 | 0 | 28.2 ±0.1 | 86.6 ±0.7 | x1 = 1.6 ±0.2 | 28.2 ±0.3 | 1.7 | 27.9 |
| 304 | 10.1 | 25.5 | 100 | 0.3 | 0 | 12.6 ±0.1 | 100.0 ±0.1 | x2 = 2.0 ±0.2 | 31.9 ±0.1 | 2.0 | 32.3 |
| 305 | 1.7 | 9.7 | 24.4 | 100 | 0.3 | 2.6 ±0.1 | 34.9 ±0.3 | x3 = 0.16 ±0.13 | 3.7 ±0.1 | 0.23 | 3.8 |
| 306 | 0 | 1.7 | 9.3 | 25.5 | 100 | 0.87 ±0.04 | 21.8 ±0.3 | x4 = 0.58 ±0.07 | 4.8 ±0.1 | 0.31 | 5.1 |
The measured unlabeled leucine fractional abundances were used to construct the A matrix. The measured isotopic abundances (y vector) are shown for the low and high enrichment mixtures with their respective standard deviations. The calculated isotopomer abundances for each mixture (x vector) for both the low and high enrichment mixtures are shown with the individual values (xi), where i = 0, 1, 2, 3, and 4 for unlabeled leucine, [1-13C]leucine, [1,2-13C2]leucine and [18O]leucine, [1-13C,18O]leucine, and [1,2-13C2,18O]leucine, respectively. Standard errors are given for each measured abundance value. The expected abundance values, based on the amount of material added to each mixture, are shown in the last two columns.
The isotopic abundance of unlabeled leucine was measured at the same time as the enrichment mixtures and was used to construct the A matrix in Table 4. The measured ion abundances for the low and high enrichment mixtures are shown as separate y vector columns in Table 4. The measured fractional abundances of the different isotopomers in the two mixtures are shown in the next two columns in Table 4. The measured fractional abundances compare very well to the expected fractional abundances for each mixture and isotopomer. The results in Table 4 demonstrate our method’s ability to determine accurately the enrichments of different isotopomers in a range of abundances. High enrichment mixtures are easily handled by our method. Small enrichment mixtures will be more sensitive to errors associated with the making of these mixtures and the limits of the instrumentation used to measure the mixture’s isotopic abundances. The low enrichment mixture results demonstrate our method’s ability to accurately and precisely measure multiple isotopomer abundances at low enrichments, as is commonly encountered in metabolic studies.
Abundance of different glucose isotopomers in a [U-13C]glucose sample
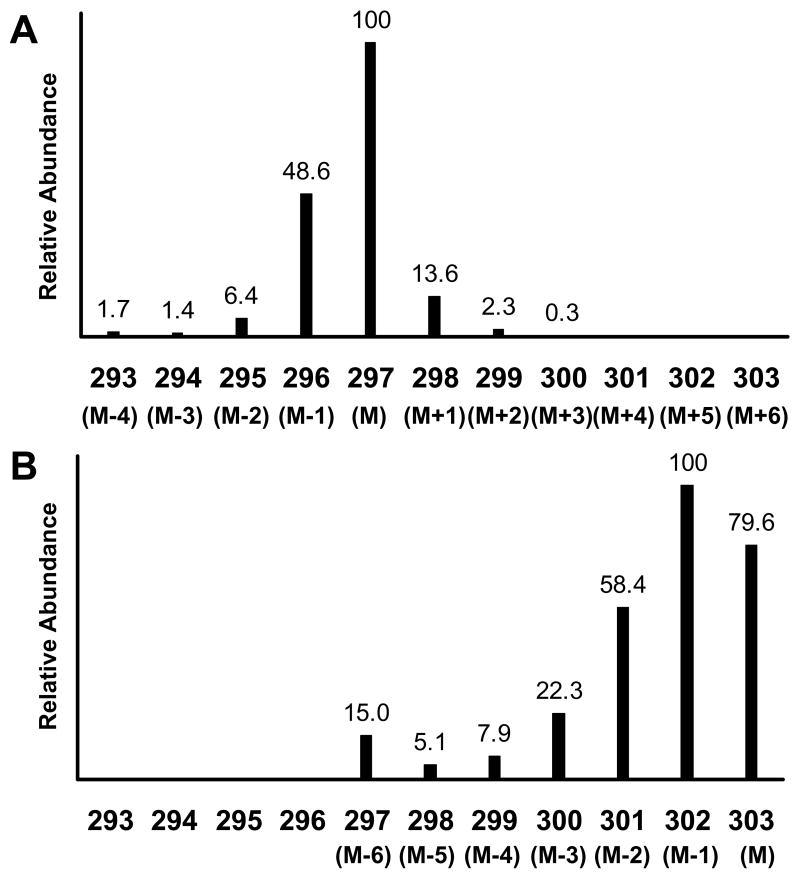
The measured normalized mass spectral intensities are shown in Figure 4 for unlabeled glucose and [U-13C]glucose samples derivatized as the bis-butylboronate acetyl derivative and measured as the [M-57]+ ion. Note in Figure 4 the large abundance of the M-1 species at m/z = 296 (49% relative intensity) in the unlabeled glucose spectrum. There are two boron atoms in the derivatized glucose molecule, and the m/z = 297 ion corresponds to the presence of two boron-11 isotopes, while the m/z = 296 ion contains a boron-10 and a boron-11 (natural boron isotope abundances are 10B = 24.8% and 11B = 75.2%). There is also a smaller M-2 ion from two 10B’s at m/z =295. Each of the ions containing boron also produces ions containing natural abundance 13C one and two masses above. Thus, the 13C correction to the (M+1)/M and (M+2)/M needs to be applied to both of the abundant ions containing boron, i.e., m/z = 296 and 297.
Figure 4.
Mass spectra of the bis-butylboronate acetyl derivative of unlabeled (panel A) and [U-13C]glucose (panel B) obtained by electron impact ionization GCMS. The presented spectra are focused on the [M-57]+ ion (defined as M for each isotopomer) and its naturally occurring isotope ions for each isotopomer. Intensities are shown as relative abundance for each ion relative to the base mass M of each isotopomer. The numerical values for the x-axis are the measured m/z values. The figure represents the data obtained from Experiment 3.
The data in Figure 4 appear in Table 5 as the y vector for the labeled glucose and as the A matrix. The possible isotopomers that were included in the A matrix were: unlabeled glucose and 13C1- to 13C6-glucose (i.e. i = 0–6). A 13C correction was applied in the A matrix to the (M+1)/M and (M+2)/M ions above both m/z = 296 and 297. The reduction in (M+1)/M ratio by 1.1% per 13C substitution can be seen in the A matrix. That is, the (M+1)/M ratio starts at 13.6% for the unlabeled isotopomer (i = 0) and decreases to 8.2% in the A matrix for the 13C5 isotopomer (i = 5). A similar correction was applied to the (M+2)/M ratio, but the magnitude of the change is much less (2.3% at i = 0 down to 1.9% at i = 4). The second to last column in Table 5 shows the individual entries of the calculated x vector, indicating the abundance of each isotopomer in the mixture with its corresponding standard error. There was a significant amount of unlabeled glucose (7.8%), as well as different 13C-labeled glucose isotopomers in the [U-13C6]glucose material. Only 41.4% of the glucose was the [U-13C]glucose isotopomer.
Table 5.
Measurement of [U-13C]Glucose and Calculation of Isotopomer Abundances
| m/z | A matrix values | [U-13C6]Glucose Isotopic | Calculated Abundances (x,%) | Isotopomer | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| i = 0 | 1 | 2 | 3 | 4 | 5 | 6 | ||||
| 297 | 100 | 48.88 | 6.43 | 1.39 | 1.76 | 0 | 0 | 14.99 ±0.43 | x0 = 7.79 ±0.18 | Unlabeled Glucose |
| 298 | 13.58 | 100 | 49.14 | 6.46 | 1.40 | 1.77 | 0 | 5.12 ±0.17 | x1 = 0.56 ±0.13 | 13C1-Glucose |
| 299 | 2.34 | 12.49 | 100 | 49.41 | 6.50 | 1.41 | 1.78 | 7.93 ±0.42 | x2 = 0.51 ±0.23 | 13C2-Glucose |
| 300 | 0.25 | 2.22 | 11.41 | 100 | 49.68 | 6.53 | 1.42 | 22.33 ±0.48 | x3 = 3.29 ±0.30 | 13C3-Glucose |
| 301 | 0 | 0.25 | 2.11 | 10.33 | 100 | 49.95 | 6.57 | 58.42 ±0.84 | x4 = 12.66 ±0.49 | 13C4-Glucose |
| 302 | 0 | 0 | 0.25 | 2.02 | 9.26 | 100 | 50.23 | 100.00 ±1.53 | x5 = 33.76 ±0.68 | 13C5-Glucose |
| 303 | 0 | 0 | 0 | 0.25 | 1.93 | 8.20 | 100 | 79.64 ±1.22 | x6 = 41.42 ±0.48 | 13C6-Glucose |
The measured unlabeled glucose isotopic abundances were used to construct the A matrix. The measured ion abundances (y vector) are shown for the [U-13C]glucose material with their respective standard deviations. The calculated isotopomer abundances (x vector) are shown for the different isotopomers, (xi). The last column indicates the different isotopomer abundances.
The [U-13C]glucose was prepared biosynthetically from yeast using a common 13C source. Assuming equal incorporation into all six carbon atoms, the incorporation of 13C into the glucose will follow a binomial distribution.12 We can use this distribution to calculate the 13C content of the starting material from the [U-13C]glucose measured isotopomer abundances. However, the high abundance of the unlabeled glucose implies that there are two a pools of glucose: (i) a residual unenriched pool and (ii) a pool of biosynthesized 13C-enriched glucose. To calculate the starting 13C enrichment of the [U-13C]glucose, we need to remove the contribution of residual unlabeled glucose and recalculate the 13C-glucose distribution. We have done so and show the recalculations in Table 6. The starting 13C content calculated from the [13C6]glucose isotopomer abundance is (45.2%)1/6 = 87.6%, based upon a binomial distribution. We then used this value to calculate the binomial distribution of the other isotopomers (13C1-13C5). These values are shown in the right-most column of Table 6. The adjusted isotopomer abundances shown in Table 6 follow the expected binomial distribution of isotopomer abundances based upon a common 13C source for glucose synthesis. These 13C labeled isotopomers make up 92.2% of the total glucose. The remainder (7.8%) is unlabeled glucose that probably was already present in preformed yeast before the biosynthetic process of 13C incorporation was begun. Our method can define isotopic distributions and distinguish between different sources of labeled isotopomers, even in the presence of significant contributions of naturally occurring isotopes, such as from boron.
Table 6.
Calculated Abundances of [U-13C]Glucose by Our Method and the Expected Abundances of [U-13C6]Glucose Based on a Predicted Binomial Distribution
| Glucose Isotopomer | Adjusted Abundance (%) | Expected Abundance (%) |
|---|---|---|
| 13C0-Glucose | 0 | 0.00 |
| 13C1-Glucose | 0 | 0.02 |
| 13C2-Glucose | 0.56 ± 0.24 | 0.27 |
| 13C3-Glucose | 3.59 ± 0.33 | 2.6 |
| 13C4-Glucose | 13.82 ± 0.53 | 13.6 |
| 13C5-Glucose | 36.84 ± 0.74 | 38.4 |
| 13C6-Glucose | 45.20 ± 0.53 | 45.2 |
The adjusted abundance removed the presence of 13C0-glucose from the isotopomer abundances shown in Table 5. The theoretical fractional abundance of the isotopomers based on the binomial distribution and a starting material 13C enrichment of 87.6% for the [U-13C]glucose material is shown in the second column.
The final point to note from this [U-13C]glucose example is the need to correct for natural abundance 13C changes with increasing 13C substitutions. At high levels of substitution (e.g. 13C5), the (M+1)/M ratio in the matrix is affected significantly. If we had not taken into consideration the correction of natural abundance 13C, the amount of 13C isotopomer defined would have been in error by about 1% times the number of 13C’s substituted minus one, i.e. ~5%. Thus, the 13C6 isotopomer abundance is changed from ~36% without a natural abundance 13C correction to the ~41% shown in Table 5.
Use of the method to define labeling of other isotopically labeled species
The examples given have focused on 13C labeling with the addition of 18O contamination. The method has been tested with a range of different possible labels including 2H, 15N and 18O, and with combinations of these labels (e.g. compounds containing both 13C and 15N). Although additional examples are not provided here, the method has been shown to be robust with any combination of isotopically labeling and be applicable to both GCMS and other methodologies, such as electrospray ionization liquid chromatography-mass spectrometry. The only caveat we have encountered is possible loss of 2H labeling in molecules highly substituted with 2H when M−1 and M−2 ions occur. In an unlabeled molecule, these ions arise from loss of one or two hydrogens. The concomitant M−1 hydrogen-loss from a highly deuterated molecule would be as a deuterium at M−2, not M−1. Under these conditions, the elements for M−1 and M−2 in the A matrix will not be correct. Because M−1 and M−2 ions are normally in relatively low abundance, the loss of a 2H from a 2H-labeled molecule by this mechanism is a relatively small correction to the calculation of 2H-labeling by the method. The reader is advised to work to minimize M−1 and M−2 ion production when highly deuterated species are being measured.
SUMMARY
We present a method that uses the measured isotopic abundance of the unlabeled material to calculate any possible combination of labeled isotopomers that could exist in a mixture of labeled species. The method is general and can be applied to any type of mass spectrometry data. It uses data as directly obtained from the instrument and uses the ion abundances measured for unlabeled material to compensate for instrumental biases as well as naturally occurring isotope biases. An example is the method’s ability to accept ions appearing below the expected fragment ion at M−1 or M−2 due to the loss of hydrogen or presence of odd isotopes (e.g. 10B). The method allows calculation of any combination of isotopomers and of significantly complicated patterns of isotopomers involving multiply labeled elements. The method can be adjusted to compensate for changes in natural abundance isotopes as labeled isotopes are substituted. The most dramatic effect is with inclusion of 13C. The method is particularly well suited for determining the isotopomer labeling pattern and abundances in highly labeled materials. The method is also capable of measuring the presence of small amounts of isotopomers above natural abundance background as is found in most applications of stable isotope tracers used to study in vivo metabolism and kinetics.
Acknowledgments
The authors thank Dr. Michael Haisch for his advice during the initial preparation of this manuscript. This work was supported in part by the National Institute of Health grants DK-38429 and RR-00109.
LITERATURE CITED
- 1.Biemann K. Mass Spectrometry: Organic Chemical Applications. McGraw-Hill; New York: 1962. [Google Scholar]
- 2.Tserng KY, Kalhan SC. Am J Physiol Endocrinol Metab. 1983;245:E308–E311. doi: 10.1152/ajpendo.1983.245.3.E308. [DOI] [PubMed] [Google Scholar]
- 3.Rosenblatt J, Chinkes D, Wolfe M, Wolfe RR. Am J Physiol Endocrinol Metab. 1992;263:E584–E596. [Google Scholar]
- 4.Lee WNP, Whiting JS, Fymat AL, Boettger HG. Biol Mass Spectrom. 1983;10:641–45. [Google Scholar]
- 5.Lee WNP. J Biol Chem. 1989;264:13002–04. [Google Scholar]
- 6.Lee WNP, Byerley LO, Bergner EA. Biol Mass Spectrom. 1991;20:451–58. doi: 10.1002/bms.1200200804. [DOI] [PubMed] [Google Scholar]
- 7.Lin YY, Cheng WB, Wright CE. Anal Biochem. 1993;209:267–73. doi: 10.1006/abio.1993.1118. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Vogt JA, Chapman TE, Wagner DA, Young VR, Burke JF. Biol Mass Spectrom. 1993;22:600–12. doi: 10.1002/bms.1200221008. [DOI] [PubMed] [Google Scholar]
- 10.Brauman JI. Anal Chem. 1966;38:607–10. [Google Scholar]
- 11.Matthews DE, Pesola G, Campbell RG. Am J Physiol Endocrinol Metab. 1990;258:E948–E956. doi: 10.1152/ajpendo.1990.258.6.E948. [DOI] [PubMed] [Google Scholar]
- 12.Pickup JF, McPherson K. Anal Chem. 1976;48:1885–90. [Google Scholar]
- 13.Lewis TO, Odell PL. Estimation in Linear Models. Prentice-Hall; Englewood Cliffs, NJ: 1971. [Google Scholar]
- 14.Mandel J. The Statistical Analysis of Experimental Data. Interscience; New York: 1964. [Google Scholar]
- 15.Mawhinney TP, Robinett RSR, Atalay A, Madson MA. J Chromatogr. 1986;358:231–42. doi: 10.1016/s0021-9673(01)90333-4. [DOI] [PubMed] [Google Scholar]
- 16.Wiecko J, Sherman WR. J Am Chem Soc. 1976;98:7631–37. [Google Scholar]