Abstract
RIC-3 is a transmembrane protein which acts as a molecular chaperone of nicotinic acetylcholine receptors (nAChRs). For some nAChR subtypes (such as homomeric α7 neuronal nAChRs), RIC-3 is required for efficient receptor folding, assembly and functional expression. In contrast, for other nAChR subtypes (such as heteromeric α4β2 neuronal nAChRs) there have been reports that RIC-3 can both enhance and reduce levels of functional expression. There is also evidence that RIC-3 can modulate maturation of the closely related 5-hydroxytryptamine (5-HT) receptor (5-HT3R). As with heteromeric nAChRs, apparently contradictory results have been reported for the influence of RIC-3 on 5-HT3R maturation in different expression systems. Recent evidence indicates that these differences in RIC-3 chaperone activity may be influenced by the host cell, suggesting that other proteins may play an important role in modulating the effects of RIC-3 as a chaperone. RIC-3 was originally identified in the nematode Caenorhabditis elegans as the protein encoded by the gene ric-3 (resistance to inhibitors of cholinesterase) and has subsequently been cloned and characterized from mammalian and insect species. This review provides a brief history of RIC-3; from the identification of the ric-3 gene in C. elegans in 1995 to the more recent demonstration of its activity as a nAChR chaperone.
Keywords: nicotinic receptor, acetylcholine receptor, molecular chaperone, RIC-3
Introduction
Resistant to inhibitors of cholinesterase (RIC)-3 is a transmembrane protein, which exerts a dramatic influence upon the maturation (folding and assembly) of neuronal nicotinic acetylcholine receptors (nAChRs). Recent evidence suggests that RIC-3 acts by interaction with unassembled receptor subunits within the endoplasmic reticulum (ER), thereby facilitating subunit folding and receptor assembly (Lansdell et al., 2005). Unlike many chaperone proteins, RIC-3 appears to be highly specific in its chaperone activity. In addition to its effect on nAChRs, RIC-3 interacts with and modulates maturation of the closely related 5-hydroxytryptamine (5-HT) type 3 receptor (5-HT3R) (Halevi et al., 2003; Cheng et al., 2005). In contrast, RIC-3 appears to have a little or no effect upon other neurotransmitter-gated ion channels, including those activated by GABA and glutamate (Miller et al., 1996; Halevi et al., 2002; Halevi et al., 2003; Lansdell et al., 2005).
Nicotinic acetylcholine receptors are members of the Cys-loop family of neurotransmitter-gated ion channels, all of which are pentameric transmembrane receptors (Lester et al., 2004; Millar, 2006). Nicotinic receptors are assembled from a diverse collection of subunits (Le Novère et al., 2002; Millar, 2003). In vertebrate species, 17 subunits (α1–α10, β1–β4, γ, δ and ɛ) have been identified, five of which (α1, β1, γ, δ and ɛ) are expressed at the neuromuscular junction. The remaining subunits (α2–α10 and β1–β4) are commonly referred to as neuronal nAChR subunits and, with some exceptions (Sharma and Vijayaraghavan, 2002), are located within the central and peripheral nervous system (McGehee and Role, 1995; Dani and Bertrand, 2007). Each subunit has a complex membrane topology, comprising a large N-terminal extracellular domain and four α-helical transmembrane domains. Compared with several other transmembrane proteins, the assembly of ion channels such as nAChRs appears to be both a slow and inefficient process (Green and Millar, 1995). Individual subunits must adopt an appropriate transmembrane topology and undergo a series of critical post-translational modifications (Green and Millar, 1995). In addition to the requirement for subunits to fold into an appropriate conformation, they must also make appropriate subunit–subunit interactions to form fully assembled pentameric receptors. The early steps of receptor folding and assembly occur within the ER, an intracellular compartment containing several proteins required for efficient protein folding and post-translational modification (Green and Millar, 1995).
There is evidence that nAChR folding, assembly and trafficking are influenced by several chaperone-type proteins. In particular, the 14-3-3η protein (Jeanclos et al., 2001) and the ER-resident chaperone proteins, BiP and calnexin (Blount and Merlie, 1991; Paulson et al., 1991; Forsayeth et al., 1992; Gelman et al., 1995; Chang et al., 1997), have been shown to associate with and/or influence trafficking of nAChR subunits. Whereas 14-3-3η, BiP and calnexin interact with a diverse range of target proteins (Shaw, 2000; Kleizen and Braakman, 2004), it appears, as will be discussed below, that RIC-3 is a chaperone protein, which acts much more selectively (on nAChRs and the closely related 5-HT3R).
Identification of a role for RIC-3 in cholinergic signalling
The gene encoding RIC-3 (ric-3) was identified in 1995 as one of several genes in Caenorhabditis elegans, which, when mutated, confers resistance to inhibitors of acetylcholinesterase (Nguyen et al., 1995). The nematode C. elegans has been used extensively in the field of neurobiology, due, in large part, to its relatively simple but well-characterized nervous system (White et al., 1986; Chalfie and Jorgensen, 1998). Several recessive mutations were identified in C. elegans, which confer resistance to aldicarb and trichlorfon (Nguyen et al., 1995; Miller et al., 1996), both of which are pesticides which act by inhibiting the enzyme acetylcholinesterase. Mutations conferring this phenotype were identified in about 20 genes. Those genes that had not previously been assigned a name in C. elegans were designated ‘ric', a term derived from the phenotype ‘resistance to inhibitors of cholinesterase'. Several of the genes identified in these studies encode synaptic proteins, such as SNAP-25 (ric-4), synaptotagmin (snt-1), synaptojanin (unc-26) and syntaxin (unc-64), or proteins involved in cholinergic signalling, such as choline acetyltransferase (cha-1), vesicular acetylcholine transporter (unc-13) and the C. elegans nAChR subunit UNC-63 (unc-63). Most of the mutations conferring resistance to inhibitors of cholinesterase have been shown to also cause deficits in GABA-dependent processes (Miller et al., 1996), suggesting that they encode proteins involved in general, rather than cholinergic-specific, synaptic transmission. In contrast, mutations in ric-3 appear to cause a more specific cholinergic deficit (Miller et al., 1996).
Further genetic screening in C. elegans revealed that mutations within the ric-3 gene also confer resistance to levamisole (Miller et al., 1996), an antiparasitic (anthelmintic) drug, which acts on nAChRs (Lewis et al., 1987). Resistance to levamisole is a phenotype that is also displayed by C. elegans strains containing mutations in genes encoding the nAChR subunits LEV-1, UNC-29 and UNC-38 (Miller et al., 1996; Fleming et al., 1997), a finding that led to the suggestion that the protein encoded by ric-3 gene might act at a postsynaptic site (Halevi et al., 2002). Interestingly, one of the mutations that was identified as conferring resistance to inhibitors of cholinesterase is located within the C. elegans gene unc-63 (Nguyen et al., 1995), which has subsequently been shown to encode a levamisole-sensitive nAChR subunit (Culetto et al., 2004). Thus, these early genetic studies provide strong evidence implicating ric-3 as a gene involved in cholinergic signalling.
Identification of a role for RIC-3 in nAChR maturation
Evidence indicating that the protein encoded by ric-3 is required for the maturation of nAChRs was obtained by screening for suppressor mutations in C. elegans (Halevi et al., 2002). Previous work had identified a missense mutation, deg-3(u662), within the C. elegans nAChR subunit DEG-3, which caused neuronal degeneration and uncoordinated movements (Treinin and Chalfie, 1995). The deg-3(u662) mutation alters an amino-acid residue, which is located within the pore-forming transmembrane domain of DEG-3 and which appears to cause neuronal degeneration due to altered nAChR function (Halevi et al., 2002). It was assumed that screening for mutations that cause suppression of the deg-3(u662) phenotype might identify genes, which are required for nAChR function. Several suppressor mutations were identified within the deg-3 gene itself (Halevi et al., 2002; Yassin et al., 2002). Other suppressor mutations were identified within des-2 (Halevi et al., 2002), which encodes a nAChR subunit (DES-2) that co-assembles with DEG-3 (Treinin et al., 1998; Yassin et al., 2001). In addition to suppressor mutations located within deg-3 and des-2, several deg-3(u662)-suppressor mutations were identified within the ric-3 gene (Halevi et al., 2002).
Electrophysiological recordings from pharyngeal and body muscles of C. elegans revealed that mutations in ric-3 are associated with deficits in cholinergic transmission, whereas glutamate and GABA currents are unaffected (Miller et al., 1996; Halevi et al., 2002). More direct evidence for an effect on nAChRs came from immunohistochemical staining, which demonstrated that C. elegans strains containing mutations within ric-3 have an altered subcellular distribution of the nAChR subunit DEG-3. In particular, reduced DEG-3 staining is observed in cell processes, a finding that led to the conclusion that RIC-3 is required for correct nAChR maturation in C. elegans (Halevi et al., 2002). More recent immunohistochemical studies in C. elegans have demonstrated that mutations in ric-3 also cause a reduction in cell-surface expression of the nAChR LEV-1 subunit (Gottschalk and Schafer, 2006). Interestingly, a similar reduction in cell-surface LEV-1 subunit expression is observed in C. elegans strains containing a mutation within unc-38 (Gottschalk and Schafer, 2006). As unc-38 encodes a nAChR subunit (UNC-38) that co-assembles with LEV-1 (Fleming et al., 1997), it appears that mutations in either RIC-3 or in nAChR subunits that co-assemble with LEV-1 can result in a similar disruption of LEV-1 maturation and cell-surface expression.
Heterologous expression of RIC-3
A common problem that has been encountered with several nAChR subtypes is that of inefficient functional expression of recombinant receptors in artificial expression systems (Millar, 1999). For some nAChR subunits, such as α7, much greater success has been achieved by expression in Xenopus oocytes, rather than in cultured mammalian cell lines (Couturier et al., 1990; Cooper and Millar, 1997). Heterologous expression studies performed in Xenopus oocytes have demonstrated that co-expression of C. elegans RIC-3 causes enhanced levels of functional expression of nAChR subtypes, such as α7 (Halevi et al., 2002), and similar results have been reported with a human homologue of RIC-3 (Halevi et al., 2003). These findings strongly support the idea that RIC-3 is able to enhance nAChR maturation.
Perhaps, the most dramatic effect of RIC-3 is its ability to facilitate the folding and functional expression of nAChRs such as α7 in host cell types, which would otherwise fail to generate functional nAChRs (Castillo et al., 2005; Lansdell et al., 2005; Williams et al., 2005). It has been known for some time that the extent to which some nAChR subunits generate functional receptors in artificial expression systems is influenced dramatically by the host cell type (Cooper and Millar, 1997; Kassner and Berg, 1997; Rangwala et al., 1997). The α7 subunit, which efficiently forms functional homomeric nAChRs when expressed in Xenopus oocytes (Couturier et al., 1990) and in some cultured cell lines (Puchacz et al., 1994; Gopalakrishnan et al., 1995; Quik et al., 1996), fails to do so when expressed in many other cell lines (Cooper and Millar, 1997; Kassner and Berg, 1997; Rangwala et al., 1997). Reverse transcription-PCR studies have demonstrated a correlation between the expression of RIC-3 mRNA in cultured cell lines and the ability of cell lines either to express endogenous α7 nAChRs or to permit the functional expression of recombinant α7 nAChRs (Lansdell et al., 2005; Williams et al., 2005).
In the absence of RIC-3, little or no specific binding of nicotinic radioligands is detected when the α7 subunit is expressed in mammalian cell lines, which lack endogenous RIC-3 (such as simian COS cells and some human HEK293-derived cell lines). In contrast, co-expression of RIC-3 with α7 facilitates both high levels of specific [125I]α-bungarotoxin binding and the expression of functional α7 nAChRs (Castillo et al., 2005; Lansdell et al., 2005; Williams et al., 2005).
In contrast to homomeric α7 nAChRs, many other nAChR subtypes (such as α3β4 and α4β2) are able to generate functional nAChRs in a wide range of cultured cell lines (Whiting et al., 1991; Wong et al., 1995; Gopalakrishnan et al., 1996; Lewis et al., 1997), including those known to lack endogenous RIC-3 (Lansdell et al., 2005). There is, however, evidence that co-expression of RIC-3 can modulate levels of functional expression of heteromeric nAChR subtypes, such as α3β4 and α4β2 (Halevi et al., 2003; Castillo et al., 2005; Lansdell et al., 2005). Whereas the influence of RIC-3 upon α7 nAChRs is consistent in all published studies (causing either enhancement or facilitation of functional expression), the influence of RIC-3 upon heteromeric nAChRs is much less consistent between studies. Co-expression of RIC-3 with heteromeric nAChRs such as α3β4 and α4β2 in a human kidney cell line has been shown to enhance levels of specific radioligand binding and functional expression (Lansdell et al., 2005). In contrast, there are reports that co-expression of RIC-3 suppresses levels of functional expression of α3β4 and α4β2 nAChRs in Xenopus oocytes (Halevi et al., 2003). Similarly, there are apparent inconsistencies between the reported effects of co-expressed RIC-3 upon 5-HT3Rs (Halevi et al., 2003; Castillo et al., 2005; Cheng et al., 2005; Cheng et al., 2007). Although co-expression of RIC-3 with the 5-HT3A subunit in Xenopus oocytes causes an almost complete suppression of functional expression of 5-HT3Rs (Halevi et al., 2003; Castillo et al., 2005), RIC-3 is reported to cause enhanced functional expression of these receptors in a human kidney cell line (Cheng et al., 2005; Cheng et al., 2007).
It is possible that the apparent differences, which have been reported concerning the influence of RIC-3 upon 5-HT3Rs and heteromeric nAChRs, may be a consequence of the different expression systems employed. Recent data to support this conclusion have come from studies with a Drosophila RIC-3 homologue. Studies in which Drosophila and human RIC-3 constructs were expressed in both insect and mammalian cultured cell lines have provided evidence that the ability of RIC-3 to act as a chaperone of nAChRs is influenced by the nature of the host cell (Lansdell et al., 2008). Specifically, it was found that Drosophila RIC-3 enhances maturation of nAChRs (containing either mammalian or Drosophila nAChR subunits) more efficiently in a Drosophila cell line, whereas human RIC-3 does so more efficiently in a human cell line (Lansdell et al., 2008). A plausible explanation for these findings might be that the extent to which RIC-3 is able to modulate the folding and maturation of nAChRs is influenced by other host-cell-specific proteins. Such interactions with host cell factors might also cause RIC-3 to suppress, rather than enhance, maturation of some receptor subtypes, thereby explaining some of the apparently contradictory results with 5-HT3Rs and heteromeric nAChRs.
An interesting finding has been the observation that, in contrast to its effect on α7, co-expression of RIC-3 with several α7/5-HT3A subunit chimeras causes a dramatic reduction in the level of receptor cell-surface expression (Castillo et al., 2006; Gee et al., 2007). Such effects have been observed consistently in both oocytes and mammalian cell expression systems. Further studies of these chimeras may help to establish the mechanism by which RIC-3 modulates receptor maturation. All published studies with recombinant nAChRs have focused on RIC-3's effects on neuronal nAChRs, which raises the question of whether RIC-3 can also modulate the maturation of muscle-type nAChRs. Recent preliminary studies indicate that this is the case. Co-expression of human RIC-3 has been found to result in a 60% enhancement in levels of α-bungarotoxin to the human muscle-type nAChR in transfected HEK-293 cells (personal communication; Andrew G. Engel, Mayo Clinic, Rochester, USA).
Identification of protein domains in RIC-3
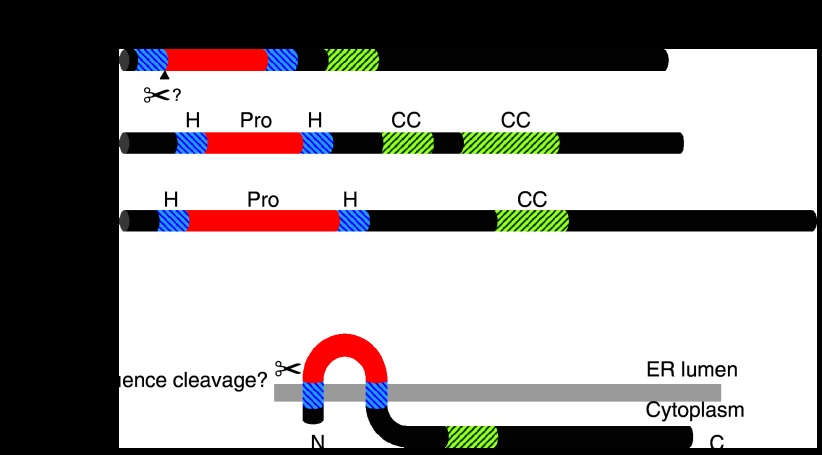
Sequence analysis of the predicted open-reading frame of the C. elegans ric-3 gene led to the proposal that the C. elegans RIC-3 protein is a membrane protein containing two transmembrane domains in which both the N- and C-terminus are located in the cytoplasm (Halevi et al., 2002) (Figure 1). The cloning of a human homologue of RIC-3 also revealed two hydrophobic domains (Halevi et al., 2003); however, analysis of the amino-acid sequence of human RIC-3 indicates that its N-terminal hydrophobic domain resembles a cleaveable signal sequence (Castillo et al., 2005; Cheng et al., 2007). Analysis of predicted RIC-3 proteins from other species suggests that RIC-3 proteins from other mammalian species contain potential cleavable signal sequences, whereas RIC-3 proteins from invertebrate species do not. For example, a RIC-3 homologue, which has been cloned from the model insect species Drosophila melanogaster, and which has been shown to have nAChR chaperone activity (Lansdell et al., 2008), resembles C. elegans RIC-3, contains two hydrophobic domains but lacks a predicted cleaved signal sequence. Further work will be required to establish whether these differences in predicted signal sequence cleavage sites reflect real differences in protein processing and transmembrane topology. Experimental evidence has been obtained which supports the presence of a cleavable signal sequence in the human RIC-3 protein (Cheng et al., 2007), but another recent study has obtained evidence which argues against proteolytic cleavage of human RIC-3 (Castelán et al., 2008).
Figure 1.
Protein domains and predicted membrane topology of RIC-3. (a) The location of hydrophopic domains (blue diagonal stripes), predicted coiled-coil domains (green diagonal stripes) and proline-rich domains (red) are shown for the RIC-3 proteins of C. elegans, D. melanogaster and human (approximately to scale). Although several alternatively spliced isoforms of RIC-3 have been reported in the literature, those illustrated here correspond to the 378 amino acid C. elegans RIC-3 isoform described by Halevi et al. (2002), accession number NM_068898; the 369 amino acid human RIC-3 isoform A (Halevi et al., 2003), accession number AY326435 and the 472 amino acid Drosophila RIC-3 isoform DmRIC-36,7,9 (Lansdell et al., 2008), accession number AM902271. The position of the predicted signal-sequence cleavage site in human RIC-3 is indicated by a scissors symbol and arrowhead. (b) The predicted transmembrane topology of RIC-3 is illustrated. Two hydrophobic regions are present in RIC-3 proteins from all species that have been examined. In all cases, it appears that the second of these hydrophobic regions is a transmembrane domain. The first of these hydrophobic regions is also predicted to be a transmembrane domain in RIC-3 proteins from invertebrate species, such as C. elegans and Drosophila. In contrast, it appears that the first hydrophobic region may be a cleaved N-terminal signal sequence in mammalian species, such as human, rat and mouse. Therefore, in invertebrate species RIC-3 is predicted to have both its N- and C-terminus located on the cytoplasmic side of the membrane. In contrast, if the postulated N-terminal signal sequence is cleaved (as is predicted in mammalian RIC-3 proteins), this would be expected to result in a single-transmembrane protein with only the C-terminus of the mature protein on the cytoplasmic side of the membrane. The position of the predicted signal-sequence cleavage site (in human RIC-3) is indicated by a scissors symbol. Predictions of protein secondary structure (such as signal sequence cleavage sites and coiled-coil domains) and of membrane topology were based on computer programs, such as COILS (Lupas et al., 1991; Lupas, 1996), Phobius (Käll et al., 2004) and PONGO (Amico et al., 2006). RIC, resistant to inhibitors of cholinesterase.
A consistent feature of RIC-3 from all species is the presence of one or more predicted coiled-coil domains located within the C-terminal region (Halevi et al., 2002; Halevi et al., 2003) (Figure 1), a motif that has been implicated in protein–protein interactions (Burkhard et al., 2001). The possible role of the coiled-coil domains in RIC-3 is unclear, although there is evidence to suggest that they are involved in aggregation of RIC-3 (Cheng et al., 2007). In contrast, there is evidence that they are not required for RIC-3's chaperone activity. Eleven alternatively spliced isoforms of Drosophila RIC-3 have been identified and characterized (Lansdell et al., 2008). Of these, those isoforms containing exon 7 contain a predicted coiled-coil domain, but those containing the alternative exon 7A lack a predicted coiled-coil motif. Despite this, no significant difference in chaperone activity has been detected between Drosophila RIC-3 isoforms containing either exon 7 or 7A (Lansdell et al., 2008). Studies with truncated RIC-3 proteins also indicates that these predicted coiled-coil domains are not essential for RIC-3 to function as a molecular chaperone (Ben-Ami et al., 2005; Cheng et al., 2007; Lansdell et al., 2008).
Another common feature of RIC-3 proteins is the presence of a proline-rich domain located between the two hydrophobic domains (Halevi et al., 2002; Halevi et al., 2003) (Figure 1). The importance of this region is unclear, but studies with alternatively spliced isoforms of Drosophila RIC-3 suggest that this region is critical to RIC-3's chaperone activity. Drosophila RIC-3 isoforms that either contain or lack a 40 amino-acid exon (exon 2), located within the proline rich domain, have been identified. Despite maintaining the open-reading frame, the presence of exon 2 (which is not rich in proline residues) dramatically reduces the nAChR-chaperone activity of Drosophila RIC-3 (Lansdell et al., 2008).
RIC-3 is a nAChR-associated protein
Evidence for an interaction between RIC-3 and the nAChR α7 subunit has been obtained by co-immunoprecipitation studies (Lansdell et al., 2005; Williams et al., 2005). Similarly, co-immunoprecipitation of RIC-3 has been demonstrated with other nAChR subunits, including α3, α4, β2 and β4 (Lansdell et al., 2005). In addition, RIC-3 has been shown to co-precipitate with 5-HT3A (Cheng et al., 2005), a subunit with close sequence similarity to nAChR subunits. In contrast, co-immunoprecipitation of RIC-3 has not been observed with subunits from other members of the Cys-loop ligand-gated ion channel family, such as the GABAA receptor α1 subunit (Lansdell et al., 2005).
Subcellular localization of RIC-3
In both vertebrate and invertebrate species, RIC-3 is expressed in muscle and nerve cells (Halevi et al., 2002; Halevi et al., 2003). RIC-3 protein is concentrated in cell bodies and appears to be located predominantly within the ER (Halevi et al., 2002; Castillo et al., 2005; Cheng et al., 2007). Immunoprecipitation studies have revealed that RIC-3 is able to associate with unassembled nAChR subunits (Lansdell et al., 2005), which are not thought to be exported from the ER to the cell surface (Green and Millar, 1995). Although there is strong evidence that RIC-3 is expressed within the ER, there is some debate about whether it is also transported to the cell surface. Studies conducted in a human kidney cell line have led to the conclusion that human RIC-3 is expressed on the cell surface (Williams et al., 2005), but other studies do not appear to support this conclusion. Experiments performed with a pH-sensitive variant of green fluorescent protein (PHluorin) have provided evidence that human RIC-3 is located predominantly within the ER, with no evidence of endosomal or cell-surface expression (Cheng et al., 2007). Similar conclusions have been made from studies using rat RIC-3 with a fluorescent protein tag, in which no evidence was obtained for expression of RIC-3 on the cell surface (Roncarati et al., 2006).
Mechanism of action
There are apparent similarities between the ability of RIC-3 to upregulate levels of cell-surface nAChRs and receptor upregulation caused by chronic exposure to nicotine. This effect of nicotine has been described as that of a ‘pharmacological chaperone' (Kuryatov et al., 2005; Sallette et al., 2005). It seems likely, however, that receptor upregulation caused by RIC-3 and by chronic exposure to nicotine may occur by different mechanisms. There is extensive evidence to indicate that chronic exposure to nicotine, as occurs during tobacco smoking, can result in an upregulation of brain nAChRs. Evidence for nicotine-induced upregulation of nAChRs in the brain has come from studies with post-mortem brain tissue from human tobacco smokers (Benwell et al., 1988) and from animal studies (Marks et al., 1985; Schwartz and Kellar, 1985). Nicotine-induced receptor upregulation occurs by a post-translational mechanism (Marks et al., 1992) and can be mimicked in cultured cell lines by exposure to low concentrations of nicotine for 24–48 h. Receptor upregulation has been observed in cultured cell lines both for α4β2 nAChRs (Peng et al., 1994; Zhang et al., 1994; Bencherif et al., 1995; Gopalakrishnan et al., 1996) and α7 nAChRs (Quik et al., 1996; Peng et al., 1997). With nAChR subtypes such as α4β2, there is evidence that chronic nicotine treatment can enhance subunit folding and induce conformational changes (Harkness and Millar, 2002). As the binding site for agonists, such as nicotine, is located at subunit interfaces (Arias, 2000; Celie et al., 2004) and is generated only after subunit co-assembly, it seems reasonable to assume that nicotine causes upregulation by an interaction with assembled or partially assembled nAChRs, rather than with unassembled subunits, a conclusion that is supported by experimental evidence (Harkness and Millar, 2002). Indeed, it has been proposed that nicotine causes receptor upregulation by acting as a pharmacological chaperone (Kuryatov et al., 2005; Sallette et al., 2005) and via a direct action at the agonist binding site (Kishi and Steinbach, 2006). In contrast, as discussed earlier, there is evidence that RIC-3 associates with unassembled nAChR subunits within the ER (Lansdell et al., 2005). As unassembled subunit do not form a nicotine-binding site, it is possible that the chaperone activity of RIC-3 is distinct from that of pharmacological chaperones, such as nicotine.
Conclusion
It is over a decade since the ric-3 mutant phenotype was first identified in C. elegans (Nguyen et al., 1995), but it is only relatively recently that the role of RIC-3 as a nAChR molecular chaperone been established (Halevi et al., 2002). In addition to studies in C. elegans, RIC-3 has now been cloned and characterized from mammalian and insect species (Halevi et al., 2003; Lansdell et al., 2008). The ability of RIC-3 to enhance the maturation of homomeric nAChRs, such as α7, is now well established. Of particular note is the ability of RIC-3 to facilitate the functional expression of α7 nAChRs in otherwise non-permissive cultured cell lines (Castillo et al., 2005; Lansdell et al., 2005; Williams et al., 2005). A considerable amount has been discovered about the role of RIC-3 in modulating the maturation of nAChRs, but several important questions are unresolved. Particularly intriguing are the apparently contradictory reports concerning the influence of RIC-3 upon heteromeric nAChR subtypes, such as α4β2 (Halevi et al., 2003; Lansdell et al., 2005). It is possible that these opposing effects (that is either enhancement or suppression of functional expression) are determined by the choice of expression system, but this remains to be confirmed and explained. It seems likely that there may be additional cell-specific proteins, which modulate the effect of RIC-3 (Lansdell et al., 2008), but these remain to be identified. Another possibility is that there are additional but currently unidentified proteins, which, themselves, have nAChR-chaperone activity. This possibility is suggested by the apparent inability of RIC-3 to resolve problems, which have been encountered in the heterologous expression of some nAChR subtypes. Of particular note are problems associated with functional expression in cultured cell lines of mammalian nAChRs subtypes such as α9α10 (Lansdell et al., 2005) and of some invertebrate nAChRs (Lansdell et al., 2008). Further work will also be required to establish more precisely the membrane topology and structure of RIC-3 and to obtain a better understanding of its mechanism of action as a molecular chaperone.
Acknowledgments
Work in the author's laboratory has received funding from the Biotechnology and Biological Sciences Research Council (UK) the Medical Research Council (UK), the Royal Society (UK) and the Wellcome Trust (UK). Additional financial support has been provided by Bayer CropScience (Germany), Bayer HealthCare (Germany), Eli Lilly (UK) and Syngenta (UK).
Glossary
- 5-HT
5-hydroxytryptamine
- ER
endoplasmic reticulum
- nAChR
nicotinic acetylcholine receptor
- RIC
resistant to inhibitors of cholinesterase
Footnotes
Conflict of interest
The author states no conflict of interest.
References
- Amico M, Finelli M, Rossi I, Zauli A, Elofsson A, Viklund H, et al. PONGO: a web server for multiple predictions of all-alpha transmembrane proteins. Nucl Acids Res. 2006;34:W169–W172. doi: 10.1093/nar/gkl208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias HR. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem Int. 2000;36:595–645. doi: 10.1016/s0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- Ben-Ami HC, Yassin L, Farah H, Michaeli A, Eshel M, Treinin M. RIC-3 affects properties and quantity of nicotinic acetylcholine receptors via a mechanism that does not require the coiled-coil domains. J Biol Chem. 2005;280:28053–28060. doi: 10.1074/jbc.M504369200. [DOI] [PubMed] [Google Scholar]
- Bencherif M, Fowler K, Lukas R, Lippiello PM. Mechanisms of up-regulation of neuronal nicotinic acetylcholine receptors in clonal cell lines and primary cultures of fetal rat brain. J Pharmacol Exp Ther. 1995;275:987–994. [PubMed] [Google Scholar]
- Benwell MEM, Balfour DJK, Anderson JM. Evidence that tobacco smoking increases the density of (−)-[3H]nicotine binding sites in human brain. J Neurochem. 1988;50:1243–1247. doi: 10.1111/j.1471-4159.1988.tb10600.x. [DOI] [PubMed] [Google Scholar]
- Blount P, Merlie JP. BIP associates with newly synthesized subunits of the mouse muscle nicotinic receptor. J Cell Biol. 1991;113:1125–1132. doi: 10.1083/jcb.113.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Castelán F, Castillo M, Mulet J, Sala S, Sala F, Domínguez del Toro E, et al. 2008Molecular characterization and localization of the RIC-3 protein, an effector of nicotinic acetylcholine receptor expression J Neurochemin press. [DOI] [PubMed]
- Castillo M, Mulet J, Gutiérrez LM, Ortiz JA, Castelán F, Gerber S, et al. Dual role of the RIC-3 protein in trafficking of serotonin and nicotinic acetylcholine receptors. J Biol Chem. 2005;280:27062–27068. doi: 10.1074/jbc.M503746200. [DOI] [PubMed] [Google Scholar]
- Castillo M, Mulet J, Gutiérrez LM, Ortiz JA, Castelán F, Gerber S, et al. Role of RIC-3 protein in trafficking of serotonin and nicotinic acetylcholine receptors. J Mol Neurosci. 2006;30:153–156. doi: 10.1385/JMN:30:1:153. [DOI] [PubMed] [Google Scholar]
- Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Jorgensen EM. C. elegans neuroscience: genetics to genome. Trends Genet. 1998;14:506–512. doi: 10.1016/s0168-9525(98)01623-0. [DOI] [PubMed] [Google Scholar]
- Chang W, Gelman MS, Prives JM. Calnexin-dependent enhancement of nicotinic acetylcholine receptor assembly and surface expression. J Biol Chem. 1997;272:28925–28932. doi: 10.1074/jbc.272.46.28925. [DOI] [PubMed] [Google Scholar]
- Cheng A, Bollan KA, Greenwood SM, Irving AJ, Connolly CN. Differential subcellular localization of RIC-3 isoforms and their role in determining 5-HT3 receptor composition. J Biol Chem. 2007;282:26158–26166. doi: 10.1074/jbc.M703899200. [DOI] [PubMed] [Google Scholar]
- Cheng A, McDonald NA, Connolly CN. Cell surface expression of 5-hydroxytryptamine type 3 receptors is promoted by RIC-3. J Biol Chem. 2005;280:22502–22507. doi: 10.1074/jbc.M414341200. [DOI] [PubMed] [Google Scholar]
- Cooper ST, Millar NS. Host cell-specific folding and assembly of the neuronal nicotinic acetylcholine receptor α7 subunit. J Neurochem. 1997;68:2140–2151. doi: 10.1046/j.1471-4159.1997.68052140.x. [DOI] [PubMed] [Google Scholar]
- Couturier S, Bertrand D, Matter JM, Hernandez MC, Bertrand S, Millar N, et al. A neuronal nicotinic acetylcholine receptor subunit (α7) is developmentally regulated and forms a homo-oligomeric channel blocked by α-BTX. Neuron. 1990;5:847–856. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- Culetto E, Baylis HA, Richmond JE, Jones AK, Fleming JT, Squire MD, et al. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor α subunit. J Biol Chem. 2004;279:42476–42483. doi: 10.1074/jbc.M404370200. [DOI] [PubMed] [Google Scholar]
- Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Ann Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- Fleming JT, Squire MD, Barnes TM, Tornoe C, Matsuda K, Ahnn J, et al. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsayeth JR, Gu Y, Hall ZW. BiP forms stable complexes with unassembled subunits of the acetylcholine receptor in transfected COS cells and in C2 muscle cells. J Cell Biol. 1992;117:841–847. doi: 10.1083/jcb.117.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee VJ, Kracun S, Cooper ST, Gibb AJ, Millar NS. Identification of domains influencing assembly and ion channel properties in α7 nicotinic receptor and 5-HT3 receptor subunit chimaeras. Br J Pharmacol. 2007;152:501–512. doi: 10.1038/sj.bjp.0707429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman MS, Chang W, Thomas DY, Bergeron JJM, Prives JM. Role of the endoplasmic reticulum chaperone calnexin in subunit folding and assembly of nicotinic acetylcholine receptors. J Biol Chem. 1995;270:15085–15092. doi: 10.1074/jbc.270.25.15085. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan M, Buisson B, Touma E, Giordano T, Campbell JE, Hu IC, et al. Stable expression and pharmacological properties of the human α7 nicotinic acetylcholine receptor. Eur J Pharmacol. 1995;290:237–246. doi: 10.1016/0922-4106(95)00083-6. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan M, Monteggia LM, Anderson DJ, Molinari EJ, Piattoni-Kaplan M, Donnelly-Roberts D, et al. Stable expression, pharmacologic properties and regulation of the human neuronal nicotinic acetylcholine α4β2 receptor. J Pharm Exp Ther. 1996;276:289–297. [PubMed] [Google Scholar]
- Gottschalk A, Schafer WR. Visualization of integral and peripheral cell surface proteins in live Caenorhabditis elegans. J Neurosci Methods. 2006;154:68–79. doi: 10.1016/j.jneumeth.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Green WN, Millar NS. Ion-channel assembly. Trends Neurosci. 1995;18:280–287. [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, et al. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevi S, Yassin L, Eshel M, Sala F, Sala S, Criado M, et al. Conservation within the RIC-3 gene family: effectors of mammalian nicotinic acetylcholine receptor expression. J Biol Chem. 2003;278:34411–34417. doi: 10.1074/jbc.M300170200. [DOI] [PubMed] [Google Scholar]
- Harkness PC, Millar NS. Changes in conformation and subcellular distribution of α4β2 nicotinic acetylcholine receptors revealed by chronic nicotine treatment and expression of subunit chimeras. J Neurosci. 2002;22:10172–10181. doi: 10.1523/JNEUROSCI.22-23-10172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14-3-3η interacts with the nicotinic acetylcholine receptor α4 subunit. J Biol Chem. 2001;276:28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kassner PD, Berg DK. Differences in the fate of neuronal acetylcholine receptor protein expressed in neurons and stably transfected cells. J Neurobiol. 1997;33:968–982. doi: 10.1002/(sici)1097-4695(199712)33:7<968::aid-neu8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Kishi M, Steinbach JH. Role of agonist binding site in up-regulation of neuronal nicotinic α4β2 receptors. Mol Pharmacol. 2006;70:2037–2044. doi: 10.1124/mol.106.029298. [DOI] [PubMed] [Google Scholar]
- Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Luo J, Cooper J, Lindstrom J. Nicotine acts as apharmacological chaperone to up-regulate human α4β2 acetylcholine receptors. Mol Pharmacol. 2005;68:1839–1851. doi: 10.1124/mol.105.012419. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Collins T, Yabe A, Gee VJ, Gibb AJ, Millar NS.2008Host-cell specific effects of the nicotinic acetylcholine receptor chaperone RIC-3 revealed by a comparison of human and Drosophila RIC-3 homologuesdoi: 10.1111/j.1471-4159.2008.05235.x [DOI] [PubMed]
- Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, et al. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol Pharmacol. 2005;68:1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- Le Novère N, Corringer P-J, Changeux J-P. The diversity of subunit composition in nAChRs: evolutionary origins, physiologic and pharmacologic consequences. J Neurobiol. 2002;53:447–456. doi: 10.1002/neu.10153. [DOI] [PubMed] [Google Scholar]
- Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys-loop receptors: new twists and turns. Trends Neurosci. 2004;27:329–336. doi: 10.1016/j.tins.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT, McLafferty S, Murphy H, Wu C. The levamisole receptor, a cholinergic receptor of the nematode Caenorhabditis elegans. Mol Pharmacol. 1987;31:185–193. [PubMed] [Google Scholar]
- Lewis TM, Harkness PC, Sivilotti LG, Colquhoun D, Millar NS. The ion channel properties of a rat recombinant neuronal nicotinic receptor are dependent on the host cell type. J Physiol. 1997;505:299–306. doi: 10.1111/j.1469-7793.1997.299bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Meth Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, Heinemann SF, et al. Nicotine binding and nicotine receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12:2765–2784. doi: 10.1523/JNEUROSCI.12-07-02765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MJ, Stitzel JA, Collins AC. Time course study of the effects of chronic nicotine infusion on drug response and brain function. J Pharmacol Exp Ther. 1985;235:619–628. [PubMed] [Google Scholar]
- McGehee DS, Role LW. Physiological diversity of nicotinic acetylcholine receptors expressed by vertebrate neurons. Annu Rev Physiol. 1995;57:521–546. doi: 10.1146/annurev.ph.57.030195.002513. [DOI] [PubMed] [Google Scholar]
- Millar NS. Heterologous expression of mammalian and insect neuronal nicotinic acetylcholine receptors in cultured cell lines. Biochem Soc Trans. 1999;27:944–950. doi: 10.1042/bst0270944. [DOI] [PubMed] [Google Scholar]
- Millar NS. Assembly and subunit diversity of nicotinic acetylcholine receptors. Biochem Soc Trans. 2003;31:869–874. doi: 10.1042/bst0310869. [DOI] [PubMed] [Google Scholar]
- Millar NS.2006Ligand-gated ion channels Encyclopedia of Life Sciences John Wiley and Sons Ltd: Chichester; , http://www.els.net/ [ doi: 10.1038/npg.els.0000154 [DOI] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, Rand JB. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Ross AF, Green WN, Claudio T. Analysis of early events in acetylcholine receptor assembly. J Cell Biol. 1991;113:1371–1384. doi: 10.1083/jcb.113.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gerzanich V, Anand R, Wang F, Lindstrom J. Chronic nicotine treatment up-regulates α3 and α7 acetylcholine receptor subtypes expressed by the human neuroblastoma cell line SH-SY5Y. Mol Pharmacol. 1997;51:776–784. doi: 10.1124/mol.51.5.776. [DOI] [PubMed] [Google Scholar]
- Peng X, Gerzanich V, Anand R, Whiting PJ, Lindstrom J. Nicotine-induced increase in neuronal nicotinic receptors results from a decrease in the rate of receptor turnover. Mol Pharmacol. 1994;46:523–530. [PubMed] [Google Scholar]
- Puchacz E, Buisson B, Bertrand D, Lukas RL. Functional expression of nicotinic acetylcholine receptors containing rat α7 subunits in human SH-SY5Y neuroblastoma cells. FEBS Lett. 1994;354:155–159. doi: 10.1016/0014-5793(94)01108-7. [DOI] [PubMed] [Google Scholar]
- Quik M, Choremis J, Komourian J, Lukas RJ, Puchacz E. Similarity between rat brain nicotinic α-bungarotoxin receptors and stably expressed α-bungarotoxin binding sites. J Neurochem. 1996;67:145–154. doi: 10.1046/j.1471-4159.1996.67010145.x. [DOI] [PubMed] [Google Scholar]
- Rangwala F, Drisdel RC, Rakhilin S, Ko E, Atluri P, Harkins AB, et al. Neuronal α-bungarotoxin receptors differ structurally from other nicotinic acetylcholine receptors. J Neurosci. 1997;17:8201–8212. doi: 10.1523/JNEUROSCI.17-21-08201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati R, Seredenina T, Kremer A, Caricasole A, Terstappen GC. Expression and subcellular localization of RIC-3, a putative alpha7-nAChR chaperone. FENS Abstr. 2006;3 A044.15:118. [Google Scholar]
- Sallette J, Pons S, Devillers-Thiery A, Soudant M, Prado de Carvalho L, Changeux J-P, et al. Nicotine upregulates its own receptors through enjanced intracellular maturation. Neuron. 2005;46:595–607. doi: 10.1016/j.neuron.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Schwartz RD, Kellar KJ. In vivo regulation of [3H]acetylcholine recognition sites in brain by nicotinic cholinergic drugs. J Neurochem. 1985;45:427–433. doi: 10.1111/j.1471-4159.1985.tb04005.x. [DOI] [PubMed] [Google Scholar]
- Sharma G, Vijayaraghavan S. Nicotinic receptor signalling in nonexcitable cells. J Neurobiol. 2002;53:524–534. doi: 10.1002/neu.10114. [DOI] [PubMed] [Google Scholar]
- Shaw A. The 14-3-3 proteins. Curr Biol. 2000;10:R400. doi: 10.1016/s0960-9822(00)00519-4. [DOI] [PubMed] [Google Scholar]
- Treinin M, Chalfie M. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Treinin M, Gillo B, Liebman L, Chalfie M. Two functionally dependent acetylcholine subunits are encoded in a single Caenorhabditis elegans operon. Proc Natl Acad Sci USA. 1998;95:15492–15495. doi: 10.1073/pnas.95.26.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Whiting PJ, Schoepfer R, Conroy WG, Gore MJ, Keyser KT, Shimasaki S, et al. Expression of nicotinic acetylcholine receptor subtypes in brain and retina. Brain Res Mol Brain Res. 1991;10:61–70. doi: 10.1016/0169-328x(91)90057-5. [DOI] [PubMed] [Google Scholar]
- Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, et al. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor α7 subunit in mammalian cells. J Biol Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- Wong ET, Holstad SG, Mennerick SJ, Hong SE, Zorumski CF, Isenberg KE. Pharmacological and physiological properties of a putative ganglionic nicotinic receptor α3β4, expressed in transfected eucaryotic cells. Mol Brain Res. 1995;28:101–109. doi: 10.1016/0169-328x(94)00189-l. [DOI] [PubMed] [Google Scholar]
- Yassin L, Gillo B, Kahan T, Halevi S, Eshel M, Treinin M. Characterization of the DEG-3/DES-2 receptor: a nicotinic acetylcholine receptor that mutates to cause neuronal degeneration. Mol Cell Neurosci. 2001;17:589–599. doi: 10.1006/mcne.2000.0944. [DOI] [PubMed] [Google Scholar]
- Yassin L, Samson AO, Halevi S, Eshel M, Treinin M. Mutations in the extracellular domain and in the membrane-spanning domains interfere with nicotinic acetylcholine receptor maturation. Biochem. 2002;41:12329–12335. doi: 10.1021/bi020193y. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gong Z-H, Hellstrom-Lindahl E, Nordberg A. Regulation of α4β2 nicotinic acetylcholine receptors in M10 cells following treatment with nicotinic agents. Neuroreport. 1994;6:313–317. doi: 10.1097/00001756-199501000-00022. [DOI] [PubMed] [Google Scholar]