Abstract
The cytochromes P450 (CYPs) comprise a vast superfamily of enzymes found in virtually all life forms. In mammals, xenobiotic metabolizing CYPs provide crucial protection from the effects of exposure to a wide variety of chemicals, including environmental toxins and therapeutic drugs. Ideally, the information on the possible metabolism by CYPs required during drug development would be obtained from crystal structures of all the CYPs of interest. For some years only crystal structures of distantly related bacterial CYPs were available and homology modelling techniques were used to bridge the gap and produce structural models of human CYPs, and thereby obtain useful functional information. A significant step forward in the reliability of these models came seven years ago with the first crystal structure of a mammalian CYP, rabbit CYP2C5, followed by the structures of six human enzymes, CYP1A2, CYP2A6, CYP2C8, CYP2C9, CYP2D6 and CYP3A4, and a second rabbit enzyme, CYP2B4. In this review we describe as a case study the evolution of a CYP2D6 model, leading to the validation of the model as an in silico tool for predicting binding and metabolism. This work has led directly to the successful design of CYP2D6 mutants with novel activity—including creating a testosterone hydroxylase, converting quinidine from inhibitor to substrate, creating a diclofenac hydroxylase and creating a dextromethorphan O-demethylase. Our modelling-derived hypothesis-driven integrated interdisciplinary studies have given key insight into the molecular determinants of CYP2D6 and other important drug metabolizing enzymes.
Keywords: comparative modelling, cytochromes P450, CYP2D6, docking, drug metabolism, homology modelling, hypothesis-driven studies
Introduction
The cytochromes P450 (CYPs) comprise a vast superfamily (>6000 known members (Nelson, 2007)) of haem-containing mono-oxygenase enzymes found in virtually all life forms. Members of this ubiquitous superfamily play an important role in the metabolism and biosynthesis of a wide range of exogenous and endogenous compounds (Nebert and Russell, 2002). In mammals, these enzymes are involved, among other things, in the metabolism of xenobiotic compounds—including environmental toxins and therapeutic drugs. One of the most interesting characteristics of the CYPs is their promiscuity. Individual isoforms are capable of interacting with a wide range of chemically diverse substrates, and some CYPs have overlapping substrate specificities. This promiscuity is useful in terms of defence of the organism against potentially harmful xenobiotics, but in some instances can lead to rapid drug clearance/inactivation, production of toxic compounds and/or adverse drug–drug interactions. The three-dimensional (3D) structure of a protein—particularly in complex with ligand(s) of interest—can provide valuable insight into its function. Therefore, it is desirable to have available structures of the drug-metabolizing CYPs. Co-crystallization of CYPs with known inhibitors and substrates can give insight into protein–ligand interactions in the active site, and from this allow the inference of likely metabolites, how modifications to the ligand and/or enzyme structure may potentially affect CYP–ligand binding, and if there is scope for adverse ligand–ligand interactions. All of this information provides a direct empirical means of assessing and predicting the potential fate of compounds. However, the structures of a substantial number of CYPs remain to be determined experimentally. In these cases, homology (comparative) modelling (Kirton et al., 2002a) can be used to give insight into the structure. The premise underpinning homology modelling arises from the observation that proteins with similar amino-acid sequences have a tendency to adopt similar 3D structures (Chothia and Lesk, 1986). Therefore, it is possible to predict the 3D structure of a protein based solely on knowledge of its amino-acid sequence and the 3D structures of proteins with similar sequences.
In humans, 90% of all of the drugs currently approved for clinical use are metabolized by one of seven CYP isoforms, CYP1A2, CYP2C9, CYP2C18, CYP2C19, CYP2D6, CYP2E1 and/or CYP3A4 (Tanaka, 1998; Guengerich, 2001; Nebert and Russell, 2002). Of these isoforms, CYP2D6 and CYP2C9 display polymorphisms that can result in the poor metabolism of drugs (Mahgoub et al., 1977; Kroemer and Eichelbaum, 1995; Sullivan-Klose et al., 1996; Aithal et al., 1999; Kidd et al., 1999, 2001; Takahashi and Echizen, 2001). Having knowledge of the structural features of the active sites of these seven isoforms in particular could lead to a tool that was able to predict whether or not a drug candidate would interact with the CYPs and, if so, which isoform the drug candidate may interact with preferentially. This would impact on the rational design of improved therapeutic drugs and target-specific inhibitors. It would also affect the risk assessment of xenobiotics and the avoidance of adverse drug–drug interactions, whereby one drug modulates the metabolism of another (Tanaka, 1998) by simple competition for the same active site, and/or by binding in an allosteric region of the same enzyme. Knowledge of the active site structure for these enzymes will significantly reduce the failure rate in clinical trials by identifying any CYP liabilities in the early stages of drug development, and reduce the amount of time and money required to bring a new pharmaceutical to the market.
Over the years, many homology models of the CYPs have appeared in the literature (de Groot et al., 2004). Until the year 2000, all structural models for the human CYPs were based on the X-ray crystal structures of distantly related bacterial CYP isoforms. In 2000, a major breakthrough was achieved with the determination of the structure of the first mammalian CYP, that of the rabbit enzyme CYP2C5 (Williams et al., 2000). This structure was more closely related to the human isoforms than the bacterial isoforms, and by incorporating the new structure into homology modelling studies, the quality and accuracy of the homology models of the human CYPs was vastly improved (Kirton et al., 2002b). Recently, the determination of X-ray crystal structures for several human isoforms important in drug metabolism (CYP1A2 (Sansen et al., 2007), CYP2A6 (Yano et al., 2005), CYP2C8 (Schoch et al., 2004), CYP2C9 (Williams et al., 2003; Wester et al., 2004), CYP2D6 (Rowland et al., 2006) and CYP3A4 (Williams et al., 2004; Yano et al., 2004)) has removed the need for homology models in some instances and has also, together with the availability of a second rabbit crystal structure (CYP2B4) (Scott et al., 2003, 2004), improved the quality of models for the other human isoforms.
We give an overview of the techniques used for modelling the 3D structures of human CYPs and CYP–drug interactions, presenting the development and validation of a model of CYP2D6, and comparison with the subsequently determined crystal structure of CYP2D6, as a case study.
CYP2D6
The CYP2D6 isoform plays a central role in drug metabolism in humans. It is responsible for the clearance of at least 20% of the compounds in current clinical use, including antiarrhythmics, antidepressants, antipsychotics, β-blockers and analgesics (Kroemer and Eichelbaum, 1995). This isoform is of particular interest to the pharmaceutical industry because it displays a genetic polymorphism, the consequence of which is large interindividual and ethnic differences in the drug metabolism mediated by CYP2D6. This is highlighted by the defect in man known as debrisoquine/sparteine polymorphism (Mahgoub et al., 1977; Eichelbaum et al., 1979). This polymorphism can arise from one of several genetic mutations and affects a significant proportion of the Caucasian population (Daly et al., 1995). It results in the defective metabolism of a number of clinical drugs, and inheritance of the ‘poor-metabolizer' phenotype has been linked with an increased susceptibility to Parkinson's disease and certain types of cancer (Eichelbaum et al., 1979; Smith et al., 1995). Consequently, many pharmaceutical companies are interested in designing drug candidates that are not metabolized by CYP2D6. Understanding the structure of CYP2D6 and potential protein–ligand interactions would aid in this rational design of potential drug candidates. Prior to the elucidation of the X-ray crystal structure, this was assisted by homology models.
Early models of CYP2D6 based on bacterial CYPs
A feature common to the vast majority of CYP2D6 substrates is the presence of a basic nitrogen atom and a planar aromatic ring. Since these features are also found in a large number of central nervous system and cardiovascular drugs (which act on G protein-coupled receptors), CYP2D6 has been a very widely studied isoform. Prior to the availability of crystal structures of mammalian CYPs, models of human CYPs were based on the structures of more distantly related bacterial CYPs—these share less than ∼25% sequence identity with CYP2D6. Based on the observation that sequence alignment becomes difficult in the ‘twilight zone' of less than 30% sequence identity (Rost, 1999), this led to the belief in some quarters that such homology models would not shed any useful light on the function of CYP2D6. However, this pessimism proved unjustified.
Many models of the active site of CYP2D6 postulated the involvement of a carboxylate group in the protein forming a salt bridge with this basic nitrogen (Meyer et al., 1986; Islam et al., 1991; Koymans et al., 1992; de Groot et al., 1996, 1999; Modi et al., 1996, 1997; Lewis et al., 1997; Lewis, 1999; Venhorst et al., 2003); this was proposed both by modelling and by mutagenesis (Mackman et al., 1996) to be Asp301, a residue in the I-helix (substrate recognition site (SRS4; Gotoh, 1992). The recently determined crystal structure of CYP2D6 (Rowland et al., 2006) has confirmed such a role for Asp301 in ligand binding (but see Discussion below).
Our early models (Modi et al., 1996, 1997) (produced using Modeller (Sali and Blundell, 1993); see Kirton et al., 2002a for more details) were also used in conjunction with sequence analysis to design a CYP2D6 mutant, F483I, based on the presence of an isoleucine in this position in testosterone-metabolizing CYP2D9, with novel specificity, able to metabolize testosterone (Smith et al., 1998). That Phe483 is in the binding site is confirmed by the crystal structure (Rowland et al., 2006). Mutagenesis data (Hayhurst et al., 2001b; Chowdry et al., 2002) suggested that, together with Phe483, Phe481 also has a detrimental impact on the oxidation of debrisoquine. While in our models (Modi et al., 1996, 1997) and in the crystal structure (Rowland et al., 2006) Phe481 is removed from the active site, constrained molecular dynamics simulations (based on the crystal structure) show that this loop can alter conformation so that both phenylalanines are in the active site (Rowland et al., 2006).
Thus, despite the low level of sequence homology between CYP2D6 and the bacterial CYP templates—and concomitant concern about the quality of the models—some useful information was accurately gleaned even from these early models.
Models of CYP2D6 based on mammalian CYPs
The determination of the first crystal structure of a mammalian CYP—rabbit CYP2C5 (Williams et al., 2000; Wester et al., 2003a, 2003b), which shares ∼40% sequence identity with CYP2D6—paved the way for more accurate CYP2D6 models. Our ‘second-generation' CYP2D6 model (Kirton et al., 2002b) (that is, including at least one mammalian CYP as a template) was generated using the rabbit CYP2C5 crystal structure as a template alongside the four bacterial templates P450cam (Poulos et al., 1986), P450BM-3 (Ravichandran et al., 1993), P450terp (Hasemann et al., 1994) and P450ery-F (Cupp-Vickery et al., 2000). To evaluate the predictive value of this CYP2D6 model, docking studies were carried out using the program GOLD (Jones et al., 1997) on known substrates of CYP2D6, including codeine (Kirton et al., 2002b), 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) (Kirton et al., 2002b), dextromethorphan (Flanagan et al., 2004) and spirosulphonamide (Kemp et al., 2004). In each case, the docking was consistent with the known site of metabolism (that is, the highest ranked docked solution positioned the known site of metabolism above the iron atom of the haem moiety).
Role of Glu216 and Asp301
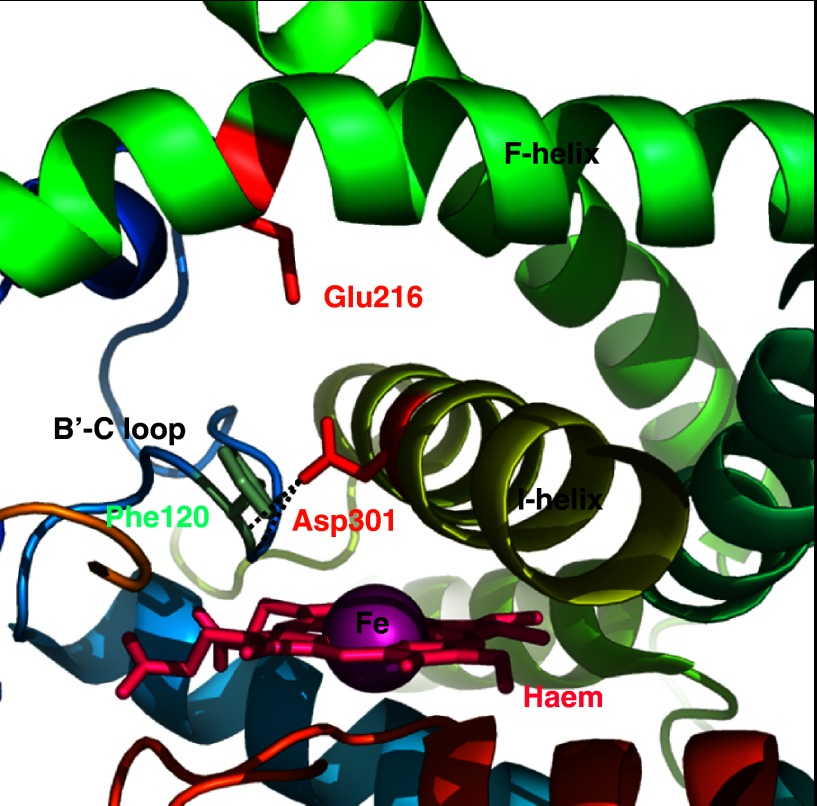
Codeine was observed to dock in the active site of the CYP2D6 model (Kirton et al., 2002b) in an orientation consistent with O-demethylation (Desmeules et al., 1991; Ladona et al., 1991; Kirton et al., 2002b). Surprisingly, the docking did not position the basic nitrogen atom of the substrate close to Asp301. Instead, the basic nitrogen was observed to interact with a second acidic residue in the active site, Glu216 (in the F-helix, SRS2); the proposed role of Glu216 was further supported by analysis of the active site using GRID (Goodford, 1985) probes. The model suggests that Asp301 is not involved directly in substrate binding but plays a structural role positioning the B-C loop (SRS1), including Phe120 (see below)—this hypothesis was subsequently verified when the crystal structure of CYP2D6 was determined (Figure 1; Rowland et al., 2006). The docking results for MPTP (Kirton et al., 2002b) and dextromethorphan (Flanagan et al., 2004) also positioned the basic nitrogen atoms of the substrates close to Glu216 and away from Asp301.
Figure 1.
Schematic representation of the crystal structure of CYP2D6 (Rowland et al., 2006), confirming the position of amino acids identified as key by model building. Hydrogen bonds identified by modelling between Asp301, and the main-chain amides of Val119 and Phe120 are denoted by dashed lines (Figure produced using Pymol; DeLano, 2002).
An independent study also suggests that Asp301 plays a structural role (Hanna et al., 2001), and an analysis of 431 CYP sequences (Kirton et al., 2002b) indicates that specificity for basic substrates requires acidic residues equivalent to both Asp301 and Glu216. The proposed key role of Glu216 was confirmed by experiment (Paine et al., 2003). Indeed, the mutation of Glu216 altered the specificity to such an extent that metabolism of testosterone was observed (Paine et al., 2003). Furthermore, the E216Q/D301N double mutant acts on ‘atypical' substrates (Paine et al., 2003), including anionic compounds such as diclofenac, ‘classic' substrates of CYP2C9. This suggests that the binding site of CYP2D6 is thus intrinsically rather promiscuous, with Glu216 and Asp301 favouring the binding of basic substrates and discriminating against acidic substrates.
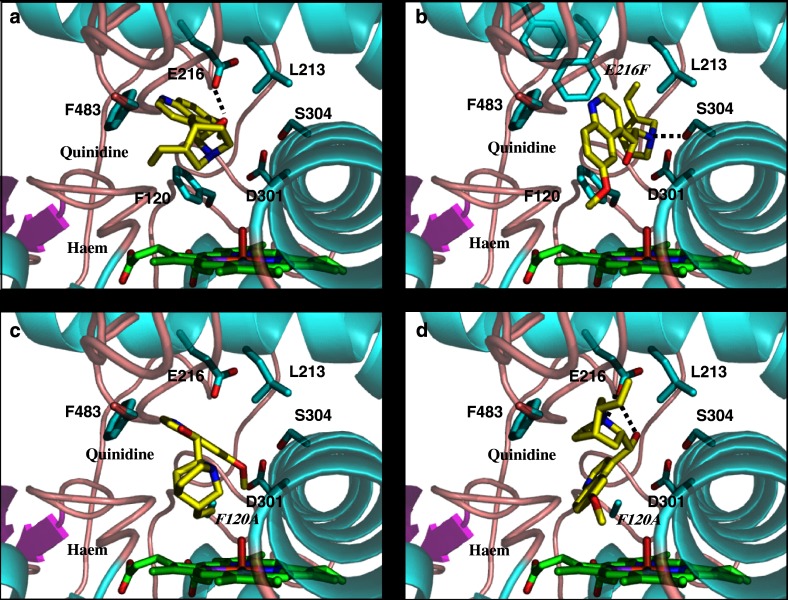
We have also shown that Glu216 and Asp301 play a key role in the action of quinidine as an inhibitor of CYP2D6 (McLaughlin et al., 2005). Quinidine is not metabolized by CYP2D6 and has long been established as a potent competitive inhibitor of this enzyme (von Bahr et al., 1985; Guengerich et al., 1986, 2002b; Otton et al., 1988; Branch et al., 2000). The fact that quinidine is an inhibitor rather than a substrate is intriguing, since it produces a classical type I binding spectrum with CYP2D6 (Hayhurst et al., 2001a) that is usually associated with the binding of substrate molecules (Schenkman et al., 1981). In addition, quinidine possesses a number of features normally associated with CYP2D6 substrates (Strobl et al., 1993). Studies of the relationship between structure and inhibitory activity for quinidine and its (less potent) stereoisomer quinine have been reported (Hutzler et al., 2003), but we have only recently established the protein–ligand interactions, which are responsible for the fact that quinidine can bind tightly but not in an orientation favourable for catalysis (McLaughlin et al., 2005). We used homology modelling and molecular docking to predict the modes of quinidine binding to wild-type and mutant enzymes (McLaughlin et al., 2005). In contrast to the wild-type enzyme (Figure 2a), the E216F mutant produced O-demethylated quinidine (Figure 2b), and E216Q/D301Q produced both O-demethylated quinidine and 3-hydroxyquinidine metabolites (McLaughlin et al., 2005).
Figure 2.
The predicted binding modes of quinidine in wild-type and mutant CYP2D6 (McLaughlin et al., 2005). (a) The best ranked docking of quinidine in the wild-type CYP2D6 model. (b) The best ranked docking into the E216F CYP2D6 model from the cluster of solutions having an orientation appropriate for formation of O-desmethyl quinidine. (c) The highest ranked docking into the F120A CYP2D6 model having an orientation appropriate for formation of 3-hydroxy quinidine. (d) The best ranked docking into the F120A CYP2D6 model from the cluster of solutions having an orientation appropriate for formation of O-desmethyl quinidine. Predicted hydrogen bonds are denoted as dashed lines (Figure produced using Pymol; DeLano, 2002).
The crystal structure of CYP2D6 (Rowland et al., 2006) supports a ligand-binding role for both Glu216 and Asp301 (Figure 1). However, it is suggested that since in this structure the trans and gauche− rotameric states of Asp301 can account for all the pharmacophoric models, Glu216 is more likely to act as a recognition residue that attracts basic ligands to the active site, where it forms an intermediate binding site prior to the ligand adopting a more ‘reactive' position in the cavity. Such a suggestion is similar to the intermediate binding pocket occupied by warfarin in the crystal structure of the S-warfarin/CYP2C9 complex (Williams et al., 2003), but apparently inconsistent with the mutation E216F transforming CYP2D6 into a quinidine demethylase. Resolution of this issue awaits co-crystallization of substrates in CYP2D6.
Role of Phe120
Examination of our model highlights an additional residue—Phe120 in the B-C loop (SRS1)—as being potentially important in ligand binding. It occupies a position close to the haem moiety, where it could have a major influence on substrate binding, possibly through π–π stacking or edge-on-face interactions with the planar aromatic ring of the substrate (Figure 2b). Using site-directed mutagenesis in conjunction with molecular modelling, the role of this residue in substrate binding and catalysis was investigated (Flanagan et al., 2004). The results show the aromatic moiety of Phe120 does indeed have a steric influence on the orientation of molecules in the active site of CYP2D6, and therefore plays a role in controlling the regioselectivity of substrate oxidation. This is further supported by our studies of quinidine metabolism by mutant CYP2D6 (see also above). Unlike wild-type CYP2D6, the F120A mutant produced both O-demethylated quinidine and 3-hydroxyquinidine metabolites (Figures 2c and d; (Flanagan et al., 2004)). Our results suggest that Phe120, positioned close to the haem iron, is a key factor in controlling access to the haem.
In accord with our modelling results, the crystal structure also suggests a role for Phe120 in controlling the orientation of substrates with respect to the haem (Rowland et al., 2006). Analysis of the CYP2D subfamily (Figure 3) reveals that CYP2D enzymes require a hydrophobic residue in this position, although an equivalent bulky residue occurs in only a small number of CYP2D enzymes. This observation suggests that most other CYP2D enzymes will exhibit different drug disposition profiles from that of CYP2D6. Interestingly, a small percentage of the southeast Asian population have a polymorphic CYP2D6 containing the mutation F120I (Solus et al., 2004), suggesting modified drug metabolism and disposition in these individuals.
Figure 3.
The B′-C region (SRS1) of CYP2D subfamily members. The residue position corresponding to Phe120 in CYP2D6 is boxed. Species and Swissprot accession codes are as follows: CYP2D6: human, P10635; CYP2D1: rabbit, P10633; CYP2D2: rat, CP2DQ; CYP2D3: rat, P12938; CYP2D4: rat, P13108; CYP2D7: human, Q6XP50; CYP2D9: mouse, P11714; CYP2D10: rat, P12938; CYP2D11: mouse, P24457; CYP2D13: mouse, Q8XC0; CYP2D14: bull, Q01361; Cyp2D15: dog, Q29473; CYP2D16: guinea-pig, Q64403; CYP2D17: macaque fascicularis, Q29488; CYP2D18: rat, Q64680; CYP2D19: marmoset, O18992; CYP2D20: golden hamster, Q9QYG5; CYP2D21: domestic pig, Q6LEL6; CYP2D23: rabbit, Q9TUJ4; CYP2D25: domestic pig, O46658; CYP2D26: mouse, Q8CIM7; CYP2D27: golden hamster, Q9QYG6; CYP2D28: golden hamster, Q9QUJ1; CYP2D30: marmoset, Q865W1.
Binding of atypical substrates
The ability of CYP2D6 homology models to predict the binding modes of substrates devoid of a basic nitrogen, for example spirosulphonamide, has been questioned (Guengerich et al., 2002a). To investigate if our CYP2D6 model is predictive with this type of substrate, the substrate spirosulphonamide was docked into our CYP2D6 model (Kemp et al., 2004). The highest ranked docked solution positioned the cyclopentyl moiety above the haem and hence correctly identified a major metabolite (Guengerich et al., 2002a), providing additional validation of our model.
Binding affinities
We have also tested the ability of our model and docking approach to predict CYP2D6 inhibition—a group of 33 compounds from the National Cancer Institute database was docked into our CYP2D6 model, and experimental IC50 values for the compounds were determined; comparison of the experimental and predicted affinities revealed a correlation with a regression coefficient of r2=0.61 (q2=0.59) (Kemp et al., 2004). This level of success is noteworthy in itself, and more so because the dockings were into a model rather than a crystal structure. Binding affinities are notoriously difficult to predict quantitatively, and a docking study on 11 different P450cam–ligand complexes (Keseru, 2001) found no clear correlation between experimental and predicted affinities. Our approach is able to discriminate between tightly and weakly binding compounds and correctly identified several novel inhibitors.
Comparison with crystal structure of CYP2D6
When the crystal structure of CYP2D6 (Rowland et al., 2006) became available, we were also able to validate our model against this. There is good agreement between residues in the substrate-binding site (as mentioned above for specific residues), with those residues within 5 Å of the haem overlaying almost perfectly. The overall Cα RMSD (root mean square deviation of alpha carbon atoms) between the model and the crystal structure is (a very reasonable) 0.8 Å. One key difference is in the F-G loop region—a region that varies in size and shape across different CYPs—which arises because this region is in a different conformation in the structural templates used for modelling to that in the crystal structure. This highlights an important limitation of homology modelling—that the structure of the model is generally limited to the structural space occupied by the templates used. Such issues can be (partially) addressed using, for example, loop modelling or molecular dynamics simulation. Additional challenges are faced by conformational changes that may occur on substrate binding, for example, the >80% increase in active site volume that can occur in CYP3A4 (Ekroos and Sjogren, 2006). Such changes are difficult to predict and difficult to address using molecular dynamics simulations.
Drug–drug interactions
Additionally, we used our model to investigate drug–drug interactions in CYP2D6 with drugs commonly taken by patients with cancer as part of a co-medication regime (Yu et al., 2006). This study uncovered the correct metabolite for metoclopramide, a drug frequently used to prevent the nausea and vomiting associated with cancer chemotherapy (Harrington et al., 1983). Our study (Yu et al., 2006) also suggested a particular CYP2D6 genotype/phenotype for those experiencing adverse reactions with metoclopramide, for example extra-pyramidal syndrome (Pall and Williams, 1987). The general applicability of our approach is illustrated by our modelling study of the interaction of anticancer co-medication drugs with CYP3A4 (Marechal et al., 2006). The commonly prescribed drugs loperamide, amitriptyline, diltiazem, domperidone, lansoprazole, omeprazole and simvastatin were correctly identified by our in silico screens as relatively potent inhibitors of CYP3A4 (Marechal et al., 2006), highlighting the likelihood of drug–drug interactions affecting chemotherapy treatment.
Concluding remarks
Using CYP2D6 as an example it is clear that a relatively ‘simple' in silico approach—combining homology modelling with molecular docking, active site characterization and bioinformatics analysis—can predict the sites of metabolism of a range of known substrates, and successfully identify key residues for substrate recognition and binding in the active site. It is also evident, initially from mutagenesis studies and more recently by comparison with the recently available crystal structure of CYP2D6, that high-quality homology models can give a range of insights into the mechanism of an enzyme, and from this infer possible metabolic consequences for compounds of interest. However, it is important to remember that caution must be exercised in the initial stages of model building by ensuring that an accurate amino-acid sequence alignment is obtained—aided by a good working knowledge of the enzyme, which can be enhanced significantly when integrated with synergistic experimental studies. The real strength of this approach is in enriching the success of experimental studies by reducing the number of non-productive ‘dead ends'.
Acknowledgments
This work was supported by the Drug Metabolism Consortium (AstraZeneca, Aventis, Boehringer-Ingelheim, Celltech Chiroscience, GlaxoSmithKline, Hoffmann-La Roche, Johnson and Johnson Pharmaceuticals, Merck Sharp and Dohme, Novartis, Novo Nordisk, Pfizer, Pharmacia and Wyeth), the Higher Education Reach-Out to Business and the Community Fund (HEROBC), and the University of Manchester.
Glossary
- CYPs
cytochromes P450
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
Footnotes
Conflict of interest
The authors state no conflict of interest.
References
- Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- Branch RA, Adedoyin A, Frye RF, Wilson JW, Romkes M. In vivo modulation of CYP enzymes by quinidine and rifampin. Clin Pharmacol Ther. 2000;68:401–411. doi: 10.1067/mcp.2000.110561. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdry J, Pucci MR, Harlow J, Tucker GT, Ellis SW. Evidence that both phenylalanine 481 and phenylalanine 483 are substrate-contact residues in the active site of cytochrome P450 2D6. Br J Clin Pharmacol. 2002;53:443P–444P. [Google Scholar]
- Cupp-Vickery J, Anderson R, Hatziris Z. Crystal structures of ligand complexes of P450eryF exhibiting homotropic cooperativity. Proc Natl Acad Sci USA. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK, Leathart JB, London SJ, Idle JR. An inactive cytochrome P450 CYP2D6 allele containing a deletion and a base substitution. Hum Genet. 1995;95:337–341. doi: 10.1007/BF00225204. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Ackland MJ, Horne VA, Alex AA, Jones BC. A novel approach to predicting P450 mediated drug metabolism. CYP2D6 catalyzed N-dealkylation reactions and qualitative metabolite predictions using a combined protein and pharmacophore model for CYP2D6. J Med Chem. 1999;42:4062–4070. doi: 10.1021/jm991058v. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Kirton SB, Sutcliffe MJ. In silico methods for predicting ligand binding determinants of cytochromes P450. Curr Top Med Chem. 2004;4:1803–1824. doi: 10.2174/1568026043387061. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Vermeulen NP, Kramer JD, van Acker FA, Donne-Op den Kelder GM. A three-dimensional protein model for human cytochrome P450 2D6 based on the crystal structures of P450 101, P450 102, and P450 108. Chem Res Toxicol. 1996;9:1079–1091. doi: 10.1021/tx960003i. [DOI] [PubMed] [Google Scholar]
- DeLano WL.2002The PyMOL Molecular Graphics System . http://www.pymol.org .
- Desmeules J, Gascon MP, Dayer P, Magistris M. Impact of environmental and genetic factors on codeine analgesia. Eur J Clin Pharmacol. 1991;41:23–26. doi: 10.1007/BF00280101. [DOI] [PubMed] [Google Scholar]
- Eichelbaum M, Spannbrucker N, Dengler HJ. Influence of the defective metabolism of sparteine on its pharmacokinetics. Eur J Clin Pharmacol. 1979;16:189–194. doi: 10.1007/BF00562060. [DOI] [PubMed] [Google Scholar]
- Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JU, Marechal JD, Ward R, Kemp CA, McLaughlin LA, Sutcliffe MJ, et al. Phe120 contributes to the regiospecificity of cytochrome P450 2D6: mutation leads to the formation of a novel dextromethorphan metabolite. Biochem J. 2004;380:353–360. doi: 10.1042/BJ20040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford PJ. A computational-procedure for determining energetically favorable binding-sites on biologically important macromolecules. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Miller GP, Hanna IH, Martin MV, Leger S, Black C, et al. Diversity in the oxidation of substrates by cytochrome P450 2D6: lack of an obligatory role of aspartate 301-substrate electrostatic bonding. Biochemistry. 2002a;41:11025–11034. doi: 10.1021/bi020341k. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Miller GP, Hanna IH, Sato H, Martin MV. Oxidation of methoxyphenethylamines by cytochrome P450 2D6. Analysis of rate-limiting steps. J Biol Chem. 2002b;277:33711–33719. doi: 10.1074/jbc.M205146200. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Muller-Enoch D, Blair IA. Oxidation of quinidine by human liver cytochrome P-450. Mol Pharmacol. 1986;30:287–295. [PubMed] [Google Scholar]
- Hanna IH, Krauser JA, Cai H, Kim MS, Guengerich FP. Diversity in mechanisms of substrate oxidation by cytochrome P450 2D6. Lack of an allosteric role of NADPH-cytochrome P450 reductase in catalytic regioselectivity. J Biol Chem. 2001;276:39553–39561. doi: 10.1074/jbc.M106841200. [DOI] [PubMed] [Google Scholar]
- Harrington RA, Hamilton CW, Brogden RN, Linkewich JA, Romankiewicz JA, Heel RC. Metoclopramide. An updated review of its pharmacological properties and clinical use. Drugs. 1983;25:451–494. doi: 10.2165/00003495-198325050-00002. [DOI] [PubMed] [Google Scholar]
- Hasemann CA, Ravichandran KG, Peterson JA, Deisenhofer J. Crystal structure and refinement of cytochrome P450terp at 2.3 Å resolution. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Harlow J, Chowdry J, Gross E, Hilton E, Lennard MS, et al. Influence of phenylalanine-481 substitutions on the catalytic activity of cytochrome P450 2D6. Biochem J. 2001a;355:373–379. doi: 10.1042/0264-6021:3550373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Harlow J, Chowdry J, Gross E, Hilton E, Lennard MS, et al. Influence of phenylalanine-481 substitutions on the catalytic activity of cytochrome P450 2D6. Biochem J. 2001b;355:373–379. doi: 10.1042/0264-6021:3550373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzler JM, Walker GS, Wienkers LC. Inhibition of cytochrome P450 2D6: structure–activity studies using a series of quinidine and quinine analogues. Chem Res Toxicol. 2003;16:450–459. doi: 10.1021/tx025674x. [DOI] [PubMed] [Google Scholar]
- Islam SA, Wolf CR, Lennard MS, Sternberg MJ. A three-dimensional molecular template for substrates of human cytochrome P450 involved in debrisoquine 4-hydroxylation. Carcinogenesis. 1991;12:2211–2219. doi: 10.1093/carcin/12.12.2211. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Flanagan JU, van Eldik AJ, Marechal JD, Wolf CR, Roberts GC, et al. Validation of model of cytochrome P450 2D6: an in silico tool for predicting metabolism and inhibition. J Med Chem. 2004;47:5340–5346. doi: 10.1021/jm049934e. [DOI] [PubMed] [Google Scholar]
- Keseru GM. A virtual high throughput screen for high affinity cytochrome P450cam substrates. Implications for in silico prediction of drug metabolism. J Comput Aided Mol Des. 2001;15:649–657. doi: 10.1023/a:1011911204383. [DOI] [PubMed] [Google Scholar]
- Kidd RS, Curry TB, Gallagher S, Edeki T, Blaisdell J, Goldstein JA. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics. 2001;11:803–808. doi: 10.1097/00008571-200112000-00008. [DOI] [PubMed] [Google Scholar]
- Kidd RS, Straughn AB, Meyer MC, Blaisdell J, Goldstein JA, Dalton JT. Pharmacokinetics of chlorpheniramine, phenytoin, glipizide and nifedipine in an individual homozygous for the CYP2C9*3 allele. Pharmacogenetics. 1999;9:71–80. doi: 10.1097/00008571-199902000-00010. [DOI] [PubMed] [Google Scholar]
- Kirton SB, Baxter CA, Sutcliffe MJ. Comparative modelling of cytochromes P450. Adv Drug Deliv Rev. 2002a;54:385–406. doi: 10.1016/s0169-409x(02)00010-8. [DOI] [PubMed] [Google Scholar]
- Kirton SB, Kemp CA, Tomkinson NP, St-Gallay S, Sutcliffe MJ. Impact of incorporating the 2C5 crystal structure into comparative models of cytochrome P450 2D6. Proteins. 2002b;49:216–231. doi: 10.1002/prot.10192. [DOI] [PubMed] [Google Scholar]
- Koymans L, Vermeulen NP, van Acker SA, te Koppele JM, Heykants JJ, Lavrijsen K, et al. A predictive model for substrates of cytochrome P450-debrisoquine (2D6) Chem Res Toxicol. 1992;5:211–219. doi: 10.1021/tx00026a010. [DOI] [PubMed] [Google Scholar]
- Kroemer HK, Eichelbaum M. ‘It's the genes, stupid'. Molecular bases and clinical consequences of genetic cytochrome P450 2D6 polymorphism. Life Sci. 1995;56:2285–2298. doi: 10.1016/0024-3205(95)00223-s. [DOI] [PubMed] [Google Scholar]
- Ladona MG, Lindstrom B, Thyr C, Dun-Ren P, Rane A. Differential foetal development of the O- and N-demethylation of codeine and dextromethorphan in man. Br J Clin Pharmacol. 1991;32:295–302. doi: 10.1111/j.1365-2125.1991.tb03902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DF. Homology modelling of human cytochromes P450 involved in xenobiotic metabolism and rationalization of substrate selectivity. Exp Toxicol Pathol. 1999;51:369–374. doi: 10.1016/S0940-2993(99)80024-4. [DOI] [PubMed] [Google Scholar]
- Lewis DF, Eddershaw PJ, Goldfarb PS, Tarbit MH. Molecular modelling of cytochrome P4502D6 (CYP2D6) based on an alignment with CYP102: structural studies on specific CYP2D6 substrate metabolism. Xenobiotica. 1997;27:319–339. doi: 10.1080/004982597240497. [DOI] [PubMed] [Google Scholar]
- Mackman R, Tschirret-Guth RA, Smith G, Hayhurst GP, Ellis SW, Lennard MS, et al. Active-site topologies of human CYP2D6 and its aspartate-301 → glutamate, asparagine, and glycine mutants. Arch Biochem Biophys. 1996;331:134–140. doi: 10.1006/abbi.1996.0291. [DOI] [PubMed] [Google Scholar]
- Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- Marechal JD, Yu J, Brown S, Kapelioukh I, Rankin EM, Wolf CR, et al. In silico and in vitro screening for inhibition of cytochrome P450 CYP3A4 by comedications commonly used by patients with cancer. Drug Metab Dispos. 2006;34:534–538. doi: 10.1124/dmd.105.007625. [DOI] [PubMed] [Google Scholar]
- McLaughlin LA, Paine MJ, Kemp CA, Marechal JD, Flanagan JU, Ward CJ, et al. Why is quinidine an inhibitor of cytochrome P450 2D6? The role of key active-site residues in quinidine binding. J Biol Chem. 2005;280:38617–38624. doi: 10.1074/jbc.M505974200. [DOI] [PubMed] [Google Scholar]
- Meyer UA, Gut J, Kronbach T, Skoda C, Meier UT, Catin T, et al. The molecular mechanisms of two common polymorphisms of drug oxidation—evidence for functional changes in cytochrome P-450 isozymes catalysing bufuralol and mephenytoin oxidation. Xenobiotica. 1986;16:449–464. doi: 10.3109/00498258609050251. [DOI] [PubMed] [Google Scholar]
- Modi S, Gilham DE, Sutcliffe MJ, Lian LY, Primrose WU, Wolf CR, et al. 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine as a substrate of cytochrome P450 2D6: allosteric effects of NADPH-cytochrome P450 reductase. Biochemistry. 1997;36:4461–4470. doi: 10.1021/bi962633p. [DOI] [PubMed] [Google Scholar]
- Modi S, Paine MJ, Sutcliffe MJ, Lian LY, Primrose WU, Wolf CR, et al. A model for human cytochrome P450 2D6 based on homology modeling and NMR studies of substrate binding. Biochemistry. 1996;35:4540–4550. doi: 10.1021/bi952742o. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- Nelson DR.2007Cytochrome P450 website . http://drnelson.utmem.edu/CytochromeP450.html
- Otton SV, Brinn RU, Gram LF. In vitro evidence against the oxidation of quinidine by the sparteine/debrisoquine monooxygenase of human liver. Drug Metab Dispos. 1988;16:15–17. [PubMed] [Google Scholar]
- Paine MJ, McLaughlin LA, Flanagan JU, Kemp CA, Sutcliffe MJ, Roberts GC, et al. Residues glutamate 216 and aspartate 301 are key determinants of substrate specificity and product regioselectivity in cytochrome P450 2D6. J Biol Chem. 2003;278:4021–4027. doi: 10.1074/jbc.M209519200. [DOI] [PubMed] [Google Scholar]
- Pall HS, Williams AC. Extrapyramidal disturbances caused by inappropriate prescribing. BMJ (Clin Res Ed) 1987;295:30–31. doi: 10.1136/bmj.295.6589.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos TL, Finzel BC, Howard AJ. Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry. 1986;25:5314–5322. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson JA, Deisenhofer J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, et al. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, et al. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Sligar SG, Cinti DL. Substrate interaction with cytochrome P-450. Pharmacol Ther. 1981;12:43–71. doi: 10.1016/0163-7258(81)90075-9. [DOI] [PubMed] [Google Scholar]
- Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, et al. An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-Å resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- Smith G, Modi S, Pillai I, Lian LY, Sutcliffe MJ, Pritchard MP, et al. Determinants of the substrate specificity of human cytochrome P-450 CYP2D6: design and construction of a mutant with testosterone hydroxylase activity. Biochem J. 1998;331:783–792. doi: 10.1042/bj3310783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Stanley LA, Sim E, Strange RC, Wolf CR. Metabolic polymorphisms and cancer susceptibility. Cancer Surv. 1995;25:27–65. [PubMed] [Google Scholar]
- Solus JF, Arietta BJ, Harris JR, Sexton DP, Steward JQ, McMunn C, et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004;5:895–931. doi: 10.1517/14622416.5.7.895. [DOI] [PubMed] [Google Scholar]
- Strobl GR, von Kruedener S, Stockigt J, Guengerich FP, Wolff T. Development of a pharmacophore for inhibition of human liver cytochrome P-450 2D6: molecular modeling and inhibition studies. J Med Chem. 1993;36:1136–1145. doi: 10.1021/jm00061a004. [DOI] [PubMed] [Google Scholar]
- Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Echizen H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin Pharmacokinet. 2001;40:587–603. doi: 10.2165/00003088-200140080-00003. [DOI] [PubMed] [Google Scholar]
- Tanaka E. Clinically important pharmacokinetic drug–drug interactions: role of cytochrome P450 enzymes. J Clin Pharm Ther. 1998;23:403–416. doi: 10.1046/j.1365-2710.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- Venhorst J, ter Laak AM, Commandeur JN, Funae Y, Hiroi T, Vermeulen NP. Homology modeling of rat and human cytochrome P450 2D (CYP2D) isoforms and computational rationalization of experimental ligand-binding specificities. J Med Chem. 2003;46:74–86. doi: 10.1021/jm0209578. [DOI] [PubMed] [Google Scholar]
- von Bahr C, Spina E, Birgersson C, Ericsson O, Goransson M, Henthorn T, et al. Inhibition of desmethylimipramine 2-hydroxylation by drugs in human liver microsomes. Biochem Pharmacol. 1985;34:2501–2505. doi: 10.1016/0006-2952(85)90533-7. [DOI] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD. Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 Å resolution: evidence for multiple substrate binding modes. Biochemistry. 2003a;42:6370–6379. doi: 10.1021/bi0273922. [DOI] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, et al. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 Å resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003b;42:9335–9345. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, et al. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-Å resolution. J Biol Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, et al. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J Biol Chem. 2004;279:38091–38104. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- Yu J, Paine MJ, Marechal JD, Kemp CA, Ward CJ, Brown S, et al. In silico prediction of drug binding to CYP2D6: identification of a new metabolite of metoclopramide. Drug Metab Dispos. 2006;34:1386–1392. doi: 10.1124/dmd.106.009852. [DOI] [PubMed] [Google Scholar]