Summary
Developmental axon pruning is widely used to refine neural circuits. We performed a mosaic screen to identify mutations affecting axon pruning of Drosophila mushroom body γ neurons. We constructed a modified piggyBac vector with improved mutagenicity and generated insertions in >2000 genes. We identified two cohesin subunits (SMC1 and SA) as essential for axon pruning. The cohesin complex maintains sister chromatid cohesion during cell division in eukaryotes. However, we show that the pruning phenotype in SMC1-/- clones is rescued by expressing SMC1 in neurons, revealing a new postmitotic function. SMC1-/- clones exhibit reduced levels of the ecdysone receptor EcR-B1, a key regulator of axon pruning. The pruning phenotype is significantly suppressed by overexpressing EcR-B1 and enhanced by reduced dosage of EcR, supporting a causal relationship. We also demonstrate a postmitotic role for SMC1 in dendrite targeting of olfactory projection neurons. We suggest that cohesin regulates diverse aspects of neuronal morphogenesis.
Keywords: Drosophila, mushroom body, γ neurons, cohesin complex, sister chromatid cohesion, ecdysone receptor, MARCM
INTRODUCTION
Developmental axon pruning is widely used for maturation and refinement of neural circuits (reviewed in Luo and O’Leary, 2005). In many well-documented cases, neurons first extend exuberant branches, and later prune away inappropriate branches with precise spatial and temporal control. Developmental axon pruning can occur by several distinct cellular mechanisms, including distal-to-proximal retraction (e.g., Liu et al., 2005; Portera-Cailliau et al., 2005), localized degeneration in which defined segments of axons break into pieces that are taken up by surrounding cells (e.g., Watts et al., 2003; Awasaki and Ito, 2004; Watts et al., 2004; Portera-Cailliau et al., 2005), and “axosome shedding” in which distal ends of retracting axons are engulfed by nearby cells (Bishop et al., 2004). Developmental axon pruning, particularly involving localized degeneration, also shares some molecular and mechanistic similarities with axon degeneration after nerve injury (Raff et al., 2002; Hoopfer et al., 2006).
Developmental axon pruning of mushroom body (MB) γ neurons in Drosophila occurs by localized degeneration (Watts et al., 2003) and is an appealing system to study mechanisms of axon pruning. During metamorphosis, MB γ neurons prune their dendrites completely and axons to a specific branch-point in a stereotypic manner (Lee et al., 1999). The initiation of pruning is regulated by the cell-autonomous action of the steroid hormone ecdysone receptor B1 (EcR-B1) and its co-receptor Ultraspiracle (Usp) (Lee et al., 2000a). For pruning to occur, γ neurons must also have a functional ubiquitin proteasome system (UPS) (Watts et al., 2003). Lastly, degenerated axon fragments are engulfed by glia (Awasaki and Ito, 2004; Watts et al., 2004). Similar molecular pathways appear to be used in other Drosophila neurons to direct developmental pruning of axons and dendrites during metamorphosis (Schubiger et al., 2003; Kuo et al., 2005; Marin et al., 2005; Williams and Truman, 2005). Despite the widespread use of these molecular pathways, our understanding of the underlying mechanisms is far from complete.
Forward genetic screens are a powerful and unbiased strategy for identifying molecules involved in complex biological processes. To study late developmental events and to identify genes that have pleiotropic functions, forward genetic screens in mosaic tissues (e.g., Xu and Rubin, 1993; Newsome et al., 2000a) have been developed. Furthermore, mosaic-labeling techniques such as the MARCM system (Mosaic Analysis with a Repressible Cell Marker; Lee and Luo, 1999) allow for visualization of only homozygous mutant cells, thereby further increasing the resolution of phenotype detection (e.g., Lee et al., 2000a). Compared to mutations induced by chemical mutagens such as EMS, transposon insertional mutagenesis permits rapid mapping of a causal mutation. However, P-element based mutagenesis is not easily adapted to FLP/FRT-based mosaic screens. Recently, the piggyBac transposon has been shown to transpose effectively in Drosophila without destabilizing P-elements (Hacker et al., 2003). We describe here a mosaic piggyBac-based insertional mutagenesis screen in Drosophila that identifies the cohesin complex as being required for axon pruning.
Cohesin is a highly conserved multisubunit complex required for sister chromatid cohesion during mitosis and meiosis. The cohesin complex is comprised of Smc1, Smc3, Scc1/Rad21 and Scc3/Stromalin (SA) (reviewed in Losada and Hirano, 2005; Nasmyth and Haering, 2005). Current data suggest a model in which Smc1, Smc3 and Rad21 form a ring that embraces sister chromatids, while SA binds to Rad21 and probably has a regulatory function (Gruber et al., 2003; Huang et al., 2005; reviewed in Nasmyth, 2005; Hirano, 2006). Cohesin is loaded onto chromosomes with the assistance of another complex comprised of Scc2/Nipped-B and Scc4/Mau-2 (Ciosk et al., 2000; reviewed in Dorsett, 2007). The cohesin complex holds sister chromatids together until the onset of anaphase, when Rad21 is cleaved by Separase to enable their separation (Uhlmann et al., 2000; Jager et al., 2001).
Using a new piggyBac mutator that is compatible with mosaic analysis and appears to efficiently disrupt genes even when inserted into introns, we have generated a large piggyBac mutant collection. Our screen in MB neurons revealed that mutations in SMC1 and SA, two subunits of the cohesin complex, disrupt axon pruning in addition to causing neuroblast proliferation defects. Postmitotic expression of a wild type (wt) SMC1 transgene is sufficient to rescue axon pruning phenotypes without rescuing the neuroblast proliferation defects. We provide evidence that this postmitotic function of SMC1 is mediated through the regulation of EcR-B1 levels. SMC1 also regulates dendrite targeting in postmitotic olfactory projection neurons. Thus, in addition to its classic function in chromosome cohesion, our study indicates that the cohesin complex also plays an essential role in neurons to regulate their morphogenesis.
RESULTS
Insertional mutagenesis using a modified piggyBac transposon
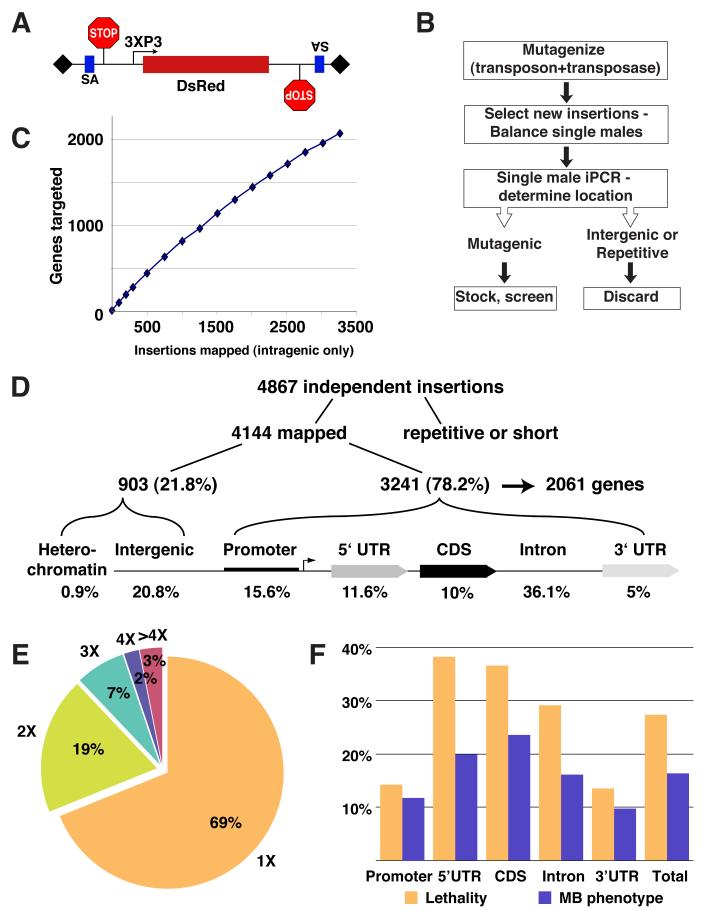
To increase mutagenicity of existing piggyBac elements and to specifically render the high proportion of intronic insertions mutagenic (Hacker et al., 2003), we added splice acceptors followed by stop codons in all three frames in both orientations of the mutator (Figure 1A). We also swapped the existing marker with a DsRed fluorescent protein to allow live screening of brains with MARCM clones expressing GFP.
Figure 1. Overview of piggyBac-based Insertional Mutagenesis.
(A) Our modified piggyBac mutator element contains, in both orientations, a splice acceptor (SA) followed by stop codons in all three reading frames; it is marked with a DsRed reporter. 3XP3 is a synthetic promoter expressed mainly in the eye (Sheng et al., 1997) and shown to effectively drive the expression of different fluorescent proteins as markers for piggyBac (Horn et al., 2000).
(B) Mutant generation scheme; see text and Supplemental Experimental Procedures for details.
(C) Number of genes targeted plotted against intragenic insertions mapped.
(D) Distribution of piggyBac transposons with regard to a generic gene structure.
(E) Frequency distribution of 3241 independent insertions that fall within transcriptional units of 2061 different genes.
(F) Rates of lethality and MB mutant phenotype for insertions in different parts of the transcriptional unit.
Mobilization of mutator elements was performed using starter insertions on the X or 2nd chromosomes (Supplemental Experimental Procedures). All insertions occurred in a quadruple FRT background (FRTs 40A, G13, 2A and 82B) such that nearly the entire 2nd and 3rd chromosomes can be subjected to mosaic screening. We developed a protocol to determine chromosomal insertion sites by inverse PCR from a single fly (Figure 1B; Supplemental Experimental Procedures). This has allowed us to map the insertion sites before a stable stock must be established and to discard insertions that were not successfully mapped (see below) or did not disrupt genes.
We sequenced 4867 insertions and mapped 4144 independent insertions, which are evenly distributed among all chromosome arms (Figure 1D). We failed to map insertions in repetitive regions, and those that yielded very short sequences. The distribution of the insertions with respect to the structure of genes is similar to what was previously reported (Hacker et al., 2003). About 80% of mapped insertions were within transcriptional units, or up to 1kb upstream of the predicted start of transcription (putative promoter). The remaining 20% were outside these areas and were considered intergenic insertions (Figure 1D). Although intergenic insertions could potentially disrupt a distant enhancer or an un-annotated gene, we discarded these insertions and those on the 4th chromosome or in centromeric regions, as they are not amenable for mosaic analysis. The largest fraction of insertions within transcriptional units was in introns (46%), justifying our effort to add splice traps to the mutator.
The 3241 independent intragenic insertions hit 2061 different genes, corresponding to ∼15% of the annotated genes in the Drosophila genome (Celniker et al., 2002). The rate at which we target new genes indicates that the screen is far from saturation (Figure 1C). 69% of the genes were hit once; 3% were hit more than 4 times; the most-hit gene was 13 times (Figure 1E). This distribution is consistent with the observations that piggyBac insertion is not random, but less biased than P-element insertions (Bellen et al., 2004; Thibault et al., 2004).
Table S1 provides a comprehensive list of 1921 insertions targeting 1803 genes that are still available and can be obtained from the Drosophila Genetic Resource Center (DGRC), Kyoto, Japan (http://www.dgrc.kit.ac.jp/en/index.html). These can be used for any mosaic or non-mosaic forward screens or candidate mutant analysis.
Mutagenicity of piggyBac insertions
To estimate the mutagenicity of our insertions, we determined the rate at which we generate homozygous lethal mutations. Our average rate is 28% (Figure 1F), which is higher than in previous studies (9-22%; Hacker et al., 2003; Thibault et al., 2004). Two factors likely contribute to this high lethality rate: the elimination of intergenic insertions and the addition of splice traps to the mutator. As expected, insertions in the 5′ UTR and coding sequence (CDS) are most mutagenic (38% and 37% lethality, respectively) while insertions in the 3′UTR are the least (13%). Intronic insertions are highly mutagenic (29% lethality), suggesting that the splice trap indeed improves the mutagenicity. As only about one-third of the genes are expected to be essential for viability (Miklos and Rubin, 1996), this high lethality rate suggests that almost every insertion is mutagenic. Higher than the one-third predicted lethality rates in 5′ UTR and CDS could be due to preferential insertions in active genes as previously shown for P-elements (Liao et al., 2000), genetic background (a result of having FRTs on 4 chromosome arms), or both. 17% of our screened mutants showed abnormal MB phenotypes. When plotted alongside the lethality rate, we observe that the distributions are similar (Figure 1F), confirming that insertions in the 5′ UTR, CDS and introns likely create strong mutant alleles.
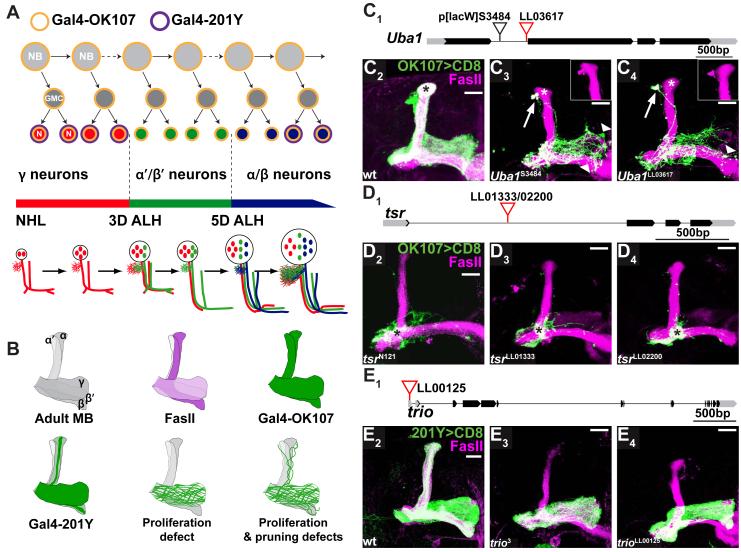
Another way to estimate the mutagenicity is to compare the defects caused by piggyBac insertions to phenotypes of known mutants in MB development using MARCM analysis (see Experimental Procedures). The MB is comprised of three major types of neurons born sequentially from four neuroblasts in each hemisphere (Figure 2A). These neuronal types can be distinguished by their distinct projections and differential expression of Gal4 lines and FasII (Figures 2A, 2B; Ito et al., 1997; Crittenden et al., 1998; Lee et al., 1999). Given the sequential birth and stereotypical projections, mutant phenotypes offer insights into the nature of the pathways affected. For example, MARCM neuroblast clones mutant in genes that affect proliferation should exhibit reduced cell numbers and should be comprised exclusively of the early born γ neurons (Figures 2A, 2B). Neuroblast clones mutant in genes that affect neuronal survival should also have reduced cell numbers but should have both early and late born neurons.
Figure 2. Phenotypic Comparison of piggyBac and Previously Characterized Alleles.
(A) Scheme of sequential generation of MB neurons. MB neurons are generated from four neuroblasts (NBs) per hemisphere. Each NB divides asymmetrically to generate another NB and a ganglion mother cell (GMC), which divides once more to generate two postmitotic neurons (N). MB neuroblasts sequentially give rise to γ neurons (red), α’/β’ (green) and α/β neurons (blue) according to depicted developmental timeline. NHL, newly hatched larvae; 3D ALH, 3 days after larval hatching. Gal4-OK107 is expressed in all MB neurons including dividing NBs (orange outline). Gal4-201Y is expressed in postmitotic γ and a subset of later born α/β neurons (purple outline). Schematic representation of the stereotypical projection of MB neurons at different developmental stages is shown in the lower part of the panel. Adapted from Lee et al., 1999.
(B) Schematic drawing shows 1) Adult MB with 5 axon lobes contributed by γ, α’/β’ and α/β neurons; 2) anti-FasII labels the γ lobe weakly and the α/β lobes strongly; 3-4) Gal4-OK107 and Gal4-201Y expression patterns in the corresponding lobes; 5) MB MARCM neuroblast clones defective in proliferation, with axons only innervating the γ lobe; 6) MB neuroblast clones defective in proliferation and pruning (as is the case for Uba1-/- and SMC1-/- clones), with residual unpruned γ axons around the dorsal lobes.
(C) piggyBac line LL03617 is inserted in the first intron of Uba1 (C1). Compared to wt MB clones (C2), homozygous UbaLL03617 MB neuroblast clones (C4) in adult exhibit neuroblast proliferation and axon pruning defects, and signs of axon degeneration, similar to the previously reported phenotype of the strong loss-of-function UbaS3484 (C3) (Watts et al., 2003). Arrows point to unpruned dorsal projections that are positive for FasII (insets). Arrowheads show blebbing and sparse axons; asterisks mark late born α/β neurons.
(D) piggyBac lines LL01333 and LL02200 are inserted in opposite orientations at the same location in the first intron of twinstar (tsr) (D1). MB neuroblast clones homozygous for either insertion contain mostly axons that fail to extend beyond the branching point (asterisks, D3,4), similar to the axon growth phenotypes previously described for null mutation tsrN121 (asterisk, D2) (Ng and Luo, 2004).
(E) piggyBac line LL00125 is inserted in the 5′ UTR of trio (E1). MB neuroblast clones homozygous for trioLL00125 do not contain late born α/β neurons (E4) as in wt (E2), indicating a neuroblast proliferation defect. This phenocopies a strong loss-of-function trio3 mutant clones (E3).
Black blocks, CDS; gray blocks, UTRs; lines, introns. All images in this and subsequent figures are confocal z-stacks of MB neurons and their axons, unless otherwise stated. Scale bars, 20μm. Genotypes are described in Supplemental Information.
MARCM analysis of insertions within three genes shows phenotypes very similar to previously reported mutants (Figures 2C-2E). 1) An intronic insertion in Uba1 (Figure 2C1) phenocopies a previously reported mutant (Watts et al., 2003). Compared with wt MB neuroblast clones (Figure 2C2), mutation in Uba1 results in a proliferation defect as no late born neurons appear in the mutant clone (asterisks in Figures 2C3,4). In addition, some γ axons fail to prune (arrows and insets in Figures 2C3,4), and show signs of degeneration (arrowheads in Figures 2C3,4). 2) Two intronic insertions in twinstar (tsr) inserted at the same site but in opposite orientation (Figure 2D1) result in an axon extension defect (asterisks in Figures 2D2-4) as previously reported (Ng and Luo, 2004; compare Figure 2D2 to 2D3,4 and to wild-type in 2C1). 3) An insertion in the 5′ UTR of trio (Figure 2E1) causes a proliferation defect of the neuroblast which lacks the last-born α/β neurons, a phenotype also observed with a previously characterized strong loss-of-function allele (Newsome et al., 2000b; compare Figure 2E3 to 2E4 and to wild-type in 2E2). These results confirm that our piggyBac insertions can create strong loss-of-function alleles.
To verify that the mutagenicity caused by intronic insertions is due to the splice trap, we performed RT-PCR experiments with appropriate primers for 5 different homozygous viable insertions (Figure S1). Semi-quantitative RT-PCR shows that in 5/5 cases we can detect the piggyBac-trapped transcript. Importantly, when comparing homozygous mutant to heterozygous flies, the endogenous transcripts were undetectable in 2/5 cases, reduced in another 2/5 cases, and exhibited no change in 1/5. These data suggest that not all intronic insertions are equally mutagenic, consistent with our finding that the lethality rate of intronic insertions is lower compared with insertions in 5′ UTRs or CDSs (Figure 1F).
Mutants can be readily screened using MARCM
Utilizing these piggyBac mutants, we have screened for defects in axon pruning of MB γ neurons. A salient advantage of performing a transposon insertional mutagenesis is the ease of mapping the entire insertion collection, which allows for a genomic perspective for a particular phenotype. For example, while screening for mutations that affect axon pruning, we observed that the most common defect in MB development is a reduction in cell number comprising a neuroblast clone, most likely due to a proliferation defect.
Table S2 provides the list of all piggyBac insertions that cause such defects. Figure S2 shows a few examples of insertions that affect clone size with different severity. As expected, genes essential for neuroblast proliferation are involved in housekeeping functions such as metabolism, transcription, translation, in addition to cell cycle progression. For example: Top2 is a topoisomerase essential for DNA replication, repair and transcription; SMC2 is a subunit of condensin important for chromosome condensation; polo is a kinase involved in cell cycle regulation; and tws is a protein phosphatase type 2A involved in cell cycle. Hereafter, we focus on studies of the cohesin complex, identified in our screen to play a role MB neuroblast proliferation as well as in γ neuron axon pruning.
SMC1 is required for axon pruning
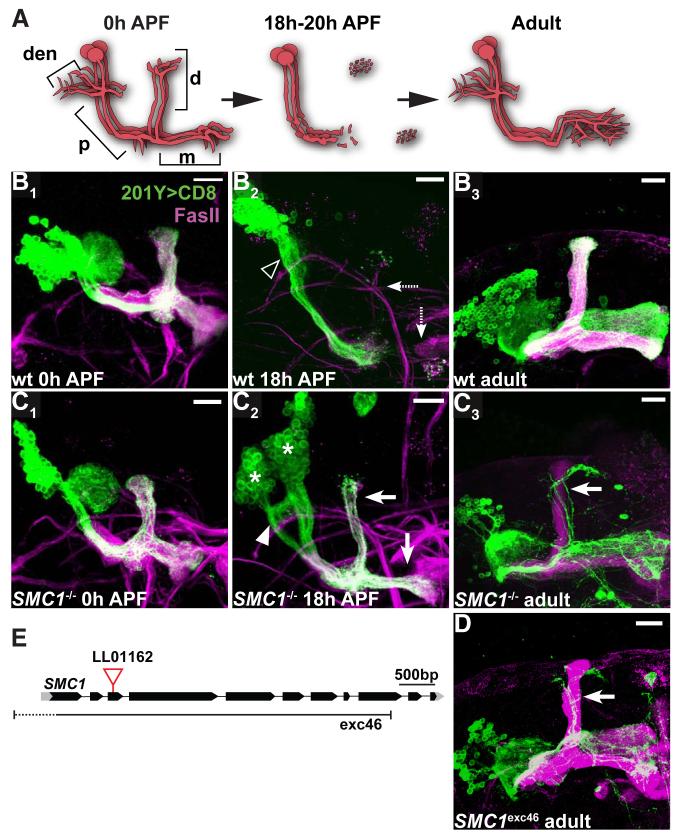
In our screen, we identified an insertion in the third exon of SMC1 (Figure 3E) that inhibits axon pruning in neuroblast clones (Figure 3). To follow the development of SMC1-/- γ neurons, we used Gal4-201Y, which is expressed in all γ and in a subset of late-pupal born α/β neurons (see Figures 2A, 2B). We find that SMC1-/- γ neurons extend axons normally during larval development, as can be seen by their axonal extension into the dorsal and medial lobes at 0h after puparium formation (APF) (compare Figure 3C1 to 3B1; diagram in Figure 3A). However, when examined at the peak of pruning (18h APF; Figure 3A), most wt γ neurons have completely pruned their axons within the dorsal and medial lobes (Lee et al., 1999; Watts et al., 2003), while SMC1-/- γ neurons retain many unpruned axons (compare Figure 3C2 to 3B2). Unpruned axons from SMC1-/- γ neurons persist in adult brains (compare Figure 3C3 to 3B3; schematized in Figure 2B). The dorsal axons in Figure 3C3 (arrows) can be distinguished as unpruned γ neurons but not α/β neurons (which are also labeled by Gal4-201Y), as they are outside the FasII fascicle (magenta) representing unlabeled, heterozygous α/β neurons derived from the other three MB neuroblasts.
Figure 3. SMC1 is Required for Axon Pruning.
(A) Scheme of developmental pruning of MB γ neurons. At 0 hr after puparium formation (0h APF), each γ neuron has a single process that gives off dendritic branches (den) near the cell body, continues an axon peduncle (p), and bifurcates to form a dorsal (d) and a medial (m) branch. The dorsal and medial axon branches, as well as dendrites, are pruned by 18h APF, leaving some fragmented axons at the tips of the lobes but an intact axon peduncle. Later, γ neurons extend axons only to the adult-specific medial lobe, with extensive branches within. Adapted from Watts et al. 2004.
(B-D) Wt (B), SMC1LL01162 (C) and SMC1Δexc46 (D) MB neuroblast MARCM clones labeled with Gal4-201Y. At 0h APF, both wt (B1) and SMC1-/- (C1) γ neurons project their axons to the dorsal and medial lobes. The cell number within SMC1-/- neuroblast clone is significantly reduced. At 18h APF, wt γ neurons have pruned their medial and dorsal axon branches (dashed arrows in B2) as well as their dendrites (open arrowhead). SMC1-/- γ neurons retain most of their axons at this stage (arrows in C2), which persist to adulthood (arrow in C3, D). In addition, SMC1-/- γ neurons retain partially unpruned dendrites at 18h APF (arrowhead in C2). Asterisks in C2 indicate two independent neuroblast clones.
(E) SMC1 gene structure. piggyBac line LL01162 is inserted in the third exon. The approximate borders of the null mutation SMC1Δexc46 (Scott Page & Scott Hawley, personal communication) are also depicted.
Green, Gal4-201Y driven mCD8::GFP; magenta, anti-FasII. Scale bars, 20μm.
In addition to the pruning defect, we observed that SMC1-/- neuroblast clones have fewer cells compared to wt clones (compare Figure 3C3 with 3B3 and Figure 3C1 with 3B1; average clone size in wt and mutant at 0h APF is 159 and 72 cells, respectively). As SMC1-/- clones never contain late-born α’/β’ or α/β neurons we conclude that the reduced clone size is caused by a neuroblast proliferation defect.
Several additional lines of evidence indicate that mutation of SMC1 causes axon pruning and neuroblast proliferation phenotypes: 1) precise excision of the piggyBac mutator reversed lethality and all mutant phenotypes (data not shown); 2) MB clones homozygous for a small deletion encompassing SMC1 (Dorsett et al., 2005) phenocopied our piggyBac insertion (Figure 3D); and 3) mutant phenotypes were rescued by transgenes expressing wt SMC1 (see below). We thus conclude that SMC1 is required for MB neuroblast proliferation and for γ neuron axon pruning.
During the piggyBac screening, we also isolated an insertion in the 5′ UTR of Stromalin (SA, the yeast Scc3 homolog), another subunit of the cohesin complex (Figure S3A). Our primary screen using a pan-MB reporter (Gal4-OK107; Figures 2A, 2B) revealed that homozygous SA-/- clones also fail to prune, in addition to exhibiting a variable neuroblast proliferation defect (Figure S3B-C). This observation suggests that other components of the cohesin complex may also function similar to SMC1 in regulating axon pruning.
SMC1 is required in postmitotic neurons for axon pruning
Both SMC1 and SA are subunits of the cohesin complex, whose main cellular role is to maintain the cohesion of duplicated sister chromatids during mitosis and meiosis (reviewed in Nasmyth and Haering, 2005). In the absence of cohesin complex function, cells experience precocious sister chromatid separation, leading to cell cycle arrest through the action of the spindle checkpoint (Vass et al., 2003). This cell cycle arrest may explain the reduced proliferation capacity of SMC1-/- neuroblasts. Perdurance of SMC1 mRNA and/or protein (see Experimental Procedures) in SMC1-/- neuroblast clones may explain why these neuroblasts can still proliferate to some extent. However, homozygous SMC1-/- clones also exhibit defective axon pruning, a process that occurs in terminally differentiated, non-dividing neurons. An important question is, therefore, whether SMC1 is required in dividing neuroblasts (as predicted if its role is solely in sister chromatid cohesion) or in postmitotic neurons for axon pruning.
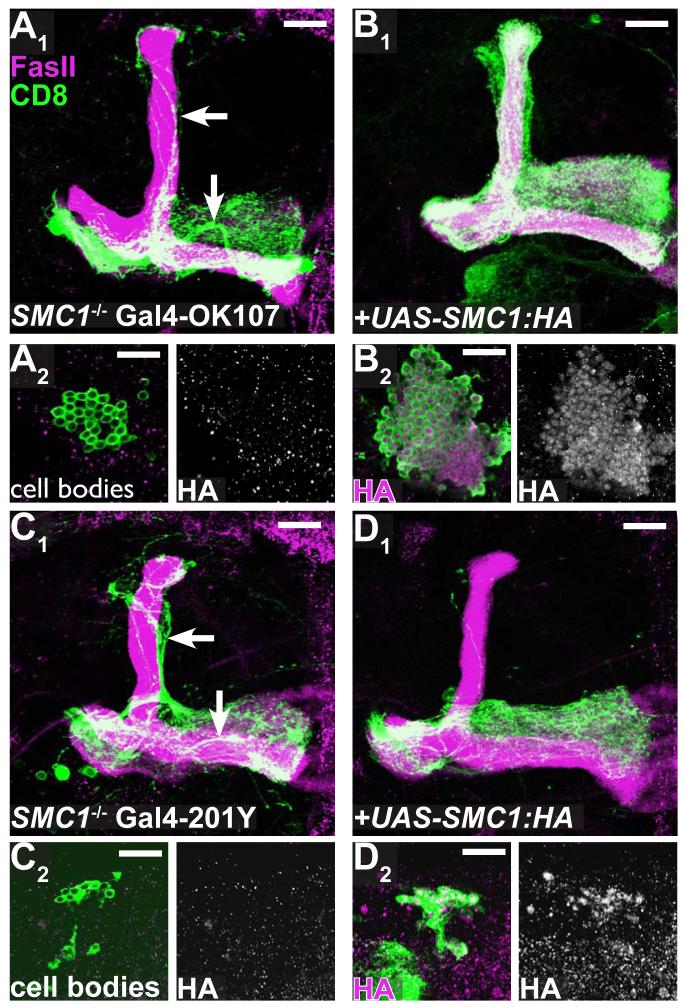
To address this question, we constructed transgenic flies expressing wt SMC1 in a Gal4-dependent manner (UAS-SMC1::HA). We crossed these flies to two lines expressing Gal4 at different stages of MB development (Gal4-OK107 and -201Y; Figures 2A, 2B) and tested whether the SMC1-/- γ neuron phenotype can be rescued by either Gal4 driver. Expression of UAS-SMC1::HA with either Gal4 lines in wt neurons does not result in any detectable gain-of-function phenotype (data not shown). As expected, driving the expression of UAS-SMC1::HA in dividing neuroblasts as well as postmitotic neurons by Gal4-OK107 fully rescued both the neuroblast proliferation and the axon pruning phenotypes of SMC1-/- neuroblast clones (compare Figure 4A1 to 4B1 for pruning phenotype and 4A2 to 4B2 for proliferation defect). It is difficult to quantify the cell number in these clones because they are densely packed. However, since all cell types are present in the rescued clone, including the latest born α/β neurons (see Figure 4B1), we conclude that the proliferation defect was mostly, if not completely, rescued.
Figure 4. Rescue of Axon Pruning and Proliferation Defect by SMC1 transgene.
(A-B) SMC1-/- MB neuroblast MARCM clones labeled with Gal4-OK107 exhibit unpruned γ neurons (arrows in A1) and reduced cell number (A2, also note the lack of α/β or α’/β’ lobes in A1). Expression of UAS-SMC1-HA transgene in mutant clones (B) rescues both phenotypes (B1, B2). SMC1-HA protein is predominantly located in the nuclei of neurons (B2 right panel).
(C-D) SMC1-/- MB neuroblast MARCM clones labeled with Gal4-201Y exhibit unpruned γ neurons (arrows in C1) and reduced cell number (C2). Expression of UAS-SMC1-HA only in mutant clones by postmitotic Gal4 driver (Gal4-201Y) either rescues pruning completely (20/27) or shows only a single unpruned γ neuron (7/27, D1). Cell number remains unchanged (D2).
Green, Gal4-OK107 (A-B) or Gal4-201Y (C-D) driven mCD8::GFP; magenta, anti-FasII (A1-D1) or anti-HA (A2-D2). Single confocal sections are shown for A2-D2. Scale bars, 20μm.
In contrast, driving the expression of SMC1 only in postmitotic neurons with Gal4-201Y (see Figures 2A, 2B; Yang et al., 1995; Lee et al., 2000b) could not rescue the neuroblast proliferation defect (compare size of clones in Figure 4C2 to 4D2; average clone size is 41.9 ± 2 cells for SMC-/- [n=10] and 43.5 ± 1.6 cells for SMC-/- + UAS-SMC1::HA [n=29]). Remarkably, the axon pruning defect in SMC1-/- MB γ neurons was rescued by postmitotic expression of SMC1 (compare Figure 4D1 to 4C1; complete rescue in 20/27 clones, one remaining dorsal axon in 7/27 clones). These results indicate that SMC1 is required in MB neuroblasts to regulate proliferation, and in postmitotic MB neurons to regulate axon pruning.
SMC1 regulates EcR-B1 expression
In addition to its well-characterized function in sister chromatid cohesion, the cohesin complex has recently been suggested to regulate gene expression through chromatin remodeling (Rollins et al., 1999; Dorsett et al., 2005; reviewed in Dorsett, 2007). A master gene that regulates developmental axon pruning in the fly is the nuclear steroid hormone receptor ecdysone receptor B1 (EcR-B1). EcR-B1 and its co-receptor Ultraspiracle (Usp), respond to a late larval ecdysone pulse by initiating cascades of gene transcription that regulate Drosophila metamorphosis, including axon pruning of MB γ neurons (Figure 5A). usp is ubiquitously expressed while the expression of EcR-B1 is highly regulated. EcR-B1 is expressed in MB γ neurons, but is absent from α’/β’ neurons that do not undergo pruning (Lee et al., 2000a). We therefore tested whether expression of EcR-B1 is altered in SMC1 mutant clones.
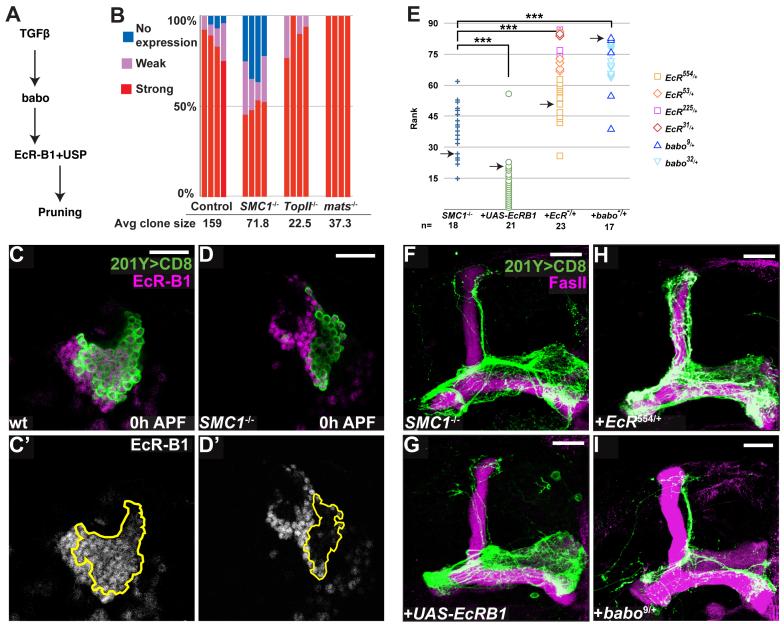
Figure 5. SMC1 Affects Pruning by Regulating the Levels of EcR-B1.
(A) Scheme of TGFβ/EcR-B1 pathway regulating MB axon pruning.
(B) Summary of EcR-B1 expression in MB neuroblasts at 0h APF. 4 neuroblast clones for each genotype were blindly analyzed; cells were classified as expressing high, low or no EcR-B1. Average clone size is also shown.
(C, D) Expression of EcR-B1 (magenta in C, D; white in C’, D’) in wt (C) and SMC1-/- (D) MB neuroblast clones. Extent of the clone is depicted in C’, D’ by a yellow line. Single confocal sections are shown.
(E) Summary of genetic interactions. To analyze the suppression or enhancement of the SMC1-/- pruning phenotype, 79 confocal Z projections from different genotypes were blindly ranked for the severity of the pruning defect. The severity was determined by comparing the unpruned dorsal γ axons to pruned γ axons in the adult-specific medial lobe; these pruned axons can be distinguished from unpruned medial axons as they branch extensively and are located at a more dorsal and posterior position. The ranks grouped by genotypes are shown. Symbols for different EcR and baboon alleles are shown on the right. Pair-wise Mann-Whitney U-tests were performed to determine significance: ***, p<0.001. Arrows indicate the examples shown in (F-I).
(F-I) Genetic interactions between SMC1 and EcR-B1. SMC1-/- MB neuroblast clone (F) with additional expression of UAS-EcR-B1 in mutant clone (G); in an EcR heterozygous background (H); and in a babo heterozygous background (I).
Green represents Gal4-201Y driven mCD8::GFP and magenta is anti-EcR-B1 (C, D) or anti-FasII (F-I). Scale bars, 20μm.
We found large differences in EcR-B1 protein levels between wt and mutant clones. In control clones the vast majority of γ neurons, visualized by Gal4-201Y at 0h APF, express high levels of EcR-B1; only 4% of the cells did not express EcR-B1 and 7% express it at reduced levels (Figure 5C; quantified in 5B). By contrast, in SMC1-/- neuroblast clones, EcR-B1 level is significantly reduced: approximately 30% of the cells express no EcR-B1, while another 20% express low levels (Figure 5D; quantified in Figure 5B).
Since the size of wt and SMC1-/- neuroblast clones differs significantly, we wanted to ascertain that our measurement of EcR-B1-positive cells was not skewed. To this end, we examined EcR-B1 expression in two piggyBac insertions in which clone size is also greatly reduced. As quantified in Figure 5B, in both cases (Top2 and mats) all cells expressed EcR-B1. Indeed, EcR-B1 negative cells in wt clones tend to be located closest to the neuroblast, suggesting that they are the youngest γ neurons that have just differentiated and are beginning to express EcR-B1. Since SMC1-/- (or Top2-/- or mats-/-) neuroblast clones most likely arrest proliferation before 3rd instar larvae judging from their small clone size, these young neurons should not be present. Therefore, in SMC1-/- neuroblast clones, non- or low-EcR-B1 expressing cells are likely early-born and their over-representation is most likely caused by failure of EcR-B1 up-regulation. Perdurance of SMC1 mRNA and protein may account for ∼50% of SMC1-/- neurons that still express high levels of EcR-B1 (see Experimental Procedures). The regulation of EcR-B1 expression appears specific, as four other proteins, including Elav, Dac, Usp and Cut did not show any change of expression in SMC1-/- compared to wt γ neurons (Figure S4).
Lack of EcR-B1 expression may account for the axon pruning defect in SMC1-/- neuroblast clones. Indeed, the percentage of EcR-B1-negative cells correlates well with around 20-30% unpruned axons in SMC1-/- neuroblast clones. To establish a causal relationship, we tested whether changing the dose of endogenous EcR-B1 would affect the pruning defect in SMC1-/- neuroblast clones. We found that reducing the dosage of EcR-B1 by generating SMC1-/- clones in an EcR-/+ background resulted in enhancement of the pruning defects (compare Figure 5H to 5F; quantified in 5E). In addition, SMC1-/- clones in a heterozygous background of babo, a TGFß receptor that regulates expression of EcR-B1 (Zheng et al., 2003), also show enhancement of the phenotype although it also affected clone size (compare Figure 5I to 5F; quantified in 5E). Importantly, overexpression of EcR-B1 markedly suppressed the pruning defects in SMC1-/- neuroblast clones (compare Figure 5G to 5F; quantified in 5E). The dosage-sensitive genetic interactions between SMC1 and EcR-B1, and in particular, the suppression of the SMC1-/- phenotype by EcR-B1 overexpression, strongly suggest that regulation of EcR-B1 expression is a major mechanism by which SMC1 regulates axon pruning. The fact that the suppression is not complete may suggest that either EcR-B1 is not the only SMC1 target gene in regulating axon pruning or the expression of the EcR-B1 transgene is not at a sufficient level, or both.
Postmitotic function of SMC1 in dendrite targeting of olfactory projection neurons
SMC1 and SA mutants were also independently identified in a parallel screen as affecting olfactory projection neuron (PN) dendrite targeting. In this screen, we used Gal4-GH146, which is expressed in ∼90 PNs originating from three different neuroblast lineages generating anterodorsal (ad), lateral (l) and ventral (v) PNs (Stocker et al., 1997; Jefferis et al., 2001). PNs are prespecified by lineage and birth order to target their dendrites to specific glomeruli of the antennal lobe and form synapses with incoming olfactory receptor neuron axons (Jefferis et al., 2001).
Wild-type neuroblast clones from all three PN lineages target stereotypic sets of glomeruli (Figures 6A-C; Jefferis et al., 2001). SMC1-/- PN neuroblast clones have a mild reduction of cell number, likely reflecting proliferation defects similar to those observed in MB neurons. Furthermore, PNs of all three neuroblast lineages show dendrite targeting defects (Figures 6D-F). For example, wt adPNs always target to the VA3 glomerulus, whereas 9 out of 12 examined SMC1-/- adPNs do not innervate VA3 (dotted outlines in Figures 6A and 6D). The failure to innervate VA3 is not a consequence of proliferation defects, since VA1d PNs are born after VA3 PNs in the adPN lineage (Jefferis et al., 2001), and the VA1d glomerulus is always innervated. The DA1 glomerulus is always innervated by wt lPN neuroblast clones but was not innervated by 12 out of 14 SMC1-/- neuroblast clones (dotted outlines in Figures 6B and 6E). In both adPNs and lPNs, lineage-inappropriate glomeruli are often innervated (asterisks in Figures 6D, E). vPNs show the most dramatic phenotype with regard to dendrite targeting. Dendrites spill outside the antennal lobe with extensive branching into the suboesophageal ganglion (SOG) and the lateral side of the antennal lobe (7/7), whereas appropriate target glomeruli (like DA1 and VA1lm) occasionally fail to be innervated (e.g., DA1: 2/7) (Figure 6F). Dendrites and axons of SMC1-/- single cell clones target appropriately (data not shown), likely due to perdurance of SMC1 wt protein. Most axons of SMC1-/- PNs branch and terminate normally in higher brain centers (data not shown); however, we cannot exclude a role of SMC1 in axon targeting due to the lack of resolution in neuroblast clones.
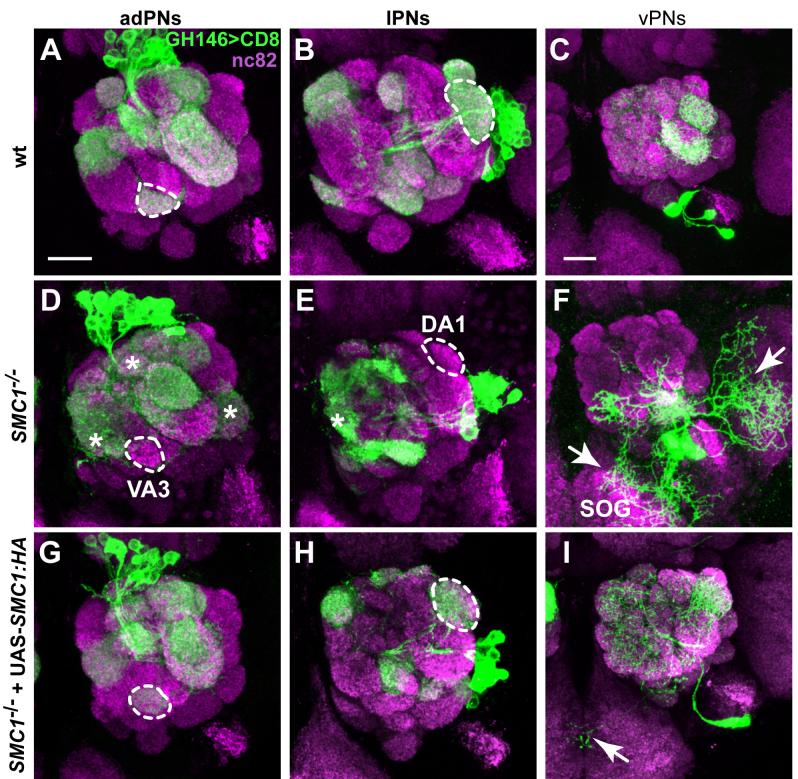
Figure 6. SMC1 Is Required for PN Dendrite Targeting.
(A-C) Stereotypic dendrite projection pattern of wt neuroblast clones that give rise to adPNs (A), lPNs (B), and vPNs (C).
(D-F) SMC1-/- PNs exhibit dendrite targeting defects. SMC1-/- adPNs do not innervate VA3 (9/12; dotted outline, D), but instead project to several inappropriate glomeruli in the medial and dorsal parts of the antennal lobe and VL2a (asterisks, D). SMC1-/- lPNs fail to target DA1 (12/14; dotted outline, E) but innervate additional medial glomeruli (asterisk, E). SMC1-/- vPNs show most severe targeting defects, with large portions of dendrites targeting to the SOG and lateral areas outside the antennal lobe (arrows, F).
(G-I) Postmitotic expression of SMC1::HA in SMC1-/- PNs fully rescues dendrite targeting targeting in adPNs (G) and lPNs (H), respectively. vPN dendrites are either indistinguishable from wt or show rare dendrites wandering to the SOG (arrow, I).
Dotted outlines represent selected glomeruli used for scoring the penetrance of SMC1 mutant phenotypes and corresponding rescues: VA3 for adPNs (in A, D and G), DA1 for lPNs (in B, E and H). Magenta, nc82 as a presynapic marker for all glomeruli; green, UAS-mCD8::GFP driven by GH146-Gal4. Scale bars, 20μm.
To distinguish whether the dendrite targeting defects are caused by the requirement of SMC1 in neuroblasts or postmitotic neurons, we performed rescue experiments by driving the expression of a UAS-SMC1::HA transgene with Gal4-GH146, which is expressed in postmitotic PNs but not in the neuroblast (Spletter et al., 2007). As expected, postmitotic expression of SMC1 does not rescue the reduction of cell number in all three types of neuroblast clones. Targeting of later born PNs is not restored since these PNs may never have been born (e.g., DM6). Nevertheless, we observe a significant rescue of dendrite targeting defects in all three PN lineages (Figures 6G-I). In SMC1-/- neuroblast clones expressing SMC1::HA, adPNs always target to VA3 (7/7) (Figure 6G, dotted outline), lPNs always target to DA1 (6/6) (Figure 6H, dotted outline), and vPNs are either rescued to complete wt innervation pattern (3/5) or show minor dendritic misrouting to the SOG (2/5) (Figure 6I, arrow) with DA1 being targeted in all examined brain hemispheres (5/5). Thus, postmitotic expression of SMC1 is sufficient to rescue PN dendrite targeting defects.
A piggyBac insertion in another cohesin subunit, SA, causes similar, though less severe, PN dendrite defects: 6/15 SA-/- adPN neuroblast clones fail to target VA3, and 2/18 SA-/- lPN neuroblast clones fail to innervate DA1. All neuroblast clones spill into additional glomeruli in medial and dorsal areas of the antennal lobe (Figures S3D-F). Dendrites of vPNs are often misrouted to the SOG (3/8; data not shown).
The mistargeting phenotype of SMC1-/- or SA-/- PNs resembles the phenotype reported for cut-/- PNs (Komiyama and Luo, 2007). As it was previously shown that SMC1 regulates the expression of cut in the developing wing (Dorsett et al., 2005), and Cut is expressed in a subset of PNs (Komiyama and Luo, 2007), we tested whether Cut expression is regulated by SMC1 in PNs. However, we did not find any change in Cut protein level in SMC1-/- clones (Figure S4D-E) nor when UAS-SMC1::HA was expressed in PNs (data not shown).
In summary, mutations in two cohesin subunits, SA and SMC1, lead to similar defects in MB axon pruning as well as similar defects in PN dendrite targeting. Rescue of the SMC1 defects using UAS-SMC1 driven by two different postmitotic Gal4 lines strongly suggests that cohesin regulates diverse aspects of morphogenesis in postmitotic neurons.
DISCUSSION
piggyBac insertional mutagenesis for mosaic screening
Forward genetic screens in mosaic animals are a powerful method to systematically identify genes that are required for complex biological processes occurring late in development. In this study, we modified existing piggyBac tools to be more mutagenic and compatible with mosaic screening.
Previous studies showed that although 80% of piggyBac insertions fall within genes, mutagenicity is decreased by its preferential targeting to introns (Hacker et al., 2003; Bellen et al., 2004). To increase the mutagenicity of intronic insertions, we added to the piggyBac mutator splice acceptors followed by stop codons in all three frames in both orientations. Indeed, the proportion of lethal mutations is significantly higher compared to previous reports, and our intronic insertions are almost as mutagenic as insertions in the 5′ UTR and CDS. According to the postulation that about one third of fly genes are essential for viability (Miklos and Rubin, 1996), our percentage of lethal mutations (∼28%) is approaching the highest possible mutagenicity with insertional mutagenesis.
Insertional mutagenesis screens are time consuming at the front end of generating mutants. To optimize the efficiency of the screen, we developed protocols to obtain the insertion data at the earliest step possible. This allowed about 30% of the insertions (intergenic plus unmapped insertions due to repetitive or short sequence) to be discarded before stock establishment. In addition, knowledge of the insertion site enabled us to use MARCM-ready flies with the appropriate FRT for mosaic screening. Although generating mutants is more time-consuming and costly than EMS mutagenesis, the ease of mapping compensates for the additional effort. Knowledge of both the phenotype and gene identity enables experimenters to make well-informed decisions as to which mutants to pursue further. Moreover, the large collection of molecularly defined piggyBac insertions describe here should be useful to researchers conducting additional mosaic screens or candidate mutant analysis.
Postmitotic neuronal functions of the cohesin complex
Developmental axon pruning of MB γ neurons is a complex process that incorporates cell specificity, spatial restrictions and temporal precision. We identified two subunits of the cohesin complex as essential for pruning. SMC1-/- neuroblast clones also revealed a clear defect in neuroblast proliferation. This provides in vivo support to previous studies showing that precocious sister chromatid separation leads to cell cycle arrest, most likely by triggering the spindle checkpoint mechanism (Vass et al., 2003). A second phenotype in SMC1-/- neuroblast clones is that the postmitotic progeny exhibit a defect in developmental axon pruning.
A possible explanation of the role of SMC1 in axon pruning is that the pruning defect is an indirect consequence of precocious sister chromatid separation. We ruled out this hypothesis by showing that postmitotic expression of SMC1 is sufficient to rescue the pruning phenotype, thus uncovering a novel function for SMC1 in postmitotic neurons. By contrast, the neuroblast proliferation defect could only be rescued when SMC1 was expressed in the dividing neuroblast.
A more mechanistic understanding of the effects of SMC1 comes with the observation that there is a significant reduction of EcR-B1 protein levels in SMC1-/- neuroblast clones of MB γ neurons. In addition, reducing levels of EcR, or the TGF-β receptor Baboon essential for EcR-B1 upregulation, enhances the pruning defect of SMC1-/- neuroblast clones. Moreover, forced expression of EcR-B1 in SMC1-/- neuroblast clones partially suppresses axon pruning defects. Since EcR-B1 is a major regulator of MB axon pruning (Lee et al., 2000), it is likely that regulation of its levels accounts for the function of SMC in axon pruning. Although our study does not establish how SMC1 regulates EcR-B1, a recent study shows that the cohesin complex binds to the EcR locus (Misulovin et al., 2007), strongly suggesting a direct regulation at the transcriptional level.
The mechanism of gene regulation by SMC1 is not well understood. It therefore remains unclear whether SMC1 has a separate function unrelated to its participation in the cohesin complex or that part or the entire cohesin machinery is utilized for postmitotic gene regulation. Our data support the latter hypothesis. We find that a regulatory subunit of cohesin, SA, which does not directly bind SMC1, is also essential for axon pruning. We therefore suggest that SMC1 acts to regulate gene expression as part of the cohesin complex that includes SA.
Our study also suggests that the cohesin complex regulates diverse morphogenesis aspects of postmitotic neurons. We show that both SMC1 and SA are essential for correct dendrite targeting of PNs. Postmitotic expression of wt SMC1 rescues dendrite targeting defects of SMC1 mutant clones. We postulate that cohesin has additional functions in postmitotic neurons as inactivation of the cohesin complex in all postmitotic neurons results in lethality (A. Pauli and K. Nasmyth, personal communication). As complete ablation of the MB (de Belle and Heisenberg, 1994) or large subsets of PNs (Berdnik et al., 2006) does not result in lethality, cohesin must also be needed in other postmitotic neurons. Interestingly, the C. elegans Scc4 homolog, Mau-2, which is part of the cohesin loading complex, is involved in neuronal migration (Takagi et al., 1997; Seitan et al., 2006), again suggesting that cohesin has a wide role in neuronal morphogenesis.
Previous studies have suggested that cohesin inhibits the expression of Cut in the dividing wing disc by interfering with the communication between a distant enhancer and the promoter (Dorsett, 2007). We show that in postmitotic MB neurons SMC1 positively regulates EcR-B1 levels. However, we did not observe any change in Cut levels, both in loss-of-function or overexpression of SMC1 in MB γ neurons or PNs. These observations suggest that cohesin regulates different genes in different developmental contexts.
Mutations in several cohesin subunits such as Smc1a and Smc3, or proteins necessary for its function, such as NIPBL, a homolog of Nipped-B/Scc2, are the causes for a large fraction of a rare disorder called Cornelia de Lange Syndrome (CdLS)(Strachan, 2005; Musio et al., 2006; Deardorff et al., 2007). CdLS patients exhibit multiple physical and neurodevelopmental deficits. Patients with mutations in Smc1a exhibit mild forms of the syndrome, but a mild-to-moderate mental retardation is the most penetrant symptom for these patients. Our finding raises the possibility that in addition to chromatid cohesion, defective SMC1 function in postmitotic neurons may contribute to the neurodevelopmental and mental deficits of CdLS patients.
EXPERIMENTAL PROCEDURES
piggyBac Vector and Mutant Generation
The piggyBac mutator pXL-BacII-SAstopDsRed was generated by cloning an EcoRI-EcoRV fragment from the vector described in Genbank accession #XXXXX into EcoRI-EcoRV restricted pXL-BacII-ECFP (Li et al., 2005). Germline transformation was performed using routine methods by co-injecting the piggyBac-transposase-encoding plasmid phspBac (a gift from A. Handler). The genetic scheme for generating mutants is presented in the Supplemental Experimental Procedures. Flies containing piggyBac transposase were described previously (Hacker et al., 2003). Insertion sites were obtained by performing inverse PCR using a protocol presented in the Supplemental Experimental Procedures. Sequencing and alignments were done by Quintara (http://www.quintarabio.com/).
Mosaic Analyses, Fly Stocks and Transgene Construction
Mosaic analyses using MARCM to generate MB and PN neuroblast clones were performed as previously described (Lee et al., 2000a; Wu and Luo, 2006) using Gal4s - 201Y, -OK107 and -GH146. An intrinsic feature of mosaic analysis including MARCM is that from the moment a clone is generated, no new functional mRNA or protein is made in mutant cells. Nevertheless, pre-existing mRNAs and proteins inherited from heterozygous parental cells can persist and function normally for a certain period of time resulting in perdurance.
To generate UAS-SMC1, cDNA containing the whole coding region was amplified using the following primers: CACCATGACCGAAGAGGACGAC and CGTGTCCTCGAACGTTGTCAAGTC. The product was subcloned into pENTR-D/TOPO (Invitrogen) and then recombined into pTWH (Gateway Collection, Drosophila Genomics Resource Center, Bloomington, IN) using Gateway LR clonase II enzyme mix (Invitrogen). UAS-EcR-B1 was described previously (Lee et al., 2000a); to obtain a second chromosome insertion, we mobilized the transgene by routine P-element transposition. SMC1Δexc46 fly was obtained from S.Page and S. Hawley. All other alleles used in this manuscript were obtained from the Bloomington stock center.
Immunostaining
Fly brains were dissected, fixed and processed for whole-mount immunostaining as previously described (Lee et al., 1999; Lee and Luo, 1999). Conditions for primary antibodies are in the Supplemental Experimental Procedures.
RT-PCR
For semi-quantitative RT-PCR, cDNA equivalent to ∼1-0.5 ng RNA was used for amplification at 64°c for 27 cycles using phusion taq (Finnzymes, Espoo, Finland). For α-tubulin and β-actin controls, a sample diluted by 4 was used. Primer sequences can be obtained upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank U. Hacker, D. Dorsett, A. Handler, S. Page, S. Howley, E. Buchner, R. Barrio and the Bloomington stock center for reagents; K. Nasmyth and A. Pauli for sharing unpublished data and for comments and discussion; T. Clandinin, M. Schuldiner and members of the Luo lab, especially E. Hoopfer and B. Tasic, for comments and discussion; and C. Guo, N. Woodling, W. Hong and A. Fan for assistance. This work was supported by fellowships from EMBO (OS), HFSP (DB), and an NIH grant R37-NS041044 (LL). LL is an HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Chihara T, Couto A, Luo L. Wiring stability of the adult Drosophila olfactory circuit after lesion. J Neurosci. 2006;26:3367–3376. doi: 10.1523/JNEUROSCI.4941-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0079. RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Sloulakis EMC, Han K-A, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Memory. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative Odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nystedt S, Barmchi MP, Horn C, Wimmer EA. piggyBac-based insertional mutagenesis in the presence of stably integrated P elements in Drosophila. Proc Natl Acad Sci USA. 2003;100:7720–7725. doi: 10.1073/pnas.1230526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O’Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Horn C, Jaunich B, Wimmer EA. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000;210:623–629. doi: 10.1007/s004270000111. [DOI] [PubMed] [Google Scholar]
- Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurons and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jager H, Herzig A, Lehner CF, Heidmann S. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 2001;15:2572–2584. doi: 10.1101/gad.207301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000a;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000b;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Li X, Harrell RA, Handler AM, Beam T, Hennessy K, Fraser MJ., Jr. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XB, Low LK, Jones EG, Cheng HJ. Stereotyped axon pruning via plexin signaling is associated with synaptic complex elimination in the hippocampus. J Neurosci. 2005;25:9124–9134. doi: 10.1523/JNEUROSCI.2648-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Luo L, O’Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- Miklos GL, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li X, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2007 doi: 10.1007/s00412-007-0129-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. How might cohesin hold sister chromatids together? Philos Trans R Soc Lond B Biol Sci. 2005;360:483–496. doi: 10.1098/rstb.2004.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000a;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson BJ. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000b;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–793. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Whitmore AV, Finn JT. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M, Tomita S, Sung C, Robinow S, Truman JW. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev. 2003;120:909–918. doi: 10.1016/s0925-4773(03)00134-5. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Develop. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Strachan T. Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr Opin Genet Dev. 2005;15:258–264. doi: 10.1016/j.gde.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Takagi S, Benard C, Pak J, Livingstone D, Hekimi S. Cellular and axonal migrations are misguided along both body axes in the maternal-effect mau-2 mutants of Caenorhabditis elegans. Development. 1997;124:5115–5126. doi: 10.1242/dev.124.24.5115. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–218. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nature protocols. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O’Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.