Abstract
Proteins that share common ancestry may differ in structure and function because of divergent evolution of their amino acid sequences. For a typical diverse protein superfamily, the properties of a few scattered members are known from experiment. A satisfying picture of functional and structural evolution in relation to sequence changes, however, may require characterization of a larger, well chosen subset. Here, we employ a “stepping-stone” method, based on transitive homology, to target sequences intermediate between two related proteins with known divergent properties. We apply the approach to the question of how new protein folds can evolve from preexisting folds and, in particular, to an evolutionary change in secondary structure and oligomeric state in the Cro family of bacteriophage transcription factors, initially identified by sequence-structure comparison of distant homologs from phages P22 and λ. We report crystal structures of two Cro proteins, Xfaso 1 and Pfl 6, with sequences intermediate between those of P22 and λ. The domains show 40% sequence identity but differ by switching of α-helix to β-sheet in a C-terminal region spanning ≈25 residues. Sedimentation analysis also suggests a correlation between helix-to-sheet conversion and strengthened dimerization.
Keywords: conformational switching, structural evolution, transitive homology, x-ray crystallography
The amino acid sequences of proteins evolve faster than the structures and functions encoded by these sequences. This neutral sequence drift allows annotation of an uncharacterized protein-coding gene based on common ancestry (homology) with a characterized gene, even if the protein sequences are quite different. Conservation of structure and function may hold even for homology so distant that no clear sequence similarity has survived evolutionary divergence. Yet there are limits: the structural and functional evolution of proteins is not completely static, and the likelihood of two proteins evolving divergent properties increases with the extent of sequence change separating them. Remote homology detection methods [for example, PSI-BLAST (1), COMPASS (2), and HHpred (3)] thus yield diminishing returns for gene annotation by grouping distantly related proteins into superfamilies that encompass diverse properties and biological roles. Simultaneously, however, the excavation of distant relationships opens a rich field for experimental studies of protein evolution, with the promise of recovered annotation power as one elucidates how structure and function vary across the “sequence space” of a superfamily.
Transitive sequence comparison is one method for detecting distant homology between highly diverged sequences (4–8). In this approach, two dissimilar sequences, A and C, are indirectly linked if a third “intermediate” sequence B exists with sufficient similarity to both A and C to imply homology with both proteins. The relationships between A and B and between B and C combine to support distant common ancestry between A and C. Transitivity can extend to several steps with multiple intermediate sequences (6, 7). In effect, transitive homology detection entails a multistep voyage through sequence space, during which the sequence of one extant protein is gradually transmuted into that of a relative through other extant homologs that serve as stepping stones.
This feature of transitive homology also provides a targeting protocol for experimental surveys of evolutionary variation within superfamilies. If the properties of two proteins at the endpoints of a transitive pathway are known to differ because of divergent evolution, experimental characterization of extant intermediate sequences is an intuitive approach to understanding how structure and function vary with sequence. Here, we apply such a stepping-stone targeting approach to the question of how new protein folds evolve, and specifically to an evolutionary secondary structure switching event in the Cro superfamily of bacteriophage DNA-binding proteins. In the process, we discover perhaps the most dramatic case of similar homologous protein sequences with different folds. In Results and Discussion, we compare the approach used here to the nice recent work of Lupas and coworkers (9), who studied β-barrel evolution, using a version of intermediate sequence targeting based on profile comparisons rather than on pairwise relationships.
Results and Discussion
Cro proteins (Fig. 1) are a salient case of remote homology detection revealing a radical evolutionary change (10). Cro homologs from bacteriophages P22 and λ have only 25% sequence identity, but several arguments indicate their distant homology (specifically, orthology): (i) a conserved gene context in the two different bacteriophage species (11–13); (ii) strong parallelism in DNA-binding function and biological role (14–16); (iii) partial structural similarity, consisting of three α-helices in a similar arrangement (10, 17, 18); and (iv) remote sequence homology suggested by PSI-BLAST and transitive homology analyses (10). Despite common ancestry, the two proteins have radically different structures (Fig. 1): P22 Cro adopts an all-α helical fold and is monomeric in solution, whereas λ Cro has a mixed α-helix/β-sheet fold and dimerizes at low micromolar concentrations or below (19–21).
Fig. 1.
Transitive homology chain connecting P22 Cro and λ Cro, which have different domain folds (subunit ribbon diagrams in green) and oligomeric states (ribbon diagram of second λ subunit shown in gray). Each “link” represents a pairwise comparison of two Cro sequences with ≈40% sequence identity, measured either across the length of the structured domain (green) or across the local region aligned in a BLAST comparison (red). For each linked pair, a BLAST E value is also shown, obtained by using a compositionally adjusted BLOSUM80 matrix with gap opening and extension penalties of 11 and 1, respectively. Green lines show approximate boundaries of the folded domain, and red lines show boundaries of each local BLAST alignment. For each BLAST alignment, red letters show identical residues, and plus signs show similar residues.
How do these structural properties vary across the Cro superfamily? In previous studies, newly sequenced microbial genomes enabled assembly of a database of 56 bona fide Cro superfamily members, some deriving from free bacteriophage and some from bacterial prophages (10). Three prophage Cro proteins in this database, originally called AF01p, XA02p, and PF01p [here renamed Afe01, Xfaso 1, and Pfl 6, respectively; see supporting information (SI) Methods] were used as sequence intermediates in a transitive homology analysis connecting P22 Cro to λ Cro (Fig. 1) (10). The chain links consist of four overlapping pairwise sequence comparisons, each of which shows ≈40% sequence identity. To investigate how Cro structural properties vary across the transitive chain, we cloned, expressed, and purified each stepping-stone protein.
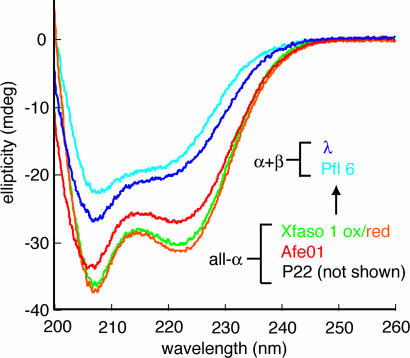
We first characterized the secondary structure, using far-UV circular dichroism (CD) (Fig. 2). For Xfaso 1, the presence of two cysteines (Cys 42 and Cys 55) suggested disulfide bonding. Purified Xfaso 1 was indeed found to be capable of forming intrasubunit cystine linkages, and we decided to study both reduced and oxidized forms. Afe01 and Xfaso 1 (both oxidized and reduced) showed strong negative peaks at both 208 and 222 nm, indicative of α-helical secondary structure. Pfl 6 gave a weaker signal with an overall spectral shape similar to that of λ Cro and no minimum at 222 nm. These results suggest that Pfl 6 shares similar secondary structure, and perhaps the same fold, as λ Cro, whereas Afe01 and Xfaso 1 have higher helical content and perhaps share the fold of P22 Cro. Thus, the data suggest structural crossover between Xfaso 1 and Pfl 6 on the pathway (Fig. 1).
Fig. 2.
Variation of secondary structure for stepping-stone Cro proteins. Far UV CD spectra are shown for Afe01, Xfaso 1, Pfl 6, and λ Cro at 15°C under comparable conditions (see Methods). A spectrum of P22 Cro is not shown because of contributions of aromatic side chains that mask helical content.
We next characterized the oligomeric state, using sedimentation equilibrium (Table 1). Fits of equilibrium concentration curves at 50–250 μM for Afe01 and Xfaso 1 (both oxidized and reduced) gave apparent molecular weights within 10% of theoretical values for monomers. Data for Afe01 suggested some weak tendency to self-associate, based on an apparent molecular weight that was 7% higher than the true monomer molecular weight at 250 μM, but fits at lower concentrations showed values within 2%, suggesting a clean monomer. Pfl 6 showed at least 15% higher values than the theoretical monomer weight at all concentrations between 100 and 250 μM. Fits to a monomer-dimer equilibrium model yielded dissociation constants between 0.7 and 1.3 mM at three concentrations and rotor speeds. Overall, current and previous data suggest essentially no dimerization for P22 Cro (10), Afe01, and Xfaso 1, weak dimerization for Pfl 6 (Kd ≈1 mM), and stronger self-association for λ Cro (Kd ≈3 μM) (19, 20).
Table 1.
Sedimentation equilibrium of stepping-stone Cro proteins
| Protein | Mapp | Mmon | Ratio | Kd, mon/dim |
|---|---|---|---|---|
| Afe01 | 9,100 | 8,470 | 1.075 | ≈5,000 μM* |
| Xfaso 1 | 9,330 | |||
| Oxidized | 9,350 | 1.002 | n.d. | |
| Reduced | 9,488 | 1.017 | n.d. | |
| Pfl 6 | 11,080 | 8,370 | 1.323 | 917 ± 64 μM† |
n.d., not determined; Mapp, apparent molecular weight from fits to single ideal species at 220–250 μM; Mmon, true monomer molecular weight.
*Calculated from average of monomer–dimer (mon/dim) fits at 23,000, 30,000, and 37,000 rpm and 250 μM protein. The number reported is an estimate, because oligomerization was not apparent at lower concentrations.
†Calculated from average of monomer-dimer fits at 23,000, 30,000, 37,000 rpm and three concentrations between 50 and 250 μM.
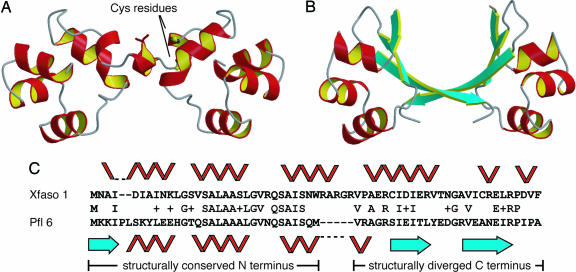
To confirm the structural transition between Xfaso 1 and Pfl 6 suggested by CD, we solved their crystal structures (1.4 Å resolution for reduced Xfaso 1; 1.7 Å for SeMet-labeled Pfl 6; see Fig. 3 and SI Tables 2 and 3). Subunit structures in the two proteins represent different folds, with reduced Xfaso 1 having an all α-helical fold similar to P22 Cro and Pfl 6 having an α+β fold like λ Cro. The asymmetric unit of Pfl 6 contains a pseudosymmetrical dimer qualitatively similar to that of λ Cro in its DNA complex (compare Figs. 1 and 3B; see also Fig. 4) (17). The asymmetric unit of Xfaso 1 contains three subunits, two of which form a putatively biological dimer (Fig. 3A) with helix–turn–helix motifs oriented like those in Pfl 6 and λ Cro. The C-terminal regions of the Pfl 6 and Xfaso 1 sequences (last 27 residues of the alignment in Fig. 3C) adopt completely different backbone structures and participate in completely distinct protein–protein interfaces within the dimers (Fig. 3 A and B). The third subunit of Xfaso 1 (data not shown) does not associate with the other two in any obviously functional manner. The crystallization of Xfaso 1 as a mixture of an apparent monomer and a biological dimer is consistent with its weaker solution dimerization relative to Pfl 6 and λ Cro. In all Xfaso 1 subunits, residues Cys 42 and Cys 55 are adjacent (Fig. 3), suggesting that a disulfide bond could form within the helical fold, in agreement with CD data (Fig. 2).
Fig. 3.
Comparison of Xfaso 1 and Pfl 6. (A and B) Crystal structures of Xfaso 1 (A) and Pfl 6 (B) with ribbon diagrams for the biological dimers shown. The Xfaso 1 asymmetric unit has a third subunit (data not shown). Cys 42 and Cys 55 are indicated for one subunit of Xfaso 1 to show spatial proximity in the reduced form. (C) One possible sequence alignment of Xfaso 1 and Pfl 6, annotated with secondary structures. This alignment gives 40% sequence identity across 65 residues, with two gaps. The unstructured C termini of Xfaso 1 (16 residues) and Pfl 6 (7 residues) are not included in the alignment.
Fig. 4.
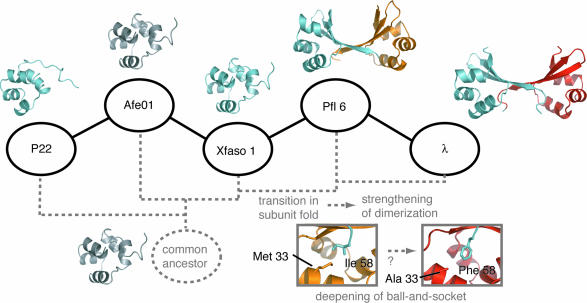
Summary of stepping-stone results and working model for Cro structural evolution. Hypothetical or qualitative aspects are gray. For example, structures of the common ancestor and Afe01 are known only at the level of general fold either from previous outgroup analysis (ancestor) or from low-resolution data in the present study (Afe01). One possible phylogenetic tree topology is indicated by dashed gray lines. The different colors of the second subunits shown for Pfl 6 Cro (gold) and λ Cro (red) are intended to indicate the stronger dimerization of the latter. (Insets) Residues in the ball-and-socket region of Pfl 6 Cro and λ Cro.
Fig. 4 summarizes our results. Perhaps our most striking finding is the sudden crossover from an all-α to an α+β fold between the third and fourth stepping stones, Xfaso 1 and Pfl 6. No intermediate folds are observed; instead, Xfaso 1 and Pfl 6 recapitulate the structural differences seen between P22 Cro and λ Cro but have more similar sequences (≈40% identity vs. 25% identity). This crossover also reveals that sequence similarity and structural similarity are not strictly correlated in Cro proteins: P22 Cro and Xfasa 1 have 26% sequence identity across the stacked domain alignment shown in Fig. 1, but have the same fold and a 3.0 Å backbone rmsd across this region; Xfasa 1 and Pfl 6, which have ≈40% sequence identity, have different folds and a 6.2 Å backbone rmsd. Our second major finding is that fold and oligomerization show some relationship. Although Cros must dimerize to bind their full DNA sites, only the α+β Cros show obvious solution dimerization among the proteins studied here. Interestingly, however, Pfl 6 dimerizes less strongly than λ Cro despite sharing its α+β fold and a similar dimer structure. Moreover, the all-α helical Xfaso 1 can form dimers within crystals, confirming that it can self-associate, albeit weakly, and allowing visualization of how the secondary structure switch remodels the dimer interface (Fig. 3).
The crossover in fold between Xfaso 1 and Pfl 6 has important implications. Most obviously, the sequence similarity between the two proteins buttresses the hypothesis that Cro proteins with different folds are truly homologous. Further, the similarity appears global, spanning both the structurally conserved N-terminal region (48% identity in Fig. 3) and the structurally diverged C terminus (37% identity). This confirms that the homology is global and implies that the C-terminal change from helix to sheet arose from conformational switching induced by simple substitutions or small indels, not from en bloc nonhomologous sequence replacements. In our previous study, we used distant homology analysis of P22 Cro and λ Cro to support such a “homologous switching” mechanism (10). The direct pairwise similarity of Xfaso 1 and Pfl 6 now offers a stronger prima facie case. In SI Methods, we offer additional evidence for homology, including PSI-BLAST and gene context arguments (SI Figs. 5 and 6). The greater sequence similarity between Xfaso 1 and Pfl 6 compared with P22 and λ also suggests that evolution of the Cro fold could have been relatively recent, because more similar sequences on average will share a more recent common ancestor. Finally, the absence of intermediate forms on the pathway is consistent with, but does not prove, the model that Cro fold evolution is an all-or-nothing switch in secondary structure rather than a multistep structural transformation.
As similar sequences with different folds, Xfaso 1 and Pfl 6 are a rarity, and perhaps the most dramatic counterexample to the textbook view that global sequence similarity between two natural proteins implies global structural similarity. We know of no other case in which two natural protein domains with ≈40% sequence identity have different folds, although pairs of differently folded domains with 30–35% identity have been described (22, 23). Other cases of apparent evolutionary switching between α-helix and β-sheet are known (24), most dramatically the recent comparison of RfaH and NusG, suggesting wholesale α-helix to β-sheet conversion in a 45-residue aligned region with slightly <20% pairwise sequence identity (25). To our knowledge, no examples exhibit the combination of sequence identity and radical topological change seen in the Cro domains.
Evolution of stronger dimerization may have accompanied changes in Cro fold. Stronger dimerization of Pfl 6 and λ Cro relative to all-α Cro proteins accords with this hypothesis, and comparison of Pfl 6 and Xfaso 1 dimers (Fig. 3) illustrates that the β-sheet interface is more intertwined. Still, Pfl 6 dimerizes less strongly than λ Cro. On the basis of site-directed mutagenesis, we proposed previously that the low micromolar dimerization of λ Cro did not result directly from its β-sheet fold but derived in part from specific side-chain mutations yielding a hydrophobic “ball and socket” at both ends of the interface (19, 26). These previous studies implicated Ala 33 and Phe 58 as key players in dimer evolution (Right Inset, Fig. 4). Notably, these residues differ in Pfl 6 (Met 33 and Ile 58) and form a shallower ball and socket (Left Inset, Fig. 4). An alternative model therefore emerges, involving incremental ball and socket development and strengthened dimerization after the secondary structure switch (see Fig. 4).
Models of Cro evolution may be further tested and developed by mapping the newly gained data onto a robust molecular phylogeny. Fig. 4 shows one of many possible phylogenetic trees for the Cros on our transitive path, constructed under the crude assumption that branching order approximately follows sequence similarity. Construction of a rigorous global Cro tree is in progress, but it is a nontrivial undertaking due both to extreme sequence diversity and to the lack of reference organismal trees for bacteriophage, which have highly mosaic genomes (27). In some cases, reliability of phylogenetic reconstructions may be enhanced by eliminating divergent taxa that lead to long branches (28); toward this end, the improved mapping of structure to sequence gained here may guide selection of an evolutionarily relevant subset of Cro sequences that share a relatively recent common ancestor. Thus, the stepping-stone analysis should initiate an iterative dialogue between experimental characterization and phylogenetic reconstruction. It may also enhance accuracy of structural annotation and modeling for uncharacterized cro genes.
Stepping stone methods based on pairwise transitive homology are practical approaches to investigating natural structural and functional variation in proteins in relation to sequence variation. We note, however, that mathematically speaking the natural sequences in a transitive homology chain certainly do not form the shortest possible route between the endpoint homologs in sequence space. One can imagine and investigate shorter pathways by designing artificial chimeric proteins, and studies of this can be very informative (29); in our work on Cro evolution, we recently investigated designed sequences with up to ≈50% identity to both P22 Cro and λ Cro (30). What transitive homology uniquely offers is a route involving relatively short steps between existing populated centers in sequence space, much as one might fly from New York to Los Angeles via Houston rather than via Kankakee, which lacks a major airport. Naturally occurring sequences have the virtue of being readily cloned, probably stable and functional, and reflective of natural evolution.
Transitive pathways can be generated by many approaches such as iterated similarity searches (6, 7) from a single homolog and clustering of databases of known homologs (10). For many protein superfamilies, numerous nearly “degenerate” routes through sequence space will exist, and targeting of multiple paths may be indicated. The validity of transitive paths as a targeting protocol should not depend critically on the similarity cutoff used to establish links, provided some evidence establishes homology between linked proteins. For example, in the Cro transitive path, some E-values are of borderline statistical significance (see SI Methods), but PSI-BLAST and gene context arguments offer further support for homology (SI Figs. 5 and 6). In the extreme, there are approaches to identifying intermediate sequences that do not rely on pairwise sequence similarity at all but instead use profile comparisons to establish more remote linkages. Lupas and coworkers recently used Hidden Markov model comparisons to identify sequences with distant similarity to both of two groups of proteins with different β-barrel folds (9). Solving the structure of one of these intermediate sequences led to insights on the origin of the two different barrel topologies. Such profile-based approaches could be considered variations on the stepping-stone theme, in which individual steps may represent longer evolutionary distances.
Methods
Cloning.
Genes for Cro proteins from prophages Pfl 6 and Xfaso 1 were obtained by PCR amplification from purified genomic DNA of host bacterial strains. Genomic DNA of Pseudomonas fluorescens Pf-5 was purified from cultures grown from a freeze-dried sample purchased from the American Type Culture collection. Genomic DNA for Xylella fastidiosa Ann-1 was obtained as a gift from B. Feil and H. Feil (University of California, Berkeley, CA). For prophage Afe01 Cro, a synthetic gene was constructed from two pairs of mutually priming oligonucleotides. Genes were tagged with NdeI and PaeR7I restriction sites to allow ligation into a pET21b NdeI-PaeR7I fragment (Novagen). Ligation yielded constructs for expression of each protein with a C-terminal LEHHHHHH tag. Expression constructs for untagged proteins were later obtained by introduction of stop codons, using QuikChange mutagenesis (Stratagene).
Protein Expression and Purification.
Histidine-tagged Afe01 and Pfl 6 proteins were overexpressed in Escherichia coli strain BL21(λDE3) and purified by denaturing Ni-NTA agarose affinity chromatography as described in ref. 19, followed by dialysis into SB250 buffer [50 mM Tris (pH 7.5), 250 mM KCl, 0.2 mM EDTA] and size-exclusion chromatography on a Sephacryl S-100 26/60 column. Purification of tagged oxidized Xfaso 1 included 15 mM β-mercaptoethanol (BME) in lysis and affinity column buffers and involved two additional steps. First, Ni-NTA eluate was dialyzed into buffer B [10 mM Tris (pH 7.5), 0.2 mM EDTA] containing 3 mM BME and chromatographed on a Mono S HR 10/10 column, using a 0–1 M sodium chloride gradient. Mono S fractions were combined, diluted to 80 μM, and dialyzed into 50 mM ammonium bicarbonate then further diluted to 25 μM in the same buffer, with addition of sodium azide to a concentration of 0.01%. Disulfide bond formation was then achieved by stirring in air for 2 days at ambient temperature. The resulting mixture was centrifuged at 12,000 × g for 30 min to remove precipitates, concentrated to 4 ml, recentrifuged for 30 min at 12,000 × g, and loaded onto the size-exclusion column. Reduced Xfaso 1 was generated by dialysis of oxidized Xfaso 1 into reducing buffer [100 mM sodium phosphate (pH 6.0), 5 mM EDTA], mixing with an equal volume of reducing agent (50 mM DTT, 100 mM BME), and incubating the mixture for 1 h at 37°C. Quantitative air oxidation and rereduction of Xfaso 1 samples could be verified by using Ellman's reagent and one-dimensional NMR spectra. Afe01 and Xfaso 1 concentrations were estimated from A280 values, using an ε280 of 5,559 cm−1 (for a single tryptophan residue) plus a 125 cm−1 correction for the cystine linkage in oxidized Xfaso 1 (31). For Pfl 6 and λ, the absence of tryptophan residues reduces accuracy of predicted ε280 values (31), so concentrations were determined by using ε280 values of 3,119 and 4,040 cm−1, respectively, measured by the Edelhoch method (32).
Untagged Pfl 6 and Xfaso 1 were overexpressed in LB medium in a similar manner as the tagged variants, using 2-h induction periods at 37°C. For SeMet-labeled untagged Pfl 6, LB was replaced by M9T medium, supplemented just before induction with l-selenomethionine at 60 mg/liter and other amino acids as described in ref. 33. Cells harvested from 4 liters of culture were lysed by sonication and the lysate subjected to selective precipitation essentially as described for P22 Cro (10), using a 46% ammonium sulfate cut for Xfaso 1 and a 55% cut for Pfl 6. Proteins remaining in solution were collected by precipitation in 90–97% saturated ammonium sulfate, resuspended in 10 ml of PC buffer [20 mM Tris (pH 8.0), 0.1 mM EDTA, 5% glycerol, and 1.4 mM BME], dialyzed extensively against PC buffer at 4°C, and centrifuged to remove precipitates. For Pfl 6, this dialysate was applied to a 20-ml HiPrep 16/10 Heparin FF column equilibrated with PC buffer, using a sodium chloride gradient. Pooled fractions containing Pfl 6 Cro (which eluted at 400–500 mM sodium chloride) were concentrated to a volume of 4 ml, dialyzed extensively against PC buffer plus 200 mM NaCl, and chromatographed in the same buffer on a Sephacryl S-100 HiPrep 26/60 size-exclusion column. Appropriate fractions were dialyzed against Buffer B [10 mM Tris (pH 7.5) and 0.2 mM EDTA] and concentrated to 18 mg/ml (9 mg/ml for the selenomethionine-labeled preparation). Purified Pfl 6 was stored at −80°C in aliquots containing sodium azide (0.01% wt/vol). For reduced Xfaso 1, the crude dialysate was chromatographed on a Mono S HR 10/10 column equilibrated with PC buffer, using a sodium chloride gradient. Pooled fractions containing Xfaso 1 Cro (which eluted at ≈200 mM salt) were concentrated to 2 ml and chromatographed on a Sephacryl S-100 26/60 size-exclusion column as with Pfl 6. Xfaso 1 Cro showed an elution volume of ≈220 ml. Pooled fractions containing Xfaso 1 Cro were concentrated to 5 ml and dialyzed into reducing buffer. The resulting solution was mixed with an equal volume of reducing agent and mixed for 1 h at 37°C. After reduction, Xfaso 1 was extensively dialyzed into crystallization buffer [10 mM Hepes (pH 7.5), 200 mM NaCl, 1 mM EDTA, and 1 mM DTT] and concentrated to 24 mg/ml.
Biophysical Characterization.
CD wavelength scans were obtained with 50 μM tagged protein in 100 mM sodium phosphate (pH 7, with 1 mM BME added in the case of reduced Xfaso 1) at a 0.05 cm pathlength in a Jasco J-710 CD spectropolarimeter at 15°C. Thermal denaturation curves (data not shown) gave midpoints of at least 45°C for all proteins, indicating that wavelength scans at 15°C should reflect native structure. Sedimentation equilibrium experiments were performed at 20°C on a Beckman XL-I analytical ultracentrifuge, on samples dialyzed extensively in SB250 buffer. BME (1.5 mM) was included in the case of reduced Xfaso 1. Radial distribution curves were measured at three concentrations from 50 to 250 μM and three rotor speeds from 23,000 to 37,000 rpm. Data were measured as averages of 10–25 replicate radial scans at wavelengths of 280–305 nm, with a radial spacing of 0.001 cm. Sedimentation curves were fit by using Kaleidagraph (Synergy Software) to single-species models to obtain apparent molecular weights, and to monomer–dimer models where warranted to obtain dissociation constants. Relevant parameters were estimated by using SEDNTERP (J. Philo, Thousand Oaks, CA, and Reversible Associations in Structural and Molecular Biology).
Crystallography.
Untagged reduced Xfaso 1 was crystallized by the hanging drop method. Drops contained 2 μl of protein solution and 2 μl of buffered precipitant [0.1 M Tris (pH 8.5), 1.5 M ammonium sulfate, 16% glycerol, and 1 mM tris(2-carboxyethyl)phosphine·HCl). Trigonal crystals in space group P32 formed within 3–4 days as tapered or clustered rods up to 1 mM long with triangular cross sections measuring 0.05 mM on each side. Crystals were flash-frozen in liquid nitrogen after being transferred to reservoir solution in which ammonium sulfate was replaced with lithium sulfate. Data were collected on crystals cooled at 100K at APS beamline 14 BM-C, using an ADSC Quantum-Q315 detector, and processed and scaled by using d*trek (34). The phase problem was solved with ACORN (35), using residues 12–35 of chain B of the structure of N15 Cro (PDB 2HIN, M. S. Dubrava, W.M.I., S.A.R., and M.H.J.C., unpublished work) as a “seed,” with sidechains removed except for Val 22, Val 27, and Trp 30, which were left intact, and Asp 12, Glu 14, and Tyr 28, which were converted to Ser residues. Solvent flattening was performed by using DM (36) followed by automatic building with RESOLVE (37), resulting in an 18-residue initial model. ARP/WARP (38) was used to extend the model to 27 residues and produced a map in which density for all three subunits was evident. Manual building in COOT (39) extended the fragment to a full subunit, and molecular replacement with Molrep (40) was then used to place the two remaining subunits into the map. Refinement, using anisotropic temperature factors, was carried out with Refmac5 (41), and manual rebuilding was carried out in COOT. Most programs were accessed through the CCP4 suite (42). Statistics are reported in SI Table 2.
Untagged selenomethionine-labeled Pfl 6 was crystallized by the hanging drop method. Single hexagonal bipyramidal crystals of SeMet-Pfl 6 in space group P3121 grew in 3–6 days from drops initially containing 2 μl of protein (9 mg/ml) and 2 μl of buffered precipitant [100 mM Tris (pH 9.5) and 2.2 M ammonium sulfate]. Native crystals of Pfl 6 grew under similar conditions and were isomorphous. Multiple wavelength data were collected at the Stanford Synchrotron Radiation Laboratory, Beam line 9-2. Heavy atom positions were obtained and phases calculated by using SOLVE (43), which gave a mean figure of merit of 0.46, using data to 1.7 Å. Density modification and model building was performed by using RESOLVE (44, 45). Standard refinement, using Refmac5 (41), yielded an R-value of 26% and and Rfree value of 30%. TLS analysis of the structure was then performed by using the TLS motion determination server (46, 47). Ten rounds of TLS refinement were conducted by using 10 TLS groups per chain followed by 10 rounds of coordinate refinement, yielding improvements of 3–4% in R and Rfree. Statistics are reported in SI Table 3.
Structure figures were generated by using PyMol (48) (Figs. 1 and 4) or Molscript (49) (Fig. 3).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sherwood Casjens, Joyce Loper, Robert Dodson, and Natalia Ivanova for communications and discussion concerning prophage nomenclature and gene annotation; Bill Feil, Helene Feil, and Alexander Purcell (University of California, Berkeley, CA) for gifts of DNA; Annie Heroux, Angela Amoia, and Andrzej Weichsel for assistance and advice; and Vahe Bandarian for helpful discussions. This work was supported by National Institutes of Health Grants GM066806 (to M.H.J.C.) and HL62969 (to W.R.M.). Parts of this work were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under Grant RR07707.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3BD1 (Xfaso 1 Cro) and 2PIJ (Pfl 6 Cro)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711589105/DC1.
References
- 1.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadreyev RI, Baker D, Grishin NV. Profile-profile comparisons by COMPASS predict intricate homologies between protein families. Protein Sci. 2003;12:2262–2272. doi: 10.1110/ps.03197403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolten E, Schliep A, Schneckener S, Schomburg D, Schrader R. Clustering protein sequences–structure prediction by transitive homology. Bioinformatics. 2001;17:935–941. doi: 10.1093/bioinformatics/17.10.935. [DOI] [PubMed] [Google Scholar]
- 5.Gerstein M. Measurement of the effectiveness of transitive sequence comparison, through a third “intermediate” sequence. Bioinformatics. 1998;14:707–714. doi: 10.1093/bioinformatics/14.8.707. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Pio F, Pawlowski K, Godzik A. Saturated BLAST: an automated multiple intermediate sequence search used to detect distant homology. Bioinformatics. 2000;16:1105–1110. doi: 10.1093/bioinformatics/16.12.1105. [DOI] [PubMed] [Google Scholar]
- 7.Salamov AA, Suwa M, Orengo CA, Swindells MB. Combining sensitive database searches with multiple intermediates to detect distant homologues. Protein Eng. 1999;12:95–100. doi: 10.1093/protein/12.2.95. [DOI] [PubMed] [Google Scholar]
- 8.Park J, Teichmann SA, Hubbard T, Chothia C. Intermediate sequences increase the detection of homology between sequences. J Mol Biol. 1997;273:349–354. doi: 10.1006/jmbi.1997.1288. [DOI] [PubMed] [Google Scholar]
- 9.Coles M, et al. Common evolutionary origin of swapped-hairpin and double-psi beta barrels. Structure (London) 2006;14:1489–1498. doi: 10.1016/j.str.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Newlove T, Konieczka JH, Cordes MH. Secondary structure switching in Cro protein evolution. Structure (Cambridge) 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 11.Vander Byl C, Kropinski AM. Sequence of the genome of Salmonella bacteriophage P22. J Bacteriol. 2000;182:6472–6481. doi: 10.1128/jb.182.22.6472-6481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedulla ML, et al. Corrected sequence of the bacteriophage p22 genome. J Bacteriol. 2003;185:1475–1477. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 14.Johnson AD, Meyer BJ, Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci USA. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AD, Pabo CO, Sauer RT. Bacteriophage lambda repressor and cro protein: Interactions with operator DNA. Methods Enzymol. 1980;65:839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- 16.Poteete AR, Hehir K, Sauer RT. Bacteriophage P22 Cro protein: Sequence, purification, and properties. Biochemistry. 1986;25:251–256. doi: 10.1021/bi00349a035. [DOI] [PubMed] [Google Scholar]
- 17.Albright RA, Matthews BW. Crystal structure of lambda-Cro bound to a consensus operator at 3.0 A resolution. J Mol Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- 18.Ohlendorf DH, Tronrud DE, Matthews BW. Refined structure of Cro repressor protein from bacteriophage lambda suggests both flexibility and plasticity. J Mol Biol. 1998;280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- 19.LeFevre KR, Cordes MH. Retroevolution of lambda Cro toward a stable monomer. Proc Natl Acad Sci USA. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia H, Satumba WJ, Bidwell GL, III, Mossing MC. Slow assembly and disassembly of lambda Cro repressor dimers. J Mol Biol. 2005;350:919–929. doi: 10.1016/j.jmb.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 21.Darling PJ, Holt JM, Ackers GK. Coupled energetics of lambda cro repressor self-assembly and site- specific DNA operator binding I: Analysis of cro dimerization from nanomolar to micromolar concentrations. Biochemistry. 2000;39:11500–11507. doi: 10.1021/bi000935s. [DOI] [PubMed] [Google Scholar]
- 22.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 24.Lauber T, Schulz A, Schweimer K, Adermann K, Marx UC. Homologous proteins with different folds: The three-dimensional structures of domains 1 and 6 of the multiple Kazal-type inhibitor LEKTI. J Mol Biol. 2003;328:205–219. doi: 10.1016/s0022-2836(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 25.Belogurov GA, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newlove T, Atkinson KR, Van Dorn LO, Cordes MH. A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Cro dimer evolution. Biochemistry. 2006;45:6379–6391. doi: 10.1021/bi052541c. [DOI] [PubMed] [Google Scholar]
- 27.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillis DM. Taxonomic sampling, phylogenetic accuracy, and investigator bias. Syst Biol. 1998;47:3–8. doi: 10.1080/106351598260987. [DOI] [PubMed] [Google Scholar]
- 29.Blanco FJ, Angrand I, Serrano L. Exploring the conformational properties of the sequence space between two proteins with different folds: an experimental study. J Mol Biol. 1999;285:741–753. doi: 10.1006/jmbi.1998.2333. [DOI] [PubMed] [Google Scholar]
- 30.Van Dorn LO, Newlove T, Chang S, Ingram WM, Cordes MH. Relationship between the sequence determinants of stability for two natural homologous proteins with different folds. Biochemistry. 2006;45:10542–10553. doi: 10.1021/bi060853p. [DOI] [PubMed] [Google Scholar]
- 31.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 33.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 34.Pflugrath JW. The finer things in x-ray diffraction data collection. Acta Crystallogr D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 35.Foadi J, et al. A flexible and efficient procedure for the solution and phase refinement of protein structures. Acta Crystallogr D. 2000;56:1137–1147. doi: 10.1107/s090744490000932x. [DOI] [PubMed] [Google Scholar]
- 36.Cowtan K. “dm”: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 37.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 38.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Vagin AA, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Terwilliger TC, Berendzen J. Automated MAD, MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 46.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 47.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 48.DeLano WL. The PyMol User's Manual. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 49.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.