Abstract
Contraction of the heart results from interaction of the myosin and actin filaments. Cardiac myosin filaments consist of the molecular motor myosin II, the sarcomeric template protein, titin, and the cardiac modulatory protein, myosin binding protein C (MyBP-C). Inherited hypertrophic cardiomyopathy (HCM) is a disease caused mainly by mutations in these proteins. The structure of cardiac myosin filaments and the alterations caused by HCM mutations are unknown. We have used electron microscopy and image analysis to determine the three-dimensional structure of myosin filaments from wild-type mouse cardiac muscle and from a MyBP-C knockout model for HCM. Three-dimensional reconstruction of the wild-type filament reveals the conformation of the myosin heads and the organization of titin and MyBP-C at 4 nm resolution. Myosin heads appear to interact with each other intramolecularly, as in off-state smooth muscle myosin [Wendt T, Taylor D, Trybus KM, Taylor K (2001) Proc Natl Acad Sci USA 98:4361–4366], suggesting that all relaxed muscle myosin IIs may adopt this conformation. Titin domains run in an elongated strand along the filament surface, where they appear to interact with part of MyBP-C and with the myosin backbone. In the knockout filament, some of the myosin head interactions are disrupted, suggesting that MyBP-C is important for normal relaxation of the filament. These observations provide key insights into the role of the myosin filament in cardiac contraction, assembly, and disease. The techniques we have developed should be useful in studying the structural basis of other myosin-related HCM diseases.
Keywords: electron microscopy, MyBP-C, thick filament, three-dimensional reconstruction
Contraction of the heart depends on interactions between the thick (myosin-containing) and thin (actin-containing) filaments that fill cardiac muscle cells. The thick filaments are polymers of myosin II together with associated proteins, including titin and myosin binding protein C (MyBP-C) (1). Mutations in myosin and MyBP-C are the most common cause of inherited hypertrophic cardiomyopathy (HCM), a relatively common and often fatal disease characterized by left ventricular hypertrophy, myocyte disarray, and interstitial fibrosis (2, 3). The molecular mechanisms by which HCM mutations lead to the pathogenesis of this disease are unclear.
Structural knowledge of vertebrate thick filaments in the relaxed state has come from x-ray diffraction and electron microscopy (EM) of striated muscle (4–9). Myosin molecules assemble into bipolar filaments, with their α-helical coiled-coil tails in the filament backbone and their paired heads on the surface, in three near-helical strands having a repeat of ≈43 nm and an axial rise between levels of heads of ≈14.3 nm (1, 4, 9). Antiparallel overlap between myosin tails at the center of the filament creates a “bare zone,” free of myosin heads, where filament polarity reverses. Three-dimensional EM studies have revealed the near-helical distribution of the myosin heads in skeletal muscle in the relaxed state. However, conformational details have been limited by the low resolution of the reconstructions, and neither MyBP-C nor titin have been resolved (10–12). In addition, three-dimensional reconstructions of cardiac filaments have not been reported.
MyBP-C (13) is located at seven to eight axial sites, 43 nm apart (the same as the myosin repeat), in the middle third of each half of the filament (14–16), where it plays a role in modulating cardiac contraction (15). Cardiac MyBP-C consists of 11 globular Ig-like (Ig) and fibronectin-like (Fn) domains, linearly arranged, but its detailed organization on the filament is unknown. Some observations suggest that the C-terminal of MyBP-C wraps around the filament backbone (15, 17), whereas others suggest that its C-terminal runs parallel to the filament axis with its N-terminal extending out toward the surrounding thin filaments (15, 18). Titin is a giant (3 MDa) protein extending from the center of the filament to the Z line (14). In the thick filament region, it consists of a long string of Ig and Fn modules arranged in repeating patterns that bind to myosin and MyBP-C and appear to correlate with the ≈43-nm periodicity of these components (19). These and other observations suggest that titin is a developmental template for sarcomere assembly (14, 19, 20), but its organization on the filament (whether it is located in the filament core or on its surface, and whether helically or linearly organized) remains unknown. In addition to our ignorance of the detailed molecular organization of myosin, titin and MyBP-C that underlies normal functioning of the thick filament, there is little information on how mutations in these proteins cause HCM.
Here, we present a three-dimensional reconstruction of a cardiac thick filament, revealing the structure of the myosin heads in the relaxed state and the organization of titin and MyBP-C. Comparison with a reconstruction of filaments from a MyBP-C knockout mouse model for HCM (21) provides additional evidence for the location of MyBP-C and reveals the impact of its loss on thick filament structure. The results provide important insights into normal thick filament function and assembly, and into how alterations in thick filament structure may affect cardiac muscle function.
Results
Three-Dimensional Reconstruction Reveals the Near-Helical Array of Myosin Heads.
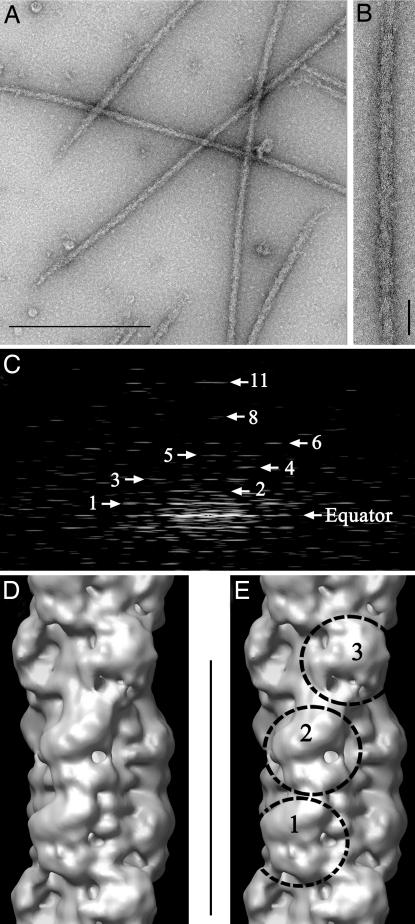
Thick filaments isolated from wild-type mouse ventricle appeared intact by negative staining electron microscopy (Fig. 1 A and B). Fourier transforms indicated good preservation of the ordered array of myosin heads (Fig. 1C), with reflections at the expected positions for a perturbed (8, 9, 22, 23) helical structure with a 42.9-nm repeat. Three-dimensional reconstruction of the MyBP-C region of the filament (approximately the middle third of each half) was carried out by single particle methods (10, 24) [see Materials and Methods and supporting information (SI) Methods]. The reconstruction, based on 2564 segments, had a resolution of 3.2–4.0 nm according to the Fourier shell correlation (FSC; SI Methods and SI Fig. 7), a factor of two better than previous vertebrate thick filament reconstructions, all from skeletal muscle [6.5 nm (10), according to the same FSC criterion, and ≈7 nm (12)]. At this resolution, key new structural features become visible. The main surface features are arranged in three “crowns” within each 42.9-nm repeat, and each crown has 3-fold rotational symmetry (Fig. 1 D and E and SI Movie 1). These features must represent myosin heads, the main globular components in the thick filament. The head array exhibits a perturbed helical structure, consistent with previous data from vertebrate striated muscle (9, 10, 12, 23). Crowns 1 and 3 are close to their correct axial (14.3 nm) and azimuthal helical positions. However, crown 2 is axially closer to crown 3 (13.2 nm) than to crown 1 (15.4 nm), and is at almost the same azimuthal position as crown 1 (instead of being rotated by 40°, as expected for a perfect helix; SI Fig. 8). The axial perturbations are similar to those seen in a reconstruction of fish skeletal filaments (10) and in a 2D analysis of rabbit cardiac filaments (23).
Fig. 1.
Isolation and 3D reconstruction of mouse cardiac thick filaments. (A) Electron micrograph of negatively stained filaments. (Scale bar, 500 nm.) (B) Portion of isolated filament, with bare zone at top. (Scale bar, 43 nm.) (C) Fourier transform of B showing layer lines at orders (numbered) of 42.9-nm repeat. (D and E) Surface views of thick filament reconstruction (filtered to 4.0-nm resolution), showing a single 42.9-nm repeat (scale bar), oriented so that bare zone would be at top. Myosin heads project from the surface at three levels (crowns 1, 2, and 3) in each repeat.
Atomic Fitting Reveals Intramolecular Interaction Between Myosin Heads.
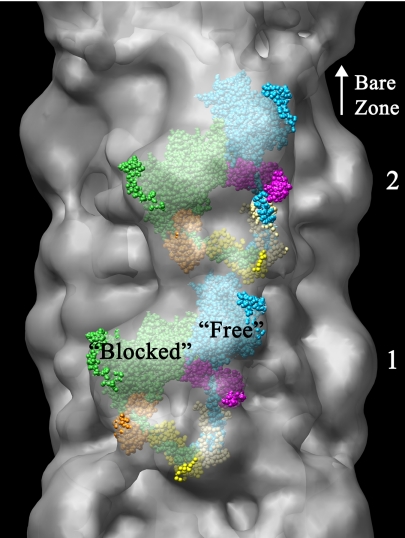
The enhanced resolution of the reconstruction makes possible a relatively precise fitting of myosin head atomic models. The striking visual resemblance, in two dimensions, between the motifs at crowns 1 and 2 and the atomic model of regulated myosin in the enzymatically inactive (“off”) state (24, 25) (SI Fig. 9) suggested that this was an appropriate model for 3D fitting. In this model, the heads are in the ADP.Pi state, as are the myosin filaments under the conditions of our experiments. We found that this model gave an excellent and unambiguous 3D fit into the globular features in crowns 1 and 2 (Fig. 2 and SI Movie 2). The myosin heads appear to interact asymmetrically, with their motor domains pointing toward the filament bare zone. This conclusion is supported by the observation of similar asymmetric head-head interactions in isolated, negatively stained mouse cardiac myosin molecules under relaxing conditions (H. Jung and R.C., unpublished data), and by the observation of a folded conformation (suggestive of head–head interaction) in rotary shadowed cardiac myosin molecules (26). Attempts to dock the two-headed model (as well as individual ADP.Pi-like S1 molecules) in other orientations were unsuccessful. Thus cardiac myosin, which is thought to have no intrinsic regulation and therefore to lack an off-state (27), adopts a similar conformation to the off-state of regulated myosin (see discussion in ref. 24).
Fig. 2.
Fitting of atomic model of myosin heads in off-state conformation (24) to head motif in crowns 1 and 2 of reconstruction. The 3D envelope has been made translucent to aid visualization of enclosed atomic model. Small regions of atomic model outside the envelope appear bright, whereas the majority, enclosed within it, appears dull (see also SI Movie 2). The heads are labeled “blocked” and “free” according to terminology proposed to describe the actin-binding capability of each head for regulated myosin (24, 25). “Blocked” head: motor domain, green; essential light chain, orange; regulatory light chain, yellow. “Free” head: motor domain, cyan; essential light chain, pink; regulatory light chain, beige. Bare zone direction toward top.
Myosin heads in crown 3 show a different conformation, which cannot be determined unambiguously from the reconstruction. Attempts to fit the two-headed off-state model, or atomic models of individual myosin heads (e.g., ref. 28), were unsuccessful because the reconstructed volume is greater than that required for one head, but too small for two. The smaller apparent volume of crown 3 heads suggests that they may have greater mobility than those in crowns 1 and 2.
The Reconstruction Reveals the Organization of Titin and MyBP-C on the Filament Surface.
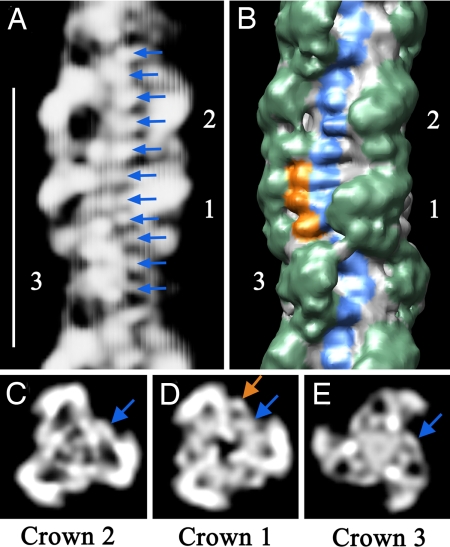
The reconstruction also has features that cannot be attributed to myosin heads. These are therefore likely to be myosin-binding proteins, whose molecular organization on the filament has not been previously visualized. In the space between azimuthally adjacent pairs of heads are small transverse bands that are especially evident in crowns 1 and 2 (blue coloring in Fig. 3B; SI Movies 1 and 3). Eleven such densities (arrows in Fig. 3A), ≈4 nm apart axially, are seen in each 42.9-nm repeat. These appear to form a longitudinal strand running approximately parallel to the filament axis, at a radius of 7–8 nm (blue arrows, Fig. 3 C–E), on the surface of the backbone. A similar periodicity has also been observed in a 2D analysis of cardiac myosin filaments (22). The arrangement of these 4 nm “beads” is consistent with the linear organization of the globular, 4 nm diameter, Fn and Ig domains in titin (14), whose organization has previously been inferred only from sequence data. Six molecules of titin are expected for each half of the thick filament, possibly arranged in pairs (29). However, our data do not allow us to distinguish any particular arrangement of the two putative titin molecules within a pair. The near azimuthal alignment of crowns 1 and 2 allows titin to follow a linear course in this region (Fig. 3B). The marked curvature around crown 3 might contribute to the different properties of the myosin heads in this crown.
Fig. 3.
Non-myosin proteins in the thick filament. (A) Projection of front side of reconstruction (rear side removed for clarity). Arrows point to 11 densities spaced 4 nm apart within the 42.9-nm repeat (protein white). (Scale bar, 43 nm.) (B) Surface view of same region at higher contour level, showing the three crowns of myosin heads (green). Surface features corresponding to the strand of densities in A are visible (blue, representing titin), together with three additional densities in crown 1 (orange, thought to represent MyBP-C). (C–E) Projections of transverse sections of the central regions of crowns 2, 1, and 3 (where the strand in A runs roughly parallel to the filament axis), showing the radial positions of the densities in A (blue arrows) and of the three additional beads of density in crown 1 in B (orange arrow). Note that, because the filaments have 3-fold rotational symmetry, each of the features marked in A–E is repeated at azimuthal angles of 120° and 240° with respect to those shown.
Crown 1 shows an additional density at a higher (10 nm) radius than the putative titin density (orange arrow in Fig. 3D). This density corresponds to three additional features, 4 nm apart, observed in the longitudinal surface view of the filament (orange coloring in Fig. 3B), to the left of the putative titin strand. The axial position of this additional density coincides with the location of strong density seen in striated muscle A-bands, which is known to represent MyBP-C (SI Methods and SI Fig. 10). The three 4-nm-diameter domains are consistent with the linear organization of Fn and Ig domains in MyBP-C. They may represent domains of MyBP-C known to interact with titin and/or the myosin tail (15).
Three-Dimensional Reconstruction of MyBP-C Knockout Filament.
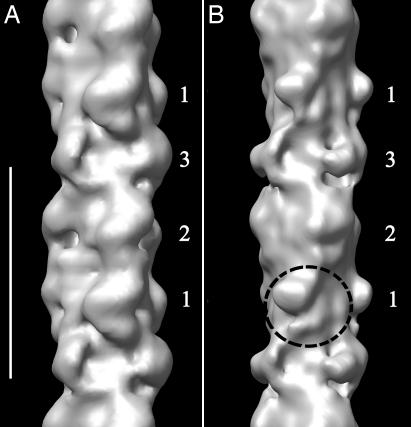
We have also carried out a 3D reconstruction of cardiac thick filaments from a MyBP-C knockout mouse model for HCM (21) (SI Methods). Although the overall appearance of wild-type and knockout filaments appears to be similar (SI Fig. 11; ref. 30), the diameter of the knockout reconstruction and the mass of the heads appear slightly smaller, and the resolution is lower (≈7 nm, compared with 4 nm for the wild-type), suggesting that the array of heads is less stable in filaments lacking MyBP-C. In addition, there is a clear structural change in crown 1 of the knockout filaments (Fig. 4 and SI Movie 4), such that the heads can no longer be fitted with the off-state atomic model of myosin. In contrast, crowns 2 and 3 appear relatively unaffected by the absence of MyBP-C. The restriction of the main structural change to crown 1 adds support to our conclusion that MyBP-C is in this crown. It also suggests that MyBP-C may be important for the establishment of the asymmetric interaction of the myosin heads in this crown.
Fig. 4.
Comparison of 3D reconstructions of MyBP-C knockout and wild-type thick filaments. (A) Wild type. (B) Knockout. The reconstructions are based on a similar number of segments in each case. Both have been filtered to 7-nm resolution (the resolution of the knockout filaments) to enable a direct comparison. Circle shows altered structure of myosin heads in crown 1 of knockout. (Scale bar, 43 nm.)
Discussion
Intramolecular Interaction Between Myosin Heads in the Cardiac Thick Filament.
The conformation of the myosin heads and putative organization of titin and MyBP-C that we have observed provide important new insights into the structure and function of vertebrate cardiac muscle thick filaments. Previous structural analysis of cardiac filaments was limited to two dimensions (8, 22, 23), whereas 3D reconstructions, carried out on vertebrate skeletal thick filaments [from frog (12) and fish (10) muscle] had considerably lower resolution (≈7 nm). The 3- to 4-nm resolution of the current work makes possible a rather precise fitting of myosin head atomic models. A surprising finding is the excellent fit of the interacting-head conformation of regulated myosin heads in the off-state (24, 25) to two of the three crowns of the 42.9-nm repeat of the filament. Head–head interaction was previously thought to be the mechanism by which ATPase and actin-binding activity were switched off in intrinsically regulated myosins (e.g., vertebrate smooth, invertebrate striated muscle myosin) (24, 25). One head (“blocked;” Fig. 2) would be prevented from binding to actin and the other head (“free”) would be unable to hydrolyze ATP (24, 25). Myosin from vertebrate-striated muscles is thought to lack an intrinsic regulatory switch and is therefore thought to be unable to switch off biochemically (these muscles are regulated primarily through the troponin–tropomyosin system on the thin filaments) (27). The observation of the interacting-head motif in cardiac filaments would therefore suggest that head-head interaction per se does not switch activity off. It may instead be a resting position or “relaxed state” conformation common to both regulated and unregulated myosin filaments. In unregulated myosins, the head-head interaction may be weak and thus may not inhibit myosin function (although it might restrict head access to actin filaments in the relaxed state; ref. 31). Additional interactions present in regulated myosins (e.g., involving the regulatory light chains, or interactions with heads in other crowns or with S2; ref. 24) may explain the ability of regulated myosins to switch off, for example by stabilizing the interacting-head structure. Alternatively, vertebrate thick filaments might possess a regulatory mechanism that has not yet been detected biochemically.
Whereas the heads in crowns 1 and 2 have a “relaxed conformation,” those in crown 3 appear to be more mobile. In the sarcomere, where actin filaments are close to the thick filaments, greater freedom of crown 3 heads may increase their probability of interaction with actin. Two populations of heads may allow fine-tuning of thick filament activity. Activity is also modulated by phosphorylation of the regulatory light chains, which increases head mobility and enhances contraction in both vertebrate skeletal and cardiac muscle (32, 33), and by MyBP-C phosphorylation (34). In the case of regulated myosin, light chain phosphorylation does not merely modulate activity, but fully activates switched off molecules. In both systems, phosphorylation probably functions by breaking the head–head (and other) interactions that maintain the relaxed conformation (25).
Titin Is Linearly Organized on the Thick Filament Surface.
The improved resolution of our reconstruction has enabled the visualization of nonmyosin thick filament proteins whose organization was previously unknown. The linear arrangement of eleven, 4-nm-diameter globular features in each 42.9-nm repeat directly suggests that we are visualizing titin (14, 35). The 11 domains are likely to correspond to the 43-nm-long superrepeat of seven Fn and four Ig domains of this size in titin in the MyBP-C region of the filament, previously identified on the basis of sequence data and thought to dictate the location of myosin and MyBP-C (14). The titin strands lie on the surface of the filament backbone (consistent with titin epitope availability in antibody labeling studies; ref. 36), oriented approximately parallel to the filament axis, where they can form longitudinal interactions with myosin tails and MyBP-C. Our observations appear to rule out models in which titin follows the helical path of the myosin heads or is buried in the filament core. They suggest instead that titin forms a simple linear scaffold for filament formation, and that myosin molecules assemble on the interior of this scaffold during sarcomere development and turnover. Interestingly, there appears to be no simple matching of the axial positions of myosin heads (spaced 15.2, 13.4, and 14.3 nm apart) with titin domains (≈3.9 nm apart), as might have been expected from the template function of titin. Thus, during thick filament assembly, a different set of titin–myosin interfaces must be used at each of the three crowns.
Organization of MyBP-C on the Filament Surface.
The molecular organization of MyBP-C in the thick filament has remained a mystery since its discovery 35 years ago (13, 15), despite its emerging importance as a modulator of cardiac contraction and as a prime target of mutations causing HCM. Our results suggest that at least three of its 11 Fn and Ig domains run along the filament surface parallel to the axis (18), interacting with titin and probably with myosin tails in crown 1. These interactions, presumably involving the C-terminal region of MyBP-C (known to interact with myosin tails and titin; ref. 15), may anchor MyBP-C to the thick filament. We see no evidence that its other domains form a collar around the backbone, as has been suggested (15), although such an arrangement should be visible if present. They may instead extend from the filament surface (18) at crown 1 where they could bind to subfragment 2 and/or actin, modulating myosin–actin interaction and force generation (15). We observe MyBP-C stripes only in sarcomeres with a very well ordered lattice (SI Fig. 10), suggesting that any such extended regions might be disordered in isolated filaments, explaining their absence from our reconstruction.
Implications from MyBP-C Knockout Filament.
MyBP-C is important for normal functioning of the heart (15) and appears to play a “cardioprotective” role in ischemic injury (37). Mouse models are being used to understand the functioning of MyBP-C (38). Knockout mice demonstrate that MyBP-C is not essential for survival, although its absence causes cardiac hypertrophy (21). The similar overall structure of MyBP-C knockout and wild type filaments (SI Fig. 11 A and B, compare Fig. 1 A and B) may explain the viability of these mice and the relatively good prognosis for most HCM patients carrying MyBP-C mutations (38). The lower resolution of the MyBP-C knockout reconstruction suggests that MyBP-C helps to stabilize the relaxed array of myosin heads in normal filaments (see also ref. 30). The decreased ability of crown 1 heads to adopt the interacting-head conformation in the absence of MyBP-C suggests that the knockout filaments may not relax as fully as wild type. Failure of the filaments to relax properly in the absence of MyBP-C may offer a molecular explanation of the compromised relaxation of the heart in MyBP-C knockout mice (21, 39) and may also be related to the abnormal diastolic function in humans with HCM (2).
Our reconstructions show that cardiac thick filaments are complex structures, where not all myosin heads behave in a similar manner, partly as a result of the arrangement of accessory proteins. This diversity suggests that cardiac thick filament function may be finely modulated. The techniques we have developed here open the way to further structural studies on HCM mouse models carrying different mutations in MyBP-C or other thick filament proteins. Such studies should reveal whether alteration in the relaxed conformation of the thick filaments is a common molecular mechanism underlying HCM.
Materials and Methods
Isolation of Filaments.
Thick filaments were isolated from ventricular muscle of mice (8–10 weeks old, strain 129 SVE, Taconic Farms, NY). Mice were injected i.p. with heparin (1,000 units/kg) and 15 min later anesthetized with sodium pentobarbital (60 units/kg). Once anesthetized, mice were killed by cervical dislocation. These procedures were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Immediately after euthanasia, the heart was harvested and placed in calcium-free Tyrode (140 mM NaCl, 5.4 mM KCl, 1.5 mM MgCl2, 10 mM Hepes, 0.33 mM Na2HPO4, 10 mM glucose, pH 7.4) plus 20 mM 2,3-butanedione monoxime (BDM, to enhance actin–myosin dissociation). The aorta was catheterized for retrograde perfusion of the coronary arteries with the following solutions: first, calcium-free Tyrode plus 20 mM BDM for 5 min; second, relaxing solution (100 mM NaCl, 2 mM EGTA, 5 mM MgCl2, 1 mM DTT, 10 mM imidazole, 5 mM MgATP, pH 7.2) plus 0.1% Saponin and 0.5% Triton X-100 for 8 min; third, relaxing solution for 5 min. After this final wash, the ventricles were cut and homogenized in relaxing solution for 10 sec at position 5 using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY). The whole procedure was carried out at room temperature.
During homogenization in relaxing solution, only a small number of filaments dissociated from the myofibrils. The yield was increased by modification of a method used for insect flight muscle (40). The homogenate was spun at 10,000 g for 2 min to obtain a pellet of myofibrils, which were resuspended and incubated with 70 μg calpain-1 (Calbiochem) in 48 μl 20 mM imidazole-HCl, 5 mM 2-mercaptoethanol, 1 mM EDTA, 1 mM EGTA, 30% glycerol, pH 6.8 plus 10 μM free Ca2+ for 1 h at room temperature. Enzymatic treatment was stopped by addition of relaxing solution, containing 100 μM blebbistatin (Toronto Research Chemicals), and the suspension was agitated to help release thick and thin filaments (blebbistatin was used to aid release of thick filaments and to stabilize the relaxed conformation of the heads (F. Zhao and R.C., unpublished data). Thin filaments were removed by fragmentation with gelsolin (41). The thick filament suspension was kept on ice and used within 24 h. For the rationale for these procedures, see SI Methods.
Electron Microscopy.
The ordering of myosin heads is rapidly lost in mammalian thick filaments at low temperature (42). Therefore, we observed filaments by negative staining at near-physiological temperature. This avoided difficulties in preserving order that we encountered using cryo-EM. Negative staining provides a very good representation of protein distribution in most macromolecular structures (43), including thick filaments (1). A drop of filament suspension was placed on an electron microscope grid coated with a thin layer of carbon supported by a thicker holey carbon film. The grid was rinsed sequentially with 6 drops of relaxing rinse (140 mM NaAc, 1 mM MgAc2, 1 mM EGTA, 5 mM imidazole, 1 mM sodium azide, 1 mM MgATP, pH 7.0) and five drops of 2% uranyl acetate. Staining was carried out at room temperature with solutions prewarmed to 37°C. Grids were observed in a Philips CM120 electron microscope (FEI, Hillsboro, OR) at 80 KV under low dose conditions. Images of filaments on thin carbon over holes were acquired at 42,000× magnification, using a 2Kx2K CCD camera (F224HD, TVIPS GmbH, Gauting, Germany) at a resolution of 0.57 nm/pixel.
Image Analysis.
Long, straight thick filaments, with a clearly visible bare zone, were chosen for analysis. Filaments were oriented vertically, with the bare zone at the top, and the region between the 3rd and 10th 42.9-nm repeats from the bare zone (where MyBP-C is present) was computationally cut (Adobe Photoshop 7.0). The presence of order in these regions was verified by computing the fast Fourier transform (Image J, v1.34s, NIH, Bethesda, MD). For single-particle analysis, we used only those regions where the Fourier transforms showed the layer lines expected for a well ordered vertebrate thick filament.
Selected filament regions were converted to SPIDER format (EM2EM; Image Science and Imperial College, London), and cut into segments 3 × 42.9 nm long in SPIDER (v11.2, Wadsworth Center, Albany, NY). Single-particle analysis was carried out by using SPIDER (see below). UCSF Chimera (beta version 1, build 2199) was used for visualization, analysis, and atomic fitting of 3D volumes (44) (see SI Methods).
Three-Dimensional Reconstruction.
Single particle analysis rather than Fourier-Bessel based helical methods were used for 3D reconstruction so that the known perturbations in the helical order of vertebrate myosin filaments would be preserved and not averaged out (11) (see SI Methods for details). Electron micrograph images of filaments are 2D projections perpendicular to the filament axis. Because filaments are expected to lie on the EM grid with different rotations about their long axes, micrographs of different filaments should represent different rotational views. If the relative rotational angle of each view is known, the 3D structure of the filaments can be determined by back projection using those angles. Relative rotations of different filament segments were determined by matching filament images against 2D projections of 3D models rotated around their long axis at known angles (see SI Methods). Each segment view was assigned the angle of the model projection that it most closely matched, and the views were back-projected to generate a 3D reconstruction. This reconstruction was used as the model for a new cycle, and the process was iterated until there were no further changes in the reconstruction (10, 24).
Possible model bias in the reconstruction was ruled out by our finding that the final reconstruction was the same with three different, independent starting models (see SI Methods; and SI Fig. 6). For true helical structures, helical refinement can improve the signal:noise ratio and resolution of the reconstruction (24, 45). We avoided helical refinement to preserve the helical perturbations present in vertebrate filaments (9, 10, 12, 23). However, C3 symmetry was imposed to take advantage of their threefold rotational symmetry (5).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Lori Nyland for suggesting the use of calpain for filament isolation. This work was supported by an American Heart Association Postdoctoral Fellowship (to M.E.Z.) and National Institutes of Health Grants AR34711 (to R.C.) and HL82900 (to R.L.M.). Electron microscopy was carried out in the Core Electron Microscopy Facility of the University of Massachusetts Medical School, supported in part by Diabetes Endocrinology Research Center Grant DK32520. Molecular graphics images were produced by using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the EBI Macromolecular Structure Database, www.ebi.ac.uk (accession no. EMD-1465).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708912105/DC1.
References
- 1.Craig R, Woodhead JL. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Marian AJ. Curr Cardiol Rev. 2005;1:53–63. [Google Scholar]
- 3.Fatkin D, Graham RM. Physiol Rev. 2002;82:945–980. doi: 10.1152/physrev.00012.2002. [DOI] [PubMed] [Google Scholar]
- 4.Huxley HE. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 5.Kensler RW, Stewart M. J Cell Biol. 1983;96:1797–1802. doi: 10.1083/jcb.96.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler RW, Stewart M. J Cell Sci. 1989;94:391–401. doi: 10.1242/jcs.94.3.391. [DOI] [PubMed] [Google Scholar]
- 7.Kensler RW, Stewart M. J Cell Sci. 1993;105:841–848. doi: 10.1242/jcs.105.3.841. [DOI] [PubMed] [Google Scholar]
- 8.Kensler RW. Biophys J. 2002;82:1497–1508. doi: 10.1016/S0006-3495(02)75503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huxley HE, Brown W. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 10.Al-Khayat HA, Morris EP, Kensler RW, Squire JM. J Struct Biol. 2006;155:202–217. doi: 10.1016/j.jsb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Eakins F, Al-Khayat HA, Kensler RW, Morris EP, Squire JM. J Struct Biol. 2002;137:154–163. doi: 10.1006/jsbi.2002.4453. [DOI] [PubMed] [Google Scholar]
- 12.Stewart M, Kensler RW. J Mol Biol. 1986;192:831–851. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- 13.Offer G, Moos C, Starr R. J Mol Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 14.Gregorio CC, Granzier H, Sorimachi H, Labeit S. Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 15.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Circ Res. 2004;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 16.Bennett P, Craig R, Starr R, Offer G. J Muscle Res Cell Motil. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 17.Moolman-Smook J, Flashman E, de Lange W, Li Z, Corfield V, Redwood C, Watkins H. Circ Res. 2002;91:704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 18.Squire JM, Luther PK, Knupp C. J Mol Biol. 2003;331:713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 19.Labeit S, Gautel M, Lakey A, Trinick J. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tskhovrebova L, Trinick J. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 21.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 22.Kensler RW. J Struct Biol. 2005;149:313–324. doi: 10.1016/j.jsb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Kensler RW. J Struct Biol. 2005;149:303–312. doi: 10.1016/j.jsb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 25.Wendt T, Taylor D, Trybus KM, Taylor K. Proc Natl Acad Sci USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi T, Fukukawa C, Naraoka C, Katoh T, Yazawa M. J Biochem (Tokyo) 1999;126:34–40. doi: 10.1093/oxfordjournals.jbchem.a022433. [DOI] [PubMed] [Google Scholar]
- 27.Lehman W, Szent-Gyorgyi AG. J Gen Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 29.Liversage AD, Holmes D, Knight PJ, Tskhovrebova L, Trinick J. J Mol Biol. 2001;305:401–409. doi: 10.1006/jmbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 30.Kensler RW, Harris SP. Biophys J. 2007;94 doi: 10.1529/biophysj.107.115899. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoghbi ME, Woodhead JL, Craig R, Padron R. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 32.Sweeney HL, Bowman BF, Stull JT. Am J Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 33.Levine RJ, Kensler RW, Yang Z, Stull JT, Sweeney HL. Biophys J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oakley CE, Chamoun J, Brown LJ, Hambly BD. Int J Biochem Cell Biol. 2007;39:2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Trinick J, Knight P, Whiting A. J Mol Biol. 1984;180:331–356. doi: 10.1016/s0022-2836(84)80007-8. [DOI] [PubMed] [Google Scholar]
- 36.Whiting A, Wardale J, Trinick J. J Mol Biol. 1989;205:263–268. doi: 10.1016/0022-2836(89)90381-1. [DOI] [PubMed] [Google Scholar]
- 37.Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J. Proc Natl Acad Sci USA. 2006;103:16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardiff JC. Heart Fail Rev. 2005;10:237–248. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 39.Pohlmann L, Kroger I, Vignier N, Schlossarek S, Kramer E, Coirault C, Sultan KR, El-Armouche A, Winegrad S, Eschenhagen T, et al. Circ Res. 2007;101:928–938. doi: 10.1161/CIRCRESAHA.107.158774. [DOI] [PubMed] [Google Scholar]
- 40.Kulke M, Neagoe C, Kolmerer B, Minajeva A, Hinssen H, Bullard B, Linke WA. J Cell Biol. 2001;154:1045–1057. doi: 10.1083/jcb.200104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hidalgo C, Padron R, Horowitz R, Zhao FQ, Craig R. Biophys J. 2001;81:2817–2826. doi: 10.1016/S0006-3495(01)75923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu S, Offer G, Gu J, White HD, Yu LC. Biochemistry. 2003;42:390–401. doi: 10.1021/bi026085t. [DOI] [PubMed] [Google Scholar]
- 43.Ohi M, Li Y, Cheng Y, Walz T. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Egelman EH. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.