Abstract
We have developed a method to detect DNA synthesis in proliferating cells, based on the incorporation of 5-ethynyl-2′-deoxyuridine (EdU) and its subsequent detection by a fluorescent azide through a Cu(I)-catalyzed [3 + 2] cycloaddition reaction (“click” chemistry). Detection of the EdU label is highly sensitive and can be accomplished in minutes. The small size of the fluorescent azides used for detection results in a high degree of specimen penetration, allowing the staining of whole-mount preparations of large tissue and organ explants. In contrast to BrdU, the method does not require sample fixation or DNA denaturation and permits good structural preservation. We demonstrate the use of the method in cultured cells and in the intestine and brain of whole animals.
Keywords: BrdU, click chemistry, DNA replication, EdU, microscopy
Detection of DNA synthesis in proliferating cells relies on the incorporation of labeled DNA precursors into cellular DNA during the S phase of the cell cycle (1–3). The labeled DNA precursors, usually pyrimidine deoxynucleosides, are added to cells during replication, and their incorporation into genomic DNA is quantified or visualized after incubation and sample staining. The same labeled deoxynucleosides can be injected into experimental animals to assay cellular proliferation in specific organs and tissues. The most common deoxynucleosides used for assaying DNA replication are [3H]thymidine and 5-bromo-2′-deoxyuridine (BrdU). [3H]Thymidine incorporated into DNA is usually detected by autoradiography, whereas detection of BrdU is accomplished immunologically, through specific anti-BrdU antibodies.
Although [3H]thymidine and BrdU labeling methods have been very useful for studying cell cycle kinetics, DNA replication, and sister chromatid exchange and for assessing cell proliferation of normal or pathological cells or tissues under different conditions, these methods exhibit several limitations. Working with [3H]thymidine is cumbersome because of its radioactivity. Autoradiography is also labor-intensive and slow (detection often lasts several months), and thus not suitable for rapid high-throughput studies. Finally, microscopic images of [3H]thymidine-labeled DNA suffer from poor resolution and low signal-to-noise ratios. In contrast to [3H]thymidine autoradiography, BrdU immunostaining is both faster (although it still lasts several hours) and allows better microscopic imaging of the labeled DNA. One major disadvantage of BrdU staining is that the complementary base pairing in double-stranded DNA blocks the access of the anti-BrdU antibody to BrdU subunits. To expose the BrdU epitope, cells and tissue samples are subjected to strong denaturing conditions such as concentrated hydrochloric acid or mixtures of methanol and acetic acid. These harsh staining conditions invariably degrade the structure of the specimen while also making the intensity of BrdU staining highly dependent on the conditions used for detection by each investigator (4). Finally, as with any immunohistological stain, the size of the tissue piece to be stained whole-mount is limited by the penetration of the antibody through the fixed tissue. This limitation can be overcome by either sectioning (which is labor-intensive and limits the size of the tissue thus surveyed) or by allowing long incubation times with the antibody (which prolongs the staining procedure to several days). Clearly, improved nucleic acid labeling techniques are still needed for the study of cell cycle kinetics, DNA synthesis, and cellular proliferation in vitro and in vivo. In particular, the development of techniques that are simple, rapid, and sensitive and do not require extensive sample preparation and/or result in the destruction of sample ultrastructure remains highly desirable.
We wanted to develop a method for labeling DNA in vivo that would allow us to image the replicated DNA in the context of well preserved cellular and chromatin ultrastructure. The resulting method uses 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analogue in which a terminal alkyne group replaces the methyl group in the 5 position, which is readily incorporated into cellular DNA during DNA replication. The terminal alkyne group is then detected through its reaction with fluorescent azides, in a Cu(I)-catalyzed [3 + 2] cycloaddition (“click” chemistry) (5, 6). This method is highly sensitive and much faster than BrdU detection. In addition, because the reagents are almost 1/500th the size of an antibody molecule, they have a much higher diffusion rate and penetrate the tissue much more effectively, which allows the rapid, whole-mount stain of large tissue and organ fragments. Finally, the reaction between ethynyl groups on DNA and fluorescent azides does not require denaturation of the specimen; this allows good structural preservation. We anticipate that this method will allow high-resolution microscopic analysis in vivo of cellular processes involving DNA, such as DNA repair, replication, recombination, and sister chromatid cohesion.
Results and Discussion
Labeling of Cellular DNA with EdU.
Cells incorporate 5-bromo-2′-deoxyuridine (BrdU) and 5-iodo-2′-deoxyuridine (IdU) into their DNA in the place of 5-methyl-2′-deoxyuridine (thymidine). We reasoned that 5-ethynyl-2′-deoxyuridine (EdU) should also be efficiently incorporated into replicating DNA and its terminal alkyne group should be available to react with organic azides in the major groove of the double helix, without steric hindrance. We synthesized EdU and fluorescent azide derivatives required for its detection in cells by using click chemistry (Fig. 1). Cells labeled in culture with 10 μM EdU overnight showed very intense nuclear staining after reaction with Alexa568-azide under Cu(I)-catalyzed click reaction conditions (Fig. 2 B, E, and H). In contrast, cells not labeled with EdU displayed no detectable staining if subjected to the same fixation and staining conditions as EdU-labeled cells (Fig. 2 A, D, and G). EdU staining was greatly reduced in cells in which DNA replication was blocked by treatment with hydroxyurea (Fig. 2 C, F, and I; see also J–M), demonstrating that the nuclear staining in EdU-labeled cells is the result of EdU incorporation during DNA synthesis. Also, in synchronized HeLa cells, the extent and intensity of DNA labeling after a pulse of EdU during S phase correlates with the EdU concentration and the length of the pulse (data not shown). EdU has been previously evaluated as an antiviral compound (7). These early studies concluded that EdU was not incorporated into bacterial and phage DNA (8); its incorporation into eukaryotic DNA, however, was not investigated. We find that EdU is strongly incorporated into the DNA of proliferating mammalian cells. EdU is also incorporated into DNA replicated in vitro in Xenopus egg extracts (data not shown, see Materials and Methods) by simple addition to the cycling extract preparation.
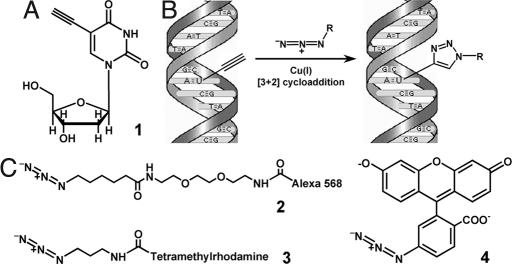
Fig. 1.
Use of 5-ethynyl-2′-deoxyuridine (EdU) to label DNA in cells. (A) Structure of 5-ethynyl-2′-deoxyuridine, a thymidine analogue that carries a terminal alkyne group instead of a methyl in the 5 position of the pyrimidine ring. (B) Schematic of the click reaction for detecting EdU incorporated into cellular DNA. The terminal alkyne group, exposed in the major groove of the DNA helix readily reacts with an organic azide (R can be any fluorophore, hapten, electron-dense particle, quantum dot, etc.) in the presence of catalytic amounts of Cu(I). (C) Structures of the fluorescent azides used in the present study. Unlike azide 2, azide 3 is cell-permeable, allowing cells to be stained while alive, without fixation and/or permeabilization. Fluorescein azide 4 can be easily prepared in large amounts by using inexpensive starting materials. See text and Materials and Methods for details.
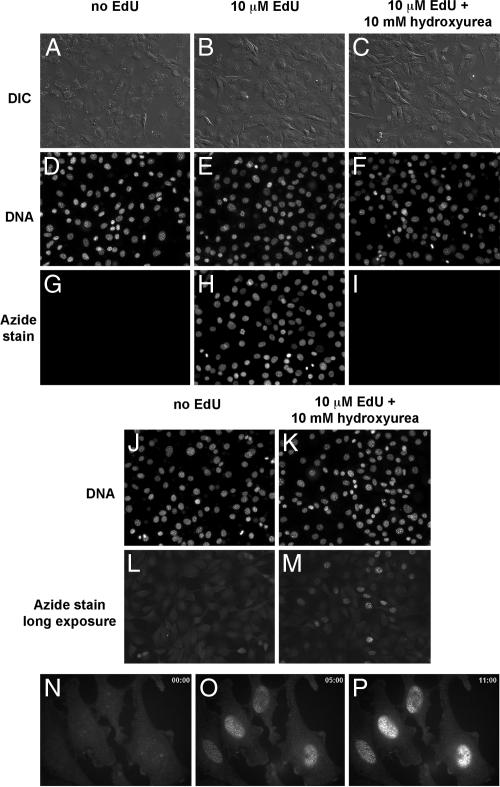
Fig. 2.
Detection of EdU incorporated into the DNA of cultured NIH 3T3 cells (A–M) and HeLa cells (N–P) by fluorescence microscopy. NIH 3T3 cells were incubated in media without EdU (A, D, G, J, and L), media supplemented with10 μM EdU (B, E, and H) or media with 10 μM EdU and 10 mM hydroxyurea to block DNA synthesis (C, F, I, K, and M). In A–M, the cells were fixed and then reacted with 10 μM Alexa568-azide (Fig. 1C, compound 2) for 10 min. The cells were then counterstained with Hoechst to reveal cellular DNA, washed, and imaged by fluorescence microscopy and differential interference contrast (DIC). Note the strong specific and low nonspecific azide stain in the presence (H) and absence (G) of EdU, respectively. Not all nuclei in H are labeled after overnight incubation with EdU, suggesting that only cells that went through S phase became labeled. Blocking DNA replication with hydroxyurea abolishes EdU incorporation almost completely (I). If cells labeled with EdU in the presence of hydroxyurea are photographed with longer exposure times (3-s exposure time in L and M, compared with 40-ms exposure time for G–I under otherwise identical illumination and camera settings), low levels of EdU incorporation can be seen in a small fraction of the hydroxyurea-treated cells. (N–P) Still images from a time-lapse recording of EdU staining of live cells by using the cell-permeable TMR-azide (Fig. 1C, compound 3). Live HeLa cells labeled with 10 μM EdU were stained with 500 nM TMR-azide in the presence of Cu(I) in PBS. Time is shown in minutes in the upper right corners of M–P. Note that Cu(I) is cytotoxic and the cells do not survive the staining reaction. See text and Materials and Methods for details.
The [3 + 2] cycloaddition reaction used to detect EdU in cells is catalyzed by Cu(I) ions. Because of the low solubility of Cu(I) salts, Cu(I) is often introduced in [3 + 2] cycloaddition reactions in a liganded form (9). We found that the best EdU staining was obtained by generating Cu(I) from Cu(II) in situ, using ascorbic acid as reducing agent. The intensity of the staining obtained was proportional to the concentration of the fluorescent azide and that of ascorbic acid in the staining reaction (data not shown). In the case of ascorbic acid, for a fixed concentration of Cu(II) of 1 mM (in the form of CuSO4), the intensity of the EdU stain increased as the ascorbic acid concentration was increased to 50–100 mM.
The staining reaction between cellular EdU-labeled DNA and fluorescent azides proceeds very quickly, being half-maximal in <5 min under the staining conditions we used (see below). This fast kinetics might be because EdU-labeled DNA is a multivalent, polymeric click chemistry substrate. The triazoles formed from the reaction between the ethynyl group and the fluorescent azide are Cu(I) chelators, increasing the local concentration of the catalyst and accelerating the reaction of neighboring ethynyl groups. This effect has been described for other multivalent [3 + 2] cycloaddition substrates (10).
EdU detection proceeds smoothly with a variety of fluorescent azides we synthesized, including aliphatic azides (compounds 2 and 3 in Fig. 1) and aromatic ones (compound 4 in Fig. 1, which can be easily synthesized in a single step in large amounts as required for some whole-mount staining procedures). The EdU detection method is also compatible with immunostaining. We recommend performing a regular immunostaining protocol after first performing the EdU stain (see below).
We find that under the conditions used (1 mM CuSO4, 100 mM ascorbic acid, and 10 μM fluorescent azide), the staining reaction does not proceed to completion and only a fraction of the ethynyl groups present on DNA is reacted before the staining mixture loses activity. As shown in Fig. 3A, if EdU-labeled cells first stained with Alexa488-azide (green) are subsequently reacted with fresh staining mixture containing Alexa594-azide (red), the DNA is strongly stained both green and red. The two staining patterns display perfect colocalization (see Fig. 3A Right, showing an overlay of the two successive EdU stains), demonstrating the high reproducibility of the EdU detection method. The intensity of the EdU stain can thus be increased through repeated incubation with fresh detection mixture, without a change in the staining pattern.
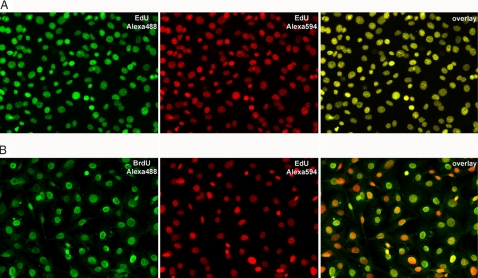
Fig. 3.
Reproducibility of EdU labeling (A) and comparison with BrdU (B). (A) NIH 3T3 cells labeled by incubation overnight with 2 μM EdU were fixed and reacted successively with 10 μM Alexa488-azide and 10 μM Alexa594-azide, respectively, as described in Materials and Methods. The cells were imaged by fluorescence microscopy. (Left) Alexa488-azide stain. (Center) Alexa594-azide stain. (Right) Overlay of the Alexa488 and Alexa594 images. The complete colocalization of the two colors indicates that the two successive azide stains detect ethynyl groups that have the same distribution within the cell nucleus. (B) NIH 3T3 cells labeled by incubation overnight with 2 μM EdU and 2 μM BrdU were fixed and reacted with 10 μM Alexa594-azide, followed by standard BrdU immunodetection by using an anti-BrdU monoclonal antibody and an Alexa488-conjugated secondary antibody. (Left) BrdU stain. (Center) EdU stain. (Right) Overlay of the BrdU and EdU images. Note that each cell that incorporated BrdU also incorporated EdU. The BrdU micrograph was taken by using a five-times longer exposure than the exposure used for EdU (500 ms compared with 100 ms) under identical digital camera settings.
We next asked how EdU compares with the conventional BrdU-labeling method. Cultured NIH 3T3 cells were labeled with both EdU and BrdU by overnight incubation in media containing 2 μM each of the two deoxynucleoside analogues. The cells were then fixed, stained with Alexa594-azid,e and then processed for BrdU immunofluorescent detection. As shown in Fig. 3B, all cells staining with BrdU also show EdU staining. In addition, the EdU staining is significantly more intense than the BrdU staining.
Detection of EdU Without Fixation.
The Alexa568-azide used earlier is not cell-permeable and required cell fixation and permeabilization to stain EdU-labeled cells. To detect EdU-labeled DNA under native conditions (i.e., without fixation and permeabilization) we developed a cell-permeable tetramethylrhodamine (TMR) azide (Fig. 1). After confirming that TMR-azide can enter live cells, we used it to stain live cells that had been labeled with EdU. The staining mixture containing TMR-azide was added to cells on a fluorescent microscope stage at time t = 0, to initiate the staining reaction, which was then followed by fluorescence confocal microscopy. As shown in Fig. 2, TMR-azide reacts rapidly with EdU-labeled DNA in cells, in the presence of Cu(I) generated in situ from Cu(II) and ascorbic acid, suggesting that the click reaction takes place efficiently on native cellular DNA. The staining reaction is half-maximal in <5 min. This experiment underestimates the staining rate, because we used only 500 nM TMR-azide in the detection reaction (as opposed to the usual 10–50 μM azide concentration we normally use for staining). The lower TMR-azide concentration used in this experiment was required to allow imaging of cellular DNA by fluorescence microscopy, against the background of unreacted TMR-azide in the staining solution.
It should be noted that Cu(I) is toxic to cells and that Cu(I)-catalyzed click reaction conditions result in cell death. We believe, however, that even if the staining conditions were nontoxic, live cell microscopy of EdU-labeled and azide-stained cells would be of limited utility. The reaction between ethynyl groups on DNA and fluorescent azide molecules results in covalent attachment of many large chemical groups to cellular DNA. If cells were to survive staining, such a chemical insult would likely trigger a massive DNA damage response and subsequent cytotoxicity. Despite Cu(I) toxicity, we envision that the ability to stain EdU-labeled cells rapidly and without fixation will be useful in a number of situations: (i) when permeabilization and fixation degrade the structure of the specimen; (ii) for assaying DNA synthesis at the end of live cell experiments, without removing the cells from the microscope stage; (iii) in high-throughput screening assays, when reducing the number of steps in the staining protocols is important. Additionally, staining cells without fixation and permeabilization can prevent cell detachment and loss from coverslips.
Use of EdU to Assay DNA Synthesis in Animals.
We next wanted to determine whether EdU can be used to detect DNA synthesis in animals. One hundred micrograms of EdU in PBS were injected i.p. into adult mice and tissues were harvested and fixed 96 h after injection. Paraffin sections were stained with Alexa568 azide for 15 min, washed, and then imaged by fluorescence microscopy. Because of the high intensity of fluorescence, large sections can be quickly scanned by using low-magnification objectives, even a simple dissecting microscope equipped with fluorescence. As shown in Fig. 4, EdU strongly labels proliferating cells in vivo (Fig. 4 A and B, showing sections of small intestine). It is well suited for detecting very low levels of cell proliferation in tissues that undergo very little to no cellular turnover (Fig. 4 C and D, showing sections of mouse brain).
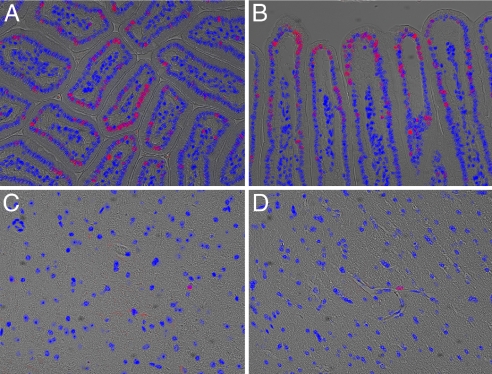
Fig. 4.
Labeling DNA in vivo by using EdU. An adult mouse was injected i.p. with 100 μg of EdU in PBS. Tissues were harvested, fixed, and sectioned 96 h later. Tissue sections on slides were then reacted for 10 min with 10 μM Alexa568-azide. The images shown are overlays of a DIC image of the sectioned tissue, a fluorescent image of cellular DNA (Hoechst stain; blue), and a fluorescent image of EdU-labeled DNA revealed by reaction with Alexa568 azide (red). (A and B) Mouse small intestine. Villi are seen in transverse section in A and in longitudinal section in B. The cells with red nuclei are descended from cells that had been in S phase during the EdU pulse and thus incorporated EdU into their DNA. (C and D) Mouse brain. The vast majority of nuclei on brain sections were not labeled with EdU, confirming that the label does not detectably incorporate into the DNA of nondividing cells. Each of the two images shows a field of cells containing an EdU-labeled nucleus, belonging to a cell of undetermined type.
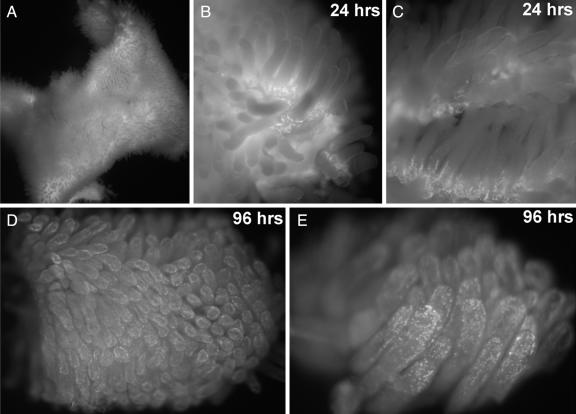
Finally, we wanted to determine whether EdU can be used to detect cell proliferation in large, fresh tissue and organ explants. Mice were injected i.p. with 100 μgrams EdU and their small intestine was harvested 24 or 96 h later. The fresh intestine was then directly stained with the cell-permeable TMR-azide, followed by fixation and washing to remove unincorporated TMR-azide. As shown in Fig. 5, after 24 h, EdU staining can be detected strongly in cells present at the base of the villi, which is the location described for the actively dividing cells of the small intestine mucosa. Ninety-six hours after the EdU pulse, the labeled cells have multiplied and migrated distally, away from the base of the villi and toward their tips. This whole-mount imaging of cellular turnover in small intestine villi is consistent with classical measurements and demonstrates that EdU can be used to assay cells proliferation in fresh tissues rapidly and with high sensitivity. In this context, EdU allows the rapid and sensitive staining of large organ fragments in whole mounts; this will make the study of organ and tissue dynamics much easier than by earlier methods.
Fig. 5.
Exploring cell proliferation and tissue dynamics in animals using EdU. Whole-mount fluorescent images of mouse small intestine, stained to detect EdU incorporation. Two adult mice were each injected i.p. with 100 μg of EdU in PBS and their small intestines were removed after 24 and 96 h, respectively. A freshly harvested 2-cm-long segment of the small intestine (A) was opened with a longitudinal cut, rinsed, and immediately stained for 10 min with 100 μM TMR-azide in the presence of Cu(I), without fixation or permeabilization. The intestine piece was then fixed, washed to remove unreacted TMR-azide, and imaged at low magnification on a dissecting microscope equipped with fluorescence. (B and C) After 24 h, the EdU shows strong incorporation in cells located at the base of each villus. (D and E) Ninety-six hours after the EdU pulse, the labeled cells have moved away from the base, near the tips of the villi. See text and Materials and Methods for details.
In some experiments it would be desirable to perform two different DNA-labeling pulses and be able to distinguish between the two. We reasoned that the alkyne and azide functionalities can be swapped between the deoxynucleoside analogue and detection reagent to allow two-color DNA staining based on click chemistry. We synthesized 5-azido-3′-deoxyuridine (AdU) as described for 5-azido-dUTP (11). Like EdU, this analogue was incorporated into cellular DNA. We detected it by reacting with a terminal alkyne conjugated to the Alexa568 fluorophore, under staining conditions identical to those used for EdU (see Materials and Methods). Although we were able to detect AdU-labeled DNA in cells by fluorescence microscopy, the background was, for unknown reasons, much higher than in the case of EdU (data not shown), as reported for other instances in which click reactions were performed on cells metabolically labeled with azides (12). The routine use of AdU as a second color for DNA labeling will have to await the development of a detection protocol that affords lower background staining. The combination of BrdU and EdU (Fig. 3B), meanwhile, offers a simple and robust way to perform double-DNA labeling.
We envision that the superior structural preservation allowed by EdU relative to BrdU staining should facilitate high-resolution electron microscopy studies of cellular chromatin. For example, EdU-labeled DNA can be imaged by electron microscopy by using an azide conjugated to eosin (or another fluorophore that generates singlet oxygen efficiently), followed by photoconversion of diaminobenzidine into an osmiophilic precipitate (13). Immunoelectron microscopy of EdU-labeled chromatin under nondenaturing conditions can also be achieved by using an azide conjugated to a good epitope (such as digoxigenin) followed by immunostaining.
A recent method called photoactivation localization microscopy (PALM) (14) generates nanometer-resolution images of biological molecules in cells by repeated cycles of photoactivation of “caged” fluorophores followed by imaging until fluorophore bleaching. The amount of fluorescent signal generated during EdU detection can be easily controlled (by varying ascorbic acid and fluorescent azide concentrations, and by the ability to perform successive stains), in a manner similar to photoactivation of fluorescence used for the PALM technique. The EdU-labeling method is thus well suited for optical imaging of cellular DNA at nanometer resolution.
Materials and Methods
EdU Labeling of Cultured Cells.
NIH 3T3 cells were grown on glass coverslips in DMEM supplemented with 10% bovine calf serum, penicillin, and streptomycin. HeLa cells were grown on glass coverslips in DMEM supplemented with 10% FBS, penicillin, and streptomycin. EdU was added to the culture media in concentrations ranging from 10 nM to 10 μM, for durations of time between 1 and 24 h. After labeling, cells were washed two to three times with PBS followed by addition of normal tissue culture media. If cells were to be analyzed immediately after labeling, the washes were omitted and cells were permeabilized and fixed instead. Cells were fixed by using a standard formaldehyde fixation protocol. Other fixation methods (formaldehyde and glutaraldehyde, or methanol) work equally well. EdU incorporated into cellular DNA is very stable and the fixed cells can be stored at 4°C and stained months later without loss of signal. To examine the effect of DNA synthesis inhibition on EdU incorporation, NIH 3T3 cells were incubated for 30 min in media supplemented with 10 mM hydroxyurea followed by overnight incubation in media supplemented with 10 μM EdU and 10 mM hydroxyurea.
EdU Staining.
After formaldehyde fixation, cells were rinsed once with TBS (although formaldehyde does not interfere with the detection reaction) and then stained by incubating for 10–30 min with 100 mM Tris (from 2M stock, pH 8.5), 0.5–1 mM CuSO4, 1–100 μM fluorescent azide (from 10 to 100 mM stocks in DMSO), and 50–100 mM ascorbic acid (added last to the mix from a 0.5 M stock in water). The staining mix was prepared fresh each time and was used for staining cells immediately after addition of ascorbate. After staining, the cells on coverslips were washed several times with TBS with 0.5% Triton X-100. EdU-stained cells can be immunostained by using standard protocols. Cells were counterstained with Hoechst, mounted in standard mounting media and imaged by fluorescence microscopy. The EdU stain is stable indefinitely at 4°C or lower temperatures.
For the experiment in Fig. 3A, NIH 3T3 cells labeled with 2 μM EdU overnight were fixed with formaldehyde, rinsed, and stained with 10 μM Alexa488-azide for 20 min. After washing several times with TBS with 0.5% Triton X-100, the cells were stained a second time with 10 μM Alexa594-azide for 20 min. The cells were washed again, mounted in standard mounting media, and imaged by fluorescence microscopy.
For the experiment in Fig. 3B, NIH 3T3 cells were labeled with 2 μM EdU and 2 μM BrdU overnight, followed by formaldehyde fixation and EdU detection by using 10 μM Alexa488-azide. The cells were then incubated in 1.5 M HCl at room temperature for 1 h with gentle rocking (to denature the DNA and expose the BrdU epitope), followed by standard immunodetection of BrdU by using a mouse anti-BrdU monoclonal antibody (clone BU-33, obtained from Sigma) and an Alexa488-conjugated anti-mouse secondary antibody (Molecular Probes). The cells were mounted in standard mounting media and imaged by fluorescence microscopy.
For staining cells alive, the coverslips were washed with medium without serum (OptiMEM) and mounted for live imaging on an inverted spinning disk confocal microscope (15) equipped with an OrcaER digital camera (Hamamatsu) and a 60× PlanApo oil objective (Nikon). The medium was then replaced with staining solution prepared fresh, which consisted of PBS supplemented with 0.5 mM CuSO4, 500 nM TMR-azide (from 100 mM stock in DMF), and 50 mM ascorbate (added immediately before use from 0.5M stock in water). The staining reaction was imaged by time-lapse fluorescence microscopy, taking one image every 30 s.
EdU Labeling of Mouse Tissues.
Mice were injected i.p. with 100–200 μg of EdU in PBS and mouse small intestines and brains were harvested at 24, 48, or 96 h after injection. For staining fixed sections, pieces of the small intestine and brain were formalin-fixed, embedded in paraffin, and sectioned. After paraffin removal, sections on glass slides were stained with 10 μM Alexa568-azide for 10–30 min, as described for fixed cells on coverslips except that the reaction and subsequent washes were performed in Coplin jars. Sections were counterstained with Hoechst and mounted for fluorescence microscopy.
For staining small intestine without fixation, 1- to 2-cm-long sections of freshly harvested small intestine were quickly rinsed in PBS, cut longitudinally to expose the mucosa, and then incubated for 30 min at room temperature in staining solution containing 100 μM TMR-azide. The small intestine explants were then fixed with formaldehyde and washed several times with PBS + 0.2% Triton X-100 by end-over-end rotation at room temperature. The stained intestine pieces were imaged on a dissecting microscope equipped with epi-fluorescence illumination.
EdU Labeling of DNA Replicated in Xenopus Egg Extracts.
Cycling Xenopus egg extracts were prepared as described in ref. 16. EdU was added to Xenopus egg extracts arrested in meiosis by cytostatic factor (CSF), at a final concentration of 20 μM. The extracts were then supplemented with permeabilized Xenopus sperm nuclei (100 nuclei per μl) and released from the CSF state by addition of calcium chloride to 0.4 mM. After allowing DNA replication to occur, the interphase extracts were returned to a CSF state by addition of CSF extract. Spindles were fixed and pelleted on polylysine-coated coverslips. EdU was stained with Alexa568-azide as described for cultured cells. Tubulin was stained with a fluorescein-labeled anti-tubulin antibody and DNA was counterstained with Hoechst dye.
Chemicals.
EdU was synthesized from 5-iodo-deoxyuridine and trimethyl-silyl-acetylene by a Sonogashira coupling reaction as described in ref. 17. The identity and purity of EdU was confirmed by TLC, NMR, and mass spectrometric analysis. EdU dissolved at 100 mM in DMSO or at 25 mM in water, was stored frozen, and was stable through multiple freezing and thawing cycles.
Compound 2 (Alexa568-azide).
2-amino-2′-[BOC-amino]ethylene glycol diethyl ether was synthesized from 2,2′-(ethylenedioxy)-bis[ethylamine] as described in ref. 18, and its identity confirmed by NMR and mass spectrometry. The NHS ester of the 6-bromo-hexanoic acid was generated in situ by reacting 1.6 g of the acid with equivalent amounts of N-hydroxysuccinimide and EDC dissolved in 20 ml of anhydrous dichloromethane, After stirring for 30 min at room temperature, an equivalent amount of 2-amino-2′-[BOC-amino]ethylene glycol diethyl ether was added and stirring was continued overnight at room temperature. The solution was extracted successively with 1 M sodium hydroxide, saturated ammonium chloride, and saturated sodium chloride, and the solvent was removed under reduced pressure. The resulting pale yellow oil was dried, then dissolved in 20 ml of dry DMF and reacted with 4 eq of sodium azide by stirring overnight at 60°C. The mixture was poured into water and was then extracted with ethyl acetate. The organic layer was evaporated and dried, yielding a pale-yellow oil. The BOC group was removed by treatment with 10 ml trifluoroacetic acid for 2 h at room temperature. After evaporation of most of the trifluoroacetic acid under reduced pressure, the remaining oil was dissolved in water. After extraction with dichloromethane (to remove any remaining BOC-protected amine), the aqueous solution was brought to pH 14 with sodium hydroxide and the amine base was extracted into dichloromethane. Removal of the organic solvent and drying afforded 2-amino-2′-[6-azido-hexanamide]ethylene glycol diethyl ether as a pale-yellow syrup. The identity of the product was confirmed by NMR and mass spectrometry. This amino azide was used to attach the azide group to a variety of fluorophores and haptens purchased as NHS esters. To obtain Alexa568-azide, the succinimidyl ester of Alexa568 carboxylic acid (mixed isomers, Molecular Probes) in anhydrous DMSO was reacted with the amino azide, according to the manufacturer's instructions.
Compound 3 (TMR-azide).
Twenty-five milligrams of carboxytetramethylrhodamine (TMR) succinimidyl ester (5,6 mixed isomers, Molecular Probes) were reacted for 2 h at room temperature in 0.5 ml of dry DMF with 2 eq of 3-bromopropylamine hydrobromide and 2 eq of triethylamine. Ten equivalents of solid sodium azide were added to the mix, which was stirred at 60°C overnight. Next day, the mixture was filtered to remove salts. The TMR-azide thus obtained was either precipitated with diethylether or used without purification, with equivalent results in staining reactions on EdU-labeled cells and tissues.
Compound 4 (5-azido-fluorescein).
Of 5-aminofluorescein, 3.5 g were dissolved in 100 ml of methanol. Four grams of sodium nitrite were added under vigorous stirring on ice, followed by the slowly dripping addition of 10 ml of a 5 M solution of hydrochloric acid. After NO stopped evolving from the stirred reaction, 5 g of solid sodium azide was added in small portions, under stirring in an open flask, in a well ventilated hood (caution: hydrazoic acid is very toxic). After nitrogen stopped evolving from the stirred mix, the mix was filtered and the solvents removed under vacuum. One hundred milliliters of a 2 M HCl solution were added to the residue and the resulting yellow-brown precipitate was washed extensively with cold water. The precipitate was then dried (dry weight: 2.5 g) and tested in a click detection reaction y by using EdU-labeled cells. No attempt was made to calculate the yield of 5-azido-fluorescein synthesis.
5-Azido-2′-deoxyuridine (AdU) was synthesized by the method used to synthesize 5-azido-dUMP (11) and was used to label cells similarly to EdU. Detection of AdU-labeled DNA was accomplished by using a protocol identical to the one used for EdU detection, using a fluorescent alkyne obtained by reacting carboxytetramethylrhodamine (TMR) succinimidyl ester (5,6 mixed isomers, Molecular Probes) in dry DMF with an equivalent amount of propargylamine, according to the manufacturer's instructions.
ACKNOWLEDGMENTS.
We thank Frank McKeon for mouse injections and dissections, Stephen C Miller and Charles Kim for chemistry advice and for many stimulating discussions, and Howard Green for helpful comments on the manuscript. This work was supported by the Leukemia and Lymphoma Society, the Smith Family Foundation, and the Rita Allen Foundation (A.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Bick MD, Davidson RL. Total substitution of bromodeoxyuridine for thymidine in the DNA of a bromodeoxyuridine-dependent cell line. Proc Natl Acad Sci USA. 1974;71:2082–2086. doi: 10.1073/pnas.71.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gratzner HG. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 3.Waldman FM, et al. A comparison between bromodeoxyuridine and 3H thymidine labeling in human breast tumors. Mod Pathol. 1991;4:718–722. [PubMed] [Google Scholar]
- 4.Rakic P. Neurogenesis in adult primate neocortex: an evaluation of the evidence. Nat Rev Neurosci. 2002;3:65–71. doi: 10.1038/nrn700. [DOI] [PubMed] [Google Scholar]
- 5.Tornoe CW, et al. Peptidotriazoles on solid phase: [1,2,3]-Triazoles by regiospecific copper(i) -catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 6.Rostovtsev VV, et al. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq E, et al. (E)-5-(2-Bromovinyl)-2′-deoxyuridine: A potent and selective anti-herpes agent. Proc Natl Acad Sci USA. 1979;76:2947–2951. doi: 10.1073/pnas.76.6.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker RT, et al. The synthesis and properties of some 5-substituted uracil derivatives. Nucleic Acids Symp Ser. 1980:s95–s102. [PubMed] [Google Scholar]
- 9.Chan TR, et al. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 10.Rodionov VO, et al. Mechanism of the ligand-free CuI-catalyzed azide-alkyne cycloaddition reaction. Angew Chem Int Ed Engl. 2005;44:2210–2215. doi: 10.1002/anie.200461496. [DOI] [PubMed] [Google Scholar]
- 11.Sunthankar P, et al. Synthesis of 5-azido-UDP-N-acetylhexosamine photoaffinity analogs and radiolabeled UDP-N-acetylhexosamines. Anal Biochem. 1998;258:195–201. doi: 10.1006/abio.1998.2600. [DOI] [PubMed] [Google Scholar]
- 12.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Deerinck TJ, et al. Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 15.Maddox PS, et al. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 2003;360:597–617. doi: 10.1016/s0076-6879(03)60130-8. [DOI] [PubMed] [Google Scholar]
- 16.Sawin KE, Mitchison TJ. Mitotic spindle assembly by two different pathways in vitro. J Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu CS, Oberdorfer F. Synthesis of (E)-5-[2-(tri-n-butylstannyl)vinyl] substituted 2′-deoxyuridine derivatives for use in halogenation and radiohalogenation reactions. Synlett. 2000;1:86–88. [Google Scholar]
- 18.Zuckermann RN, et al. Discovery of nanomolar ligands for 7-transmembrane G-protein-coupled receptors from a diverse N-(substituted)glycine peptoid library. J Med Chem. 1994;37:2678–2685. doi: 10.1021/jm00043a007. [DOI] [PubMed] [Google Scholar]