Abstract
The central questions of bacterial ecology and evolution require a method to consistently demarcate, from the vast and diverse set of bacterial cells within a natural community, the groups playing ecologically distinct roles (ecotypes). Because of a lack of theory-based guidelines, current methods in bacterial systematics fail to divide the bacterial domain of life into meaningful units of ecology and evolution. We introduce a sequence-based approach (“ecotype simulation”) to model the evolutionary dynamics of bacterial populations and to identify ecotypes within a natural community, focusing here on two Bacillus clades surveyed from the “Evolution Canyons” of Israel. This approach has identified multiple ecotypes within traditional species, with each predicted to be an ecologically distinct lineage; many such ecotypes were confirmed to be ecologically distinct, with specialization to different canyon slopes with different solar exposures. Ecotype simulation provides a long-needed natural foundation for microbial ecology and systematics.
Keywords: Bacillus, Evolution Canyon, ecotype, periodic selection, species concept
To fully understand any community's ecology, we need to identify its ecologically distinct populations and to determine their mutual interactions, because these are the units that contribute uniquely to community assembly, function, and dynamics (1). In the case of a bacterial community, identifying the ecologically distinct members is a particularly formidable task. This is due to the enormous number of bacterial species and ecological roles played within a typical community (2), our inability to cultivate more than a small fraction of these species for study within the laboratory (3), and our inability to predict the genes responsible for ecological divergence, owing to the role of horizontal genetic transfer in bacterial adaptation (4). A DNA sequence-based approach can help overcome these challenges, because the bacteria falling into sequence clusters for a given gene can correspond to ecologically distinct populations, even for genes not related to the adaptive divergence between populations (5). However, the ecological interpretation of a sequence-based phylogeny is not straightforward. Any phylogeny contains a hierarchy of subclusters within clusters, and it is generally not clear which level of sequence cluster corresponds to ecologically distinct populations. Also, a sequence-based phylogeny is complicated when factors, such as geographical isolation, genetic drift, plasmid gain and loss, or rapid speciation, result in a failure of correspondence between sequence divergence and ecological divergence (4, 6). Currently, bacterial systematics employs universal thresholds of molecular divergence values to help demarcate species (7–10), but there is no theoretical basis for identifying the thresholds that yield ecologically distinct populations, nor is there evidence to suggest that a single sequence-identity cut-off value appropriately demarcates the fundamental units of bacterial ecology and evolution (4, 6, 11). Indeed, the traditional approaches of bacterial systematics have led to species that are enormously diverse in their genome content, physiology, and ecology (4, 12).
Here, we propose and test a conceptual framework, based on the evolutionary dynamics of bacterial populations, to estimate the number of ecologically distinct populations within a given clade (a group of organisms sharing a common ancestor) and to identify the members of each such population. We present an algorithm, “ecotype simulation” (ES) (4, 13, 14), which models an ecotype as an ecologically distinct group whose diversity is limited by a force of cohesion, usually the genome-wide purging of diversity known as periodic selection but also genetic drift (4). Periodic selection occurs when a new adaptive mutant arises within an asexual or rarely sexual ecotype, and natural selection causes the mutant and its nearly clonal descendants to replace all competing variants within the ecotype (4, 15) [see supporting information (SI)]. A new ecotype is founded when an adaptive mutation (or a recombination event) allows a variant to invade a new ecological niche. Owing to ecological differences between ecotypes, a periodic selection event within one ecotype does not extinguish the diversity within other ecotypes (4). So defined, bacterial ecotypes have the quintessential properties of species recognized by many systematists outside of microbiology: They are ecologically distinct groups belonging to genetically cohesive and irreversibly separate evolutionary lineages, and they are each invented only once (4, 6, 16).
We have applied ES to examine two clades whose ecological diversity and habitats have been intensively studied. We surveyed multilocus diversity among strains of Bacillus simplex and the Bacillus subtilis–Bacillus licheniformis clade isolated from the “Evolution Canyons” of Israel, which are arid, east–west-running canyons providing three major habitat zones—north- and south-facing slopes with extremely different levels of insolation and a usually dry streambed at the canyon bottom (17–20). We will demonstrate that many of the ecotypes predicted by ES are adapted to the different microhabitats of the canyons. We show that ES is able to discern ecologically distinct populations that are invisible to the current framework of bacterial systematics. As a solution, we suggest a means to incorporate high-resolution theory-based demarcation into bacterial systematics.
Results and Discussion
Clade Sequence Diversity.
We surveyed sequence diversity at three protein-coding genes within B. simplex isolated from Evolution Canyons I and II near Haifa, Israel, and within the B. subtilis–B. licheniformis clade from Evolution Canyon III in the Negev Desert. We graphically characterized the sequence diversity for a clade by plotting the number of bins (or sequence clusters) required to encompass the sample of sequences from the clade, as increasingly stringent sequence identity criteria were used to define these clusters (21, 22) (Fig. 1). Complete linkage clustering (23) was used to bin the sequences into clusters with different levels of minimum pairwise identity (Fig. 1). The “clade sequence diversity” is the pattern of the number of bins over different sequence identity criteria for binning (Fig. 1). Because sequence divergence increases with the time since divergence, this pattern is a way of describing the evolutionary history of cladogenesis (splitting of lineages) within a clade (22).
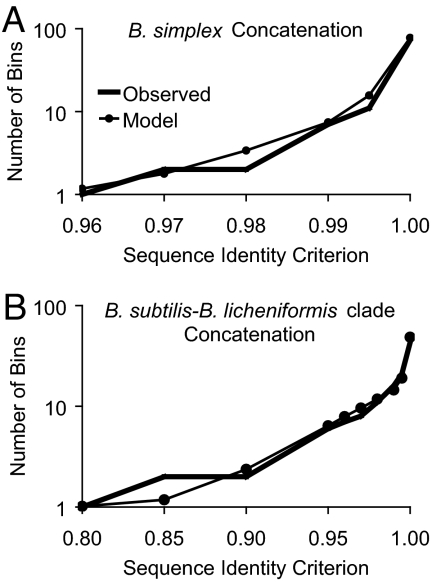
Fig. 1.
Observed and modeled clade sequence diversity patterns. Sequences for a gene (or a concatenation of genes) were binned by complete linkage clustering (23) into clusters with different levels of minimum pairwise identity. ES analysis of each clade yielded two solutions, one with low drift and one with high drift. The high-drift solutions require population sizes that are unrealistically low for these taxa (SI), so the model curves are based on the low-drift solutions. For taxa with low population sizes and/or extremely high recombination rates, high-drift solutions (with little or no periodic selection) may be the most appropriate. The individual points for each model are means based on 1,000 replications of the low-drift solution. (A) Diversity among 116 B. simplex isolates from Evolution Canyons I and II based on a concatenation of gapA, rpoB, and uvrA, with recombinant organisms removed. (B) Diversity among 73 isolates within the B. licheniformis–B. subtilis clade, primarily from Evolution Canyon III, as based on a concatenation of gapA, gyrA, and rpoB, with recombinant organisms removed.
As typically seen in clade sequence diversity patterns (21, 24), there is an approximately log-linear increase in the number of bins with increasing sequence identity, which has been interpreted as revealing a constant net rate of ecotype formation (22), and there is a flair of increased diversity at an inflection point (in this case ≈98–99% identity), previously interpreted as reflecting the ephemeral sequence divergence within populations that has not yet been purged by periodic selection or drift (21, 24). These interpretations are supported by observing the effects of ecotype formation, periodic selection, and drift rates on the shape of clade sequence diversity curves (see SI).
Quantification and Demarcation of Ecotypes.
We modeled the evolutionary history of each clade, using four parameters to yield with maximum likelihood the observed clade sequence diversity pattern (Fig. 2). In this model, genome-wide diversity within each ecotype is purged recurrently by periodic selection at rate σ. Diversity within ecotypes is also limited by genetic drift, occurring at rate d. New ecotypes are formed at net rate Ω, representing the difference between rates of ecotype formation and extinction. The model also includes a parameter for the total number of ecotypes, n, in the sample of sequences. The ES analysis evaluates different quartets of parameter values for their likelihood of yielding an evolutionary history consistent with the observed clade sequence diversity (Fig. 1). Thus, ES quantifies the ecological diversity within a community (as the number of ecotypes sampled, n) by analyzing the community's evolutionary history. ES also estimates the rates of net ecotype formation and periodic selection, allowing for future tests of how a clade's ecological and life history characteristics might determine its evolutionary rates (see Fig. 2 and SI). The ecotype simulation software is available at http://fcohan.web.wesleyan.edu/ecosim.
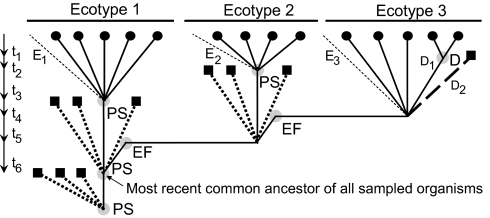
Fig. 2.
The ecotype simulation algorithm. The algorithm simulates the evolutionary history of the ν organisms sampled from nature, under different quartets of values for the net rate of ecotype formation (EF), the rates of periodic selection (PS) and drift (D), and the number of ecotypes in the sample. In the coalescence approach taken (36), the algorithm considers only the lineages that are directly ancestral to the ν sampled organisms (represented by black circles). These focal lineages are represented by solid lines; the many contemporary lineages not sampled from each ecotype are indicated by light dashed lines (E1, E2, and E3); the lineages extinguished by past PS and D are represented by bold short-dashed lines and long-dashed lines, respectively, with each extinction represented by a square. The program begins with a “backward” simulation that stochastically produces a phylogenetic representation of the history of the community, establishing nodes of coalescence of lineages (due to PS, EF, or D; indicated by gray circles) and time between nodes (t1, t2, etc.); this phylogeny is then taken as a scaffold for the forward simulation. The purpose of the forward simulation is to produce mutational nucleotide substitutions throughout the history of the clade, according to the phylogenetic scaffold. To begin a simulation, a set of ν contemporary organisms (representing the ν organisms sampled from nature) are distributed randomly (according to the canonical lognormal distribution) among n ecotypes (here, ν = 14 and n = 3). Working backward from the ν organisms in the present, the processes of EF, PS, and D occur stochastically in time according to their respective rates (Ω, σ, and d). For each such event, one or more lineages coalesce into a single ancestral lineage, as described in SI. Note that in the backward-looking view of the coalescence formulation, each PS appears as a coalescence event, in which all lineages after the PS coalesce into the survivor of the PS event. Likewise, each D event appears in this backward-looking view as the coalescence of a pair of lineages within an ecotype (e.g., two contemporary lineages coalesce into lineage D1 to reflect the increased representation of lineage D1 after the random loss by drift of lineage D2). Because Ω is the net rate of EF events, taking into account extinction, we include in the simulation only those EF events resulting in ecotypes that survive into our contemporary sample. The backward phase of the simulation ends when all of the branches have coalesced into a single node; this represents the most recent common ancestor of all of the sampled organisms. Then the forward simulation begins when a sequence (of the same length as the observed sequence data) is assigned to this most recent common ancestor. Nucleotide substitutions then occur stochastically, going forward in time, between each pair of nodes in the phylogeny derived from the backward simulation, according to the time between the events determining the nodes. This generates a matrix of pairwise sequence divergence between all ν contemporary organisms for a simulation replicate, from which a clade sequence diversity curve is calculated; the simulated clade sequence diversity curve is then compared against the observed clade sequence diversity curve (see SI).
The ES of the history of the concatenated gene sequence in B. simplex estimated the presence of 13 putative ecotypes within this named species in Evolution Canyons I and II (Table 1). The ES of the B. subtilis–B. licheniformis clade estimated 17 putative ecotypes. The more rapidly evolving gene gyrA estimated somewhat more ecotypes than the other genes in the B. subtilis–B. licheniformis clade. Also, the most rapidly evolving gene among the three used for B. simplex, rpoB, estimated considerably more ecotypes than the other genes, but the estimate based on the concatenation was not affected greatly by the outlier gene.
Table 1.
Estimates of parameter values for the low-drift solutions of each clade with 95% confidence intervals in parentheses
| Clade | Gene | Net rate of ecotype formation (Ω) | Rate of periodic selection (σ) | Rate of drift (d) | Ecotypes, no. (n) | Ratio of σ:Ω |
|---|---|---|---|---|---|---|
| B. simplex | Concat. | 0.084 (0.026, 0.18) | 0.57901 (0.083, 4.19) | 0 (0, 0.70) | 13 (5, 28) | 6.89 |
| gapA | 0.20 (0.042, 0.44) | 3.56 (0.24, ∞) | 0 (0, ∞) | 11 (3, 40) | 17.8 | |
| rpoB | 0.43 (0.19, 0.93) | 54.16 (1.17, ∞) | 0 (0, ∞) | 34 (9, 79) | 126 | |
| uvrA | 0.29 (0.060, 0.63) | 1.28 (0.08 > 100.0) | 0 (0, ∞) | 10 (3, 65) | 4.41 | |
| B. subtilis–B. licheniformis | Concat. | 0.028 (0.013, 0.041) | 0.92 (0.14, 4.55) | 0 (0, 0.70) | 17 (9, 27) | 32.9 |
| gapA | 0.083 (0.038, 0.18) | 2.10 (0.44, 31.33) | 0 (0, 5.12) | 10 (6, 20) | 25.3 | |
| gyrA | 0.049 (0.036, 0.073) | 2.54 (0.40, 27.66) | 0 (0, 1.43) | 20 (15, 30) | 51.8 | |
| rpoB | 0.10 (0.049, 0.16) | 8.47 (1.34, ∞) | 0 (0, 39.68) | 12 (8, 19) | 84.7 |
See SI for high-drift solutions. The confidence intervals were determined following the method of Felsenstein (see SI) and are indicated in parentheses. The number of ecotypes estimated here tends to be greater than the number of ecotypes demarcated in Fig. 3, because ecotype demarcation is performed conservatively—a clade is deemed an ecotype if the confidence interval of n includes 1, even if the maximum likelihood estimate for n is greater than 1 (SI). Concat., concatenation of three genes.
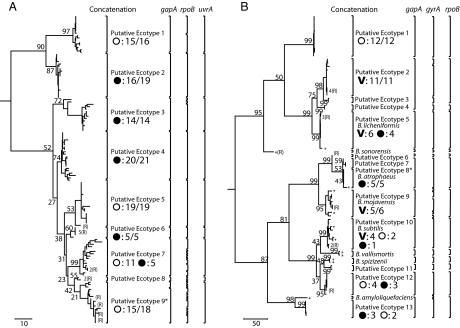
We next extended the ES approach to identify the individual ecotypes within each clade, with the ultimate aim of testing whether each putative ecotype actually corresponds to an ecologically distinct population. Our approach for ecotype demarcation was to find the most inclusive clades that are each consistent with being a single ecotype, as explained in SI. This is a conservative approach that tends to yield fewer demarcated ecotypes than indicated by the parameter estimates of Table 1 (see SI). Accordingly, nine and 13 putative ecotypes were identified in the B. simplex and B. subtilis–B. licheniformis clades' concatenated gene sets, respectively (Fig. 3). For B. simplex, the individual genes yielded the same demarcations as the concatenation, except for several cases in which rpoB split a putative ecotype into two or more ecotypes; in one case, gapA split a putative ecotype into two. Likewise, the individual gene analyses of the B. subtilis–B. licheniformis clade gave the same demarcations as the concatenation, except in two cases where putative ecotypes were split by gyrA. In general, analyses using rapidly evolving genes were more likely to discern very closely related clades into distinct ecotypes than were analyses using more slowly evolving genes. Future analyses that involve sequences of many genes, perhaps even whole genomes, should focus on a subset of genes with a history of rapid evolution and infrequent recombination (25).
Fig. 3.
Phylogeny and ecotype demarcation of the B. simplex and B. subtilis–B. licheniformis clades. Phylogenies are based on parsimony, with 100-replicate bootstrapping, using MEGA (37). In isolates for which one gene had recombined (a group of related recombinants is indicated by “R” following the number of recombinants), the recombined gene was replaced in the input for MEGA by “unknown” nucleotides; so the phylogeny estimates the recombination-free tree. Ecotypes were demarcated conservatively as the most inclusive clades that were each consistent with being a single ecotype (SI). For nearly all putative ecotypes, the maximum likelihood solution of n was equal to 1, although in some cases n = 2 with the lower confidence interval including n = 1. Ecotype demarcations are indicated by brackets, for the concatenation and each individual gene. The ecotype demarcations were similar based on the concatenation and the individual genes, except that the more rapidly evolving genes (gyrA in the case of B. subtilis–B. licheniformis and rpoB in the case of those genes analyzed in B. simplex) tended to split the ecotypes determined by analysis of the concatenation. For isolates that had recombined at one gene locus, ecotype placement was determined by ES of the two genes that had not recombined. For example, the three recombinants indicated within PE 5 of the B. subtilis–B. licheniformis clade (which had recombined at gapA) were found to be in the same ecotype as the members of PE 5 in analyses of gyrA and rpoB. With two exceptions, demarcated ecotypes were supported as monophyletic groups in at least 50% of bootstrap replications; the exceptions were PE9 of B. simplex and PE8 of the B. subtilis–B. licheniformis clade, which are asterisked to indicate that their phylogenetic status is tentative, pending additional sequence data. The median bootstrap support for putative ecotypes was 72% within B. simplex and 99% within the B. subtilis–B. licheniformis clade. Whereas the B. simplex ecotypes do not generally have high bootstrap support, owing in part to their very close relatedness, their monophyly is supported by alternative phylogenetic approaches, including Bayesian and neighbor joining algorithms (data not shown). Microhabitat sources were the south-facing slope (open circles), the north-facing slope (filled circles), and the canyon bottom (indicated by V). For each ecotype represented by at least four isolates, the principal microhabitat source(s) is indicated. If one microhabitat provided at least 80% of the isolates, the principal microhabitat source is indicated; for ecotypes not so dominated by a single source, all microhabitat sources are indicated. The number of isolates from each source is indicated. (A) The phylogeny of B. simplex rooted by B. subtilis, B. licheniformis, B. cereus, and B. halodurans. The nine putative ecotypes of this clade differ significantly in their associations with the two slopes. (B) The phylogeny of the B. subtilis–B. licheniformis clade rooted by B. halodurans. The 13 putative ecotypes differ significantly in their associations with the two slopes and the canyon bottom.
In cases where a strain had been eliminated from the concatenation analysis because of recombination (Fig. 3), the strain was classified into an ecotype based on single-gene ES analysis of the two genes that did not recombine.
Ecological Distinctness of Putative Ecotypes.
We next investigated whether the ecotypes hypothesized by ES are ecologically distinct by making use of the topography of the Evolution Canyons. As part of the Evolution Canyon paradigm developed by E. Nevo and colleagues (18), the canyons have three major habitats, a south-facing slope (SFS) with high solar insolation (and other covarying physical and chemical parameters, and/or interactions with other organisms adapted to this habitat), a north-facing slope (NFS) with less insolation, and a canyon bottom with greater access to water (18, 26).
In B. simplex, the nine putative ecotypes we identified were significantly heterogeneous in their associations with the two major habitats from which B. simplex was isolated (χ2 = 82.16; P < 0.0001; 8 df) (Fig. 3A). The clade at the top of Fig. 3A contains two putative ecotypes, one found primarily on the SFS [putative ecotype 1 (PE1)] and the other on the NFS (PE2). Note also that PE3 and PE4 represent well supported clades found nearly entirely on the NFS. The clade containing PE5–PE9 contains what appear to be specialists to the SFS (PE5 and PE9), one ecotype specialized to the NFS (PE6), and one ecotype (PE7) that is abundant on both slopes but in higher frequency on the SFS, with physiological adaptation to the SFS (20).
The ecological distinctness of the B. simplex ecotypes is further confirmed by their habitat-related physiological properties. The ecotypes associated with the hotter SFSs have greater growth rates at a stressful high temperature than do the ecotypes associated with NFS's, but the differences disappear at optimal temperature (20) (see SI). Also, the SFS-associated ecotypes constitutively produce greater amounts of isomethyl-branched fatty acids, which are beneficial for heat tolerance, than NFS-associated ecotypes (J.S., E.B., R. Kroppenstedt, and B. Tindall, unpublished data). Resistance to UV-C radiation does not appear to contribute to SFS adaptation (19). Other aspects of ecotype divergence yet to be explored in this system are differences in nutrient resources and interactions with other microorganisms (27).
Within the B. subtilis–B. licheniformis clade, the 13 hypothesized ecotypes are heterogeneous in their associations with the major habitats (χ2 = 84.25; P < 0.0001; 24 df) (Fig. 3B). PE1 appears to be specialized to the SFS; PE2 and PE9 appear to be specialized to the canyon bottom; PE5 (which includes the type strain of B. licheniformis) appears to be specialized to the canyon bottom and NFS; and PE8 (which includes the type strain of B. atrophaeus) appears to be specialized to the NFS. As seen also in SFS-adapted ecotypes in B. simplex, the SFS-adapted PE1 of the B. subtilis–B. licheniformis clade has heat-adapting isomethyl-branching fatty acids in higher abundance than closely related ecotypes not specialized to SFS (J.S., E.B.P., and F.M.C., unpublished data).
We do not currently have sufficient ecological data to discern the specific factors that may contribute to the ecological distinctness of every putative ecotype. We anticipate that in future applications of ES, microbiologists will confirm the ecological distinctness of putative ecotypes through microhabitat distribution studies and comparisons of genome content and analyses of genome-wide gene expression and comprehensive metabolic phenotype. In general, we expect closely related ecotypes to differ in their adaptations to a great diversity of ecological dimensions, including different temperatures (14), photic zones (14), sources of inorganic nutrients (28), carbon sources used (29), and host organisms (30).
Resolution of Ecotype Simulation.
Except in cases of compelling phenotypic differentiation (usually related to human disease), bacterial systematists have frequently used 16S rRNA gene divergence as a guide to species demarcation, in which divergence scores >3% between strains denote that they should be recognized as different species; recently, this value has been changed to 1% divergence (7). Although this guideline does not require clades <1% divergent to be subsumed within a single species, it provides no encouragement for systematists to discover ecologically significant diversity among such clades. We note that the 1% guideline would group together within a single species many of the most closely related ecotypes identified by ES, many of which have been confirmed to be ecologically distinct. This is particularly striking in the case of B. simplex—here, the 131 isolates comprising the nine ecotypes identified by ES (Fig. 3A) are absolutely identical in their 16S rRNA sequences (≈1350 nt were sequenced) and are thus invisible to a 1% 16S rRNA divergence criterion for species demarcation. The putative ecotypes identified within the B. subtilis–B. licheniformis clade correspond more closely to the named species, except that four ecotypes (PE2–PE5) are within 0.72% 16S rRNA divergence of the B. licheniformis type strain and would most probably be recognized by bacterial systematics as members of that species (Fig. 3B). Also, PE11 and PE12 are within 0.40% divergence of B. spizizenii and would fit within that species. One newly discovered ecotype (PE1) is just outside the 1% divergence guideline from B. licheniformis. Thus, ES has discovered ecological diversity that would probably have been ignored within the current systematics of bacteria (as well as one ecotype that would have been recognized).
This conclusion is supported also by results from preliminary versions of ES, in which 11 putative ecotypes were discovered within the species Legionella pneumophila; some ecotypes were confirmed as distinct in their host ranges and in their gene expression patterns during infection (13). Similarly, two ecotypes were identified within subclade A of hot spring Synechococcus, one of which was associated with a specific temperature (65°C); also, two ecotypes were identified within subclade A′, each of which was associated with a different depth in the photic zone (14). Within each of the Synechococcus subclades, sequences were within 0.71% divergence in 16S rRNA sequence, and so the current framework of systematics would most probably not distinguish these ecotypes.
Thus, ES promises to be an effective way to discover ecological diversity. Many of the ecotypes hypothesized by ES and confirmed as ecologically distinct would fit within the species demarcated by traditional bacterial systematics approaches. ES has an important advantage over current methods in bacterial systematics in that it does not employ a universal threshold of molecular divergence that is arbitrarily defined and subjectively applied. In ES, analysis of the evolutionary history of a particular clade yields the appropriate criteria for demarcating ecotypes of that clade. The result is that ES can identify ecotypes that are not discerned by our current framework for bacterial systematics (Fig. 3 and Table 1).
That ecotypes can be found lumped within established species suggests that bacterial systematics is failing in its fundamental mission—to precisely provide the ecological properties of any organism that is classified to species (31). The ES approach promises to rectify this by offering a general theory-based approach for identifying a multiplicity of ecotypes per taxon, even before the ecological differences among putative ecotypes can be confirmed.
Incorporating Ecology into Bacterial Systematics.
We propose a paradigm by which bacterial systematics may use ES to demarcate ecotypes, while taking into account a potential diversity of evolutionary models. The ES approach is most likely to reveal ecotypes under a “stable ecotype” model in which new ecotypes are formed only rarely and each ecotype endures many periodic selection events during its lifetime. Under these circumstances, there is time for accretion of sequence divergence between ecotypes, with recurrent purging of diversity within but not between ecotypes. The sequence clustering we observe in these systems is thus dominated by periodic selection, yielding a close correspondence between ecotypes and sequence clusters (4, 6). We note that, for each clade, ES has estimated the rate of periodic selection to be much greater than the net rate of ecotype formation—the condition promoting correspondence of ecotypes and sequence clusters (Table 1).
However, bacterial systematics must take into account that factors other than periodic selection may contribute to sequence clustering in certain lineages (4). To accommodate these additional factors, we suggest that longstanding ecotypes be demarcated as the smallest clades that show (i) a history of coexistence as separate ecologically distinct lineages, as inferred from ES (or an equivalent sequence-based approach) and supported as monophyletic groups by bootstrap or similar analysis, and (ii) a prognosis for future coexistence, as inferred from the ecological distinctness of the groups in nature (4). Ecotypes cannot be inferred by sequence clustering alone, because it is possible for one ecotype to fall into multiple sequence clusters when geography and genetic drift have been major factors in the history of the lineage (4) (see SI). We note also that showing ecological distinctness alone, even in nature, is not sufficient to infer that the groups will coexist as separate lineages, given the possibility of recurrent, plasmid-based evolution into a particular ecological niche (see SI). Also, ecological differences inferred from laboratory tests may not be relevant to coexistence in nature. Identification of ecotypes therefore requires both a sequence-based approach to formulate hypotheses about putative ecotypes and an ecological approach to confirm these hypotheses.
Ecotypes that are confirmed to have a history of coexistence as distinct lineages and a prognosis of future coexistence are the fundamental units of bacterial ecology and evolution (4, 6, 11). We recommend that they be recognized also as the fundamental units of diversity in bacterial systematics (32). We suggest that, when multiple ecotypes are discovered within the accepted phylogenetic range of an established species (e.g., with 1% divergence in 16S rRNA), the ecotypes should be recognized and named by adding an “ecovar” epithet to the species binomial, for example, by naming the ecotypes within B. simplex. When an ecotype is discovered that is outside the phylogenetic range of any established species, we propose that the ecotype should be given a new species name, for example, by naming PE1 of the B. subtilis–B. licheniformis clade as a separate species. This dual approach should enrich bacterial systematics with ecologically significant but previously ignored groups, while respecting the stability of taxon names.
By identifying taxa at the level of ecotypes, bacterial systematics will provide a long-needed, biologically meaningful, taxonomic grouping of microbial diversity, to the benefit of other biological disciplines such as ecology, evolution, biochemistry, genomics, epidemiology, and biotechnology (4). The identification of these groups will be a critical step forward in our venture to understand the myriad ecological interactions within a natural microbial community.
Materials and Methods
Diversity Within B. simplex.
The details of soil collection and isolation of B. simplex from Evolution Canyons I and II, each with a south-facing slope and a north-facing slope, are described in ref. 19. PCR and sequencing of the gapA, rpoB, uvrA, and 16S rRNA genes (GenBank accession nos. EU305743–EU306135) were as described in refs. 19, 20, and 33.
Diversity Within the B. subtilis–B. licheniformis Clade.
Evolution Canyon III is located in the southern Negev desert, at Nahal Shaharut, a tributary of Nahal Hiyyon (lat 29°55′N, long 34°58′E); EC III has an SFS (≈35° rise) and an NFS (≈30° rise), separated by ≈150 m at the bottom (26). The soil (from the top 1- to 3-cm layer, taken on March 25, 2003) was collected from three elevation stations each from the SFS and NFS habitats and from one collecting station at the canyon bottom, with three collecting sites per station. Strains from the B. subtilis–B. licheniformis clade were identified by metabolic tests (34) and confirmed by sequences of rpoB. Additional strains and species from this clade were obtained from National Center for Agricultural Utilization Research of the U.S. Department of Agriculture.
DNA extraction, PCR, and sequencing were as described in SI. The gapA, gyrA, rpoB, and 16SrRNA sequences are accessible as GenBank numbers EU304829–EU304903, EF026654–EF026744, EF015305–EF015395, and EU304904–EU304976, respectively.
Preparation of Sequences for Ecotype Simulation.
Alignment positions with gaps or indeterminate nucleotides in any sequence were removed. We compensated for PCR and sequencing error by “correcting” a random subset of the singleton nucleotides (i.e., occurring at a particular nucleotide position in only one sequence) equal in number to the expected number of PCR and sequencing errors (see SI). Each chosen singleton nucleotide was corrected to the nucleotide of its closest relative at that site as determined by pairwise sequence distance. For each clade, we conducted separate analyses for the concatenation of the three protein-coding genes and for each of these three genes individually, excluding recombinant sequences as described below.
Accommodation for Recombination.
The ES algorithm takes recombination into account by allowing that periodic selection may not be significant in some taxa with high recombination rates. ES allows for periodic selection and/or genetic drift, to constrain within-ecotype sequence diversity.
The ES algorithm does not take into account that recombination between ecotypes can introduce sequence diversity into an ecotype at a gene locus being surveyed. Therefore, we identified strains that underwent recombination and the particular gene involved in each recombination event, using the “majority rules” rationale (35): In cases where a strain had changed its phylogenetic affiliation for one among the set of genes, we interpreted the two genes showing congruence of relationship as indicating the organism's phylogenetic position, and the aberrant gene as having recombined. A recombination-free phylogeny was estimated based on the concatenation with the recombined gene of each recombinant strain represented as “unknown” nucleotides. Using the ClonalFrame algorithm (35), we were able to confirm that our method identified partial- and whole-gene recombinants.
If a strain was shown to have recombined in any of the three protein-coding genes, the strain was eliminated from ES analysis of the three-gene concatenation; such a strain was also eliminated from single-gene ES analysis for its recombined gene but not from analysis of the other two genes.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Mary Bateson for suggesting improvements in earlier manuscripts. This work was funded by National Science Foundation Frontiers in Biological Research Program Award EF-0328698 (to D.M.W. and F.C.) and the National Aeronautics and Space Administration Exobiology Program Award NAG5–8824 (to D.M.W. and F.M.C.), a grant from the Ancell–Teicher Research Foundation for Genetics and Molecular Evolution (to E.N.), Deutsche Forschungsgemeinschaft Grant 1352/1–1 (to J.S.), and research grants from Wesleyan University (to D.K. and F.M.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU305743–EU306135, EU304829–EU304976, EF026654–EF026744, and EF015305–EF015395).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712205105/DC1.
References
- 1.Mayr E. The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Cambridge, MA: Harvard Univ Press; 1982. [Google Scholar]
- 2.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 3.Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- 4.Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 5.Blaxter ML. The promise of a DNA taxonomy. Philos Trans R Soc Lond B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevers D, et al. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 7.Stackebrandt E, Ebers J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 8.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 9.Wayne LG, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 10.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward DM, et al. Genomics, environmental genomics and the issue of microbial species. Heredity. 2008;100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- 12.Staley JT. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans R Soc Lond B. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohan FM, Koeppel A, Krizanc D. In: Legionella: State of the Art 30 Years after Its Recognition. Cianciotto NP, et al., editors. Washington, DC: ASM; 2006. pp. 367–376. [Google Scholar]
- 14.Ward DM, et al. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Phil Trans Roy Soc Ser B. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohan FM. In: Selective Sweep. Nurminsky D, editor. Georgetown, TX: Landes Bioscience; 2005. pp. 78–93. [Google Scholar]
- 16.de Queiroz K. Ernst Mayr and the modern concept of species. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6600–6607. doi: 10.1073/pnas.0502030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishkan I, Nevo E, Wasser SP, Beharav A. Adaptive spatiotemporal distribution of soil microfungi in “Evolution Canyon” II, Lower Nahal Keziv, western Upper Galilee, Israel. Biol J Linnean Soc. 2003;78:527–539. [Google Scholar]
- 18.Nevo E. Asian, African and European biota meet at “Evolution Canyon”, Israel: local tests of global biodiversity and genetic diversity patterns. Proc R Soc London B. 1995;262:149–155. [Google Scholar]
- 19.Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc Natl Acad Sci USA. 2005;102:15924–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikorski J, Nevo E. Patterns of thermal adaptation of Bacillus simplex to the microclimatically contrasting slopes of “Evolution Canyons” I and II, Israel. Environ Microbiol. 2007;9:716–726. doi: 10.1111/j.1462-2920.2006.01193.x. [DOI] [PubMed] [Google Scholar]
- 21.Acinas SG, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 22.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31:264–323. [Google Scholar]
- 24.Ho SY, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 25.Zeigler DR. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol. 2003;53:1893–1900. doi: 10.1099/ijs.0.02713-0. [DOI] [PubMed] [Google Scholar]
- 26.Grishkan I, Beharav A, Kirzhner V, Nevo E. Adaptive spatiotemporal distribution of soil microfungi in “Evolution Canyon” III, Nahal Shaharut, extreme southern Negev Desert, Israel. Biol J Linnean Soc. 2007;90:263–277. [Google Scholar]
- 27.Yim G, Wang HH, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 28.Bhaya D, et al. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 2007;1:703–713. doi: 10.1038/ismej.2007.46. [DOI] [PubMed] [Google Scholar]
- 29.Höfle MG, Ziemke F, Brettar I. In: Bell CR, Brylinsky M, Johnson-Green P, editors. Microbial Biosystems: New Frontiers; Proceedings of the 8th International Symposium on Microbial Ecology; Halifax, NS, Canada: Atlantic Canada Society for Microbial Ecology; 2000. pp. 135–142. [Google Scholar]
- 30.Smith NH, et al. Ecotypes of the Mycobacterium tuberculosis complex. J Theor Biol. 2006;239:220–225. doi: 10.1016/j.jtbi.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson GE. In: Population Biology and Evolution. Lewontin RC, editor. Syracuse, NY: Syracuse Univ Press; 1968. pp. 177–186. [Google Scholar]
- 32.Sikorski J. Populations under microevolutionary scrutiny: What will we gain? Arch Microbiol. 2008;189:1–5. doi: 10.1007/s00203-007-0294-x. [DOI] [PubMed] [Google Scholar]
- 33.Rooney AP, Swezey JL, Wicklow DT, McAtee MJ. Bacterial species diversity in cigarettes linked to an investigation of severe pneumonitis in U.S. Military personnel deployed in operation Iraqi freedom. Curr Microbiol. 2005;51:46–52. doi: 10.1007/s00284-005-4491-z. [DOI] [PubMed] [Google Scholar]
- 34.Cohan FM, Roberts MS, King EC. The potential for genetic exchange by transformation within a natural population of Bacillus subtilis. Evolution. 1991;45:1393–1421. doi: 10.1111/j.1558-5646.1991.tb02644.x. [DOI] [PubMed] [Google Scholar]
- 35.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson RR. Gene genealogies and the coalescent process. Oxford Surveys in Evolutionary Biology. 1990;7:1–44. [Google Scholar]
- 37.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.