Abstract
The shikimic acid pathway is responsible for the biosynthesis of many aromatic compounds by a broad range of organisms, including bacteria, fungi, plants, and some protozoans. Animals are considered to lack this pathway, as evinced by their dietary requirement for shikimate-derived aromatic amino acids. We challenge the universality of this traditional view in this report of genes encoding enzymes for the shikimate pathway in an animal, the starlet sea anemone Nematostella vectensis. Molecular evidence establishes horizontal transfer of ancestral genes of the shikimic acid pathway into the N. vectensis genome from both bacterial and eukaryotic (dinoflagellate) donors. Bioinformatic analysis also reveals four genes that are closely related to those of Tenacibaculum sp. MED152, raising speculation for the existence of a previously unsuspected bacterial symbiont. Indeed, the genome of the holobiont (i.e., the entity consisting of the host and its symbionts) comprises a high content of Tenacibaculum-like gene orthologs, including a 16S rRNA sequence that establishes the phylogenetic position of this associate to be within the family Flavobacteriaceae. These results provide a complementary view for the biogenesis of shikimate-related metabolites in marine Cnidaria as a “shared metabolic adaptation” between the partners.
Keywords: symbiosis, Tenacibaculum, Cnidaria
The concentrations of inorganic nutrients and planktonic food are often low in certain marine environments, where many invertebrates compensate for such oligotrophic conditions by adopting phototrophic symbioses with microorganisms, especially dinoflagellates, other unicellular algae, and photosynthetic bacteria. The dinoflagellate endosymbionts of marine invertebrates, colloquially known as “zooxanthellae,” typically belong to the genus Symbiodinium. Such zooxanthellae are particularly common in association with anthozoan cnidarians, especially corals and sea anemones. In cnidarian photoautotrophic symbioses, the animal host receives organic carbon, including carbohydrates, lipids, and amino acids, from its endosymbionts, whereas the photobionts themselves are fertilized by the recycling of essential nitrogen, phosphorus, and sulfur wastes produced from catabolism in the host and by respiratory CO2 to support algal photosynthesis (1–3). Symbiotic interactions go far beyond the cycling of carbon and nutrients and must also include other metabolic adaptations for survival in marine ecosystems (4). For example, UV-absorbing mycosporine-like amino acids (MAAs) that protect against the direct and indirect damaging effects of solar UV radiation have been isolated from diverse free-living algae and marine invertebrate-algal symbioses, including zooxanthellate cnidarians. MAAs are putatively synthesized via a branch of the shikimic acid pathway at 3-dehydroquinate (5–8). Therefore, the expectation is that MAAs in phototrophic symbioses are produced by the microbial partner, or obtained from the diet, because metazoans reputedly lack the shikimate pathway to synthesize “essential” aromatic compounds (5, 9, 10).
Fragmentary evidence suggests an exception to this traditional view. For example, nonsymbiotic scleractinian corals reportedly can synthesize aromatic amino acids (11), and aposymbiotic specimens (temporarily lacking zooxanthellae owing to experimental or special environmental circumstances) of the sea anemone Aiptasia pulchella can produce other amino acids deemed essential for metazoans (albeit not aromatic amino acids produced in postchorismate metabolism; ref. 12). Aposymbiotic specimens of the sea anemone Anthopleura elegantissima contain the same complement and concentration of MAAs derived from the shikimate pathway as found in zooxanthellate conspecifics (9), a situation that is not altered by controlled diets (13). Moreover, the zooxanthellae (Symbiodinium muscatinei) freshly isolated from A. elegantissima do not contain MAAs nor do these algae produce MAAs in pure culture (14). These facts suggest that neither the zooxanthellae nor the diet are the immediate source of MAAs in these sea anemones. Symbiotic specimens of A. elegantissima may naturally harbor taxonomically different algae (chlorophytes and dinoflagellates) under different environmental circumstances, yet animals having different symbionts contain the same complement of MAAs (9); it is improbable that such phylogenetically distant algae would produce exactly the same subset of MAAs from among an identified suite of >20 such compounds, so the endosymbionts may not account for the MAAs in this case, either. Finally, some symbiotic corals harbor phylotypes of zooxanthellae that do not produce MAAs in culture, yet the symbioses do contain MAAs (15, 16), which again suggests an extra-algal origin of the MAAs or a regulated metabolism unique to the holobiont. We have chosen the shikimic acid pathway to interrogate the metabolic adaptations in marine invertebrates, which to date are poorly defined at the genetic level (4, 17, 18).
In the absence of genomic data for symbiotic anthozoans and their zooxanthellae, we have examined Nematostella vectensis, the burrowing “starlet” sea anemone found in estuaries and salt marshes along the North American Atlantic and Pacific coasts and on the southeast coast of the United Kingdom, and which has become a model system in developmental biology (19). This basal metazoan was the first to have its genome sequenced (20), which provided us with the opportunity to mine the genome (21) of a nominally symbiont-free cnidarian for evidence of genes encoding enzymes of the shikimic acid pathway.
Results and Discussion
The anabolic shikimic acid pathway has seven steps [supporting information (SI) Scheme 1], which may be catalyzed by seven different polypeptides or by fewer multifunctional polypeptides (22). The enzymes for five of the biosynthetic steps are homologous in all organisms that possess the pathway. For two of the steps, there are two different enzymes known for each, and every organism expressing the pathway has a homologue to one of these enzymes. Furthermore, there are two additional considerations in detecting genes encoding shikimic acid pathway enzymes in N. vectensis: (i) the evolutionary origin of the genes would be uncertain so that the sequences might have diverged considerably from any comparison sequences used and (ii) the genomic sequence may contain introns.
To obtain the greatest sensitivity of interrogation, the HMMER suite of programs (23) was used to search for consensus protein sequences by using hidden Markov model profiles. This method provides greater weight to evolutionary conserved residues, and local profiles reveal the protein fragments in coding exons. The genome sequence of N. vectensis was translated in all six reading frames and searched by using nine profiles covering all seven enzymes of the shikimic acid pathway obtained from the Pfam database (24). Two alignments (“hits”) were found in large scaffolds with HMMER using the aroA and aroB profiles (SI Dataset 1). The aroA hit occurred in scaffold_33 (1.4 Mbp). When the predicted protein sequence was used for a BLAST search, it aligned with the murA gene product of a variety of bacteria with ≈40% amino acid identity. This bacterial gene encodes UDP-N-acetylglucosamine 1-carboxyvinyltransferase (SI Dataset 2), an enzyme related to aroA (3-phosphoshikimate 1-carboxyvinyltransferase), whereas the MurA enzyme is involved in the biosynthesis of the peptidoglycan cell wall. This finding initially suggested that the aligned sequence might originate from bacterial contamination. However, close examination of the HMMER results showed that the predicted protein lacked ≈20 conserved amino acids at the C terminal, and that the missing amino acid sequence was located ≈1 kb downstream in the scaffold. Visual comparison of the genomic sequence with consensus sequences for vertebrate introns revealed plausible splice sites (AGGTRA and AGG, respectively) that would produce mRNA encoding a full-length murA homologue having a close fit to the search profile. The presence of introns thus eliminates the question of bacterial contaminants or symbionts as the proximal source of this gene.
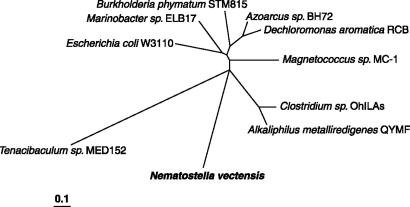
The 1.4-Mbp scaffold containing the aroA-like homolog was translated in all six reading frames and scanned by using HMMER with the entire Pfam library. This process showed the presence of a variety of typical eukaryotic domains including reverse transcriptase, EGF, calcium binding EGF domain, defensin-like peptide, actin, and the fork-head domain, again supporting the idea that the aroA-like homolog is contained in the N. vectensis genome itself. The sequence of the predicted protein was used to construct a phylogenetic tree to compare with the closest bacterial sequences found in the BLAST search and with Tenacibaculum sp. MED152 and Escherichia coli W3110 (Fig. 1). The aroA-like sequence of N. vectensis did not cluster with homologs from any group of bacteria tested, but showed a sequence divergence from bacterial sequences comparable with those of murA genes between different bacterial groups. Whether this gene in N. vectensis directs biosynthesis of peptidoglycan or shikimate pathway intermediates is yet unknown.
Fig. 1.
Phylogenetic tree showing the relationship of the predicted protein sequence of the N. vectensis aroA-like gene to the predicted murA protein sequences of the seven best hits in a BLAST analysis and to those in E. coli and Tenacibaculum. Distances were calculated from a CLUSTAL W alignment using the Jones-Taylor-Thornton matrix, and the tree was constructed by using the neighbor-joining algorithm in programs of the PHYLIP package (version 3.63). The distance is proportion of amino acid substitutions.
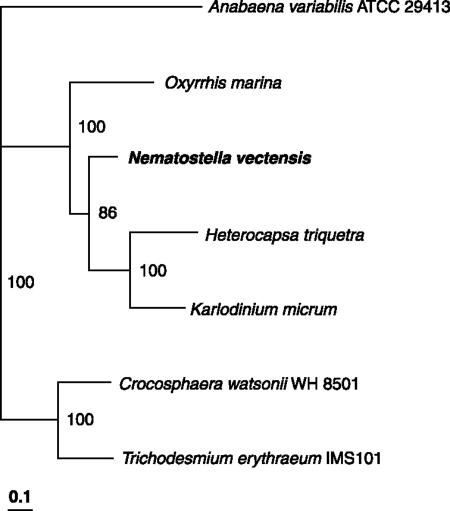
The second alignment, related to aroB, was present on scaffold-85 (0.8 Mbp). When the predicted protein sequence was used for a BLAST search (SI Dataset 3), the closest fit was with the dinoflagellate Oxyrrhis marina (66% amino acid sequence identity). In this dinoflagellate, the aroB enzyme (3-dehydroquinate synthase) is present in the chloroplast and is fused to an O-methyltransferase (25). When the complete fusion protein sequence from O. marina was used in a BLAST search against the translated DNA of N. vectensis, it was evident that a fusion protein gene was also present in N. vectensis (SI Dataset 4). This gene contains five introns. When the aroB segment of the gene was used to construct a phylogenetic tree with the closest BLAST hits (Fig. 2), the N. vectensis sequence emerged as being closest to those of two dinoflagellates (O. marina and Heterocapsa triquetra) that each possess the complete fusion gene. Again, this gene could be involved in the synthesis of precursors leading to shikimate pathway-derived secondary metabolites, most notably 3-dehydroquinate, the putative intermediate branchpoint to MAA biosynthesis (5).
Fig. 2.
Phylogenetic tree showing the relationship of the deduced protein sequence of the aroB part of the AroB-O-methyltransferase protein of N. vectensis to homologous dinoflagellate proteins. Sequences were aligned with CLUSTALW and the tree was constructed by using the neighbor-joining algorithm with distances derived from the Jones-Taylor-Thornton model (using PHYLIP version 3.63). The tree was rooted by using Anabaena variabilis as an out group. The distances are the proportion of amino acid substitutions, and the bootstrap values based on 100 samples are shown.
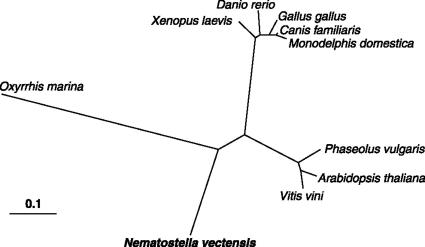
Because endosymbiotic dinoflagellates are often associated with cnidarians, the possibility had to be considered that there was an undetected dinoflagellate contaminating the N. vectensis sequence. The predicted protein sequences derived from the neighboring genes on either side of the aroB-like homolog [encoding a RuvB-like protein and a probable malate synthase (MS), respectively] were used for BLAST searches. The closest alignments were with various vertebrates and with sequences from the sea urchin Strongylocentrotus purpuratus, which makes it unlikely that these shikimate pathway genes in the host metazoan's genome are from contamination by the genome of an associated dinoflagellate (SI Dataset 5). In addition, three putative protein sequences [β-tubulin, heat shock protein 90 (HSP-90), and proliferating cell nuclear antigen (PCNA)] from O. marina were used for BLAST searches against N. vectensis. The best hits were used to construct a phylogenetic tree, and in no case were the N. vectensis and O. marina sequences closely related (Fig. 3). It must be stressed, however, that additional evidence is necessary to determine the assumed function of these genes and for proof of their acquisition by horizontal gene transfer (HGT) in N. vectensis, particularly because cnidarians reputedly have conserved genes that they inherited from nonmetazoan ancestors (26). Although the importance of HGT in eukaryotic evolution remains controversial, there is independent evidence for the occurrence of another HGT event in N. vectensis. Comparative genomic examination of glyoxylate cycle enzymes has revealed the likely transfer of a bifunctional isocitrate lyase (ICL) and a MS, encoded by a fused ICL-MS gene from a bacterial precursor, to the N. vectensis genome (27). Our findings are similar to those of others reporting evidence of gene transfer to freshwater cnidarian (Hydra) species from multiple ancestral eukaryotic partners (18, 28).
Fig. 3.
Phylogenetic tree of PCNA protein sequences. The sequence of the PCNA protein of O. marina was used for a BLAST search against the translated genomic sequences of N. vectensis. The BLAST alignments were used to assemble the protein sequence from N. vectensis. The sequences from the two species were used for BLAST searches of GenBank, and a selection of the best hits for each species was used to construct a phylogenetic tree by using the neighbor-joining algorithm in programs of the PHYLIP package (version 3.63). The distance is proportion of nucleotide substitutions.
Our genomic mining of N. vectensis revealed another surprise beyond the transfer of genes from a bacterium and a dinoflagellate to the cnidarian's genome. We found seven good sequence alignments corresponding to five potential genes of the shikimic acid pathway. Among these were four very strong alignments corresponding to the genes aroA, aroB, aroC, and aroE of E. coli (SI Dataset 6). The predicted protein sequences of these genes were used in BLAST search queries (29) against the National Center for Biotechnology Information (NCBI) GenBank database to reveal related sequences. In all four cases, the best matches were to the genes of the shikimic acid pathway in Flavobacteria, having ≈70% amino acid identity (SI Dataset 7). In most cases Tenacibaculum sp. MED152, whose genome is being sequenced (www.moore.org/marine-micro.aspx), was the best match, although a strict similarity may be influenced by database bias for this bacterium. A fifth gene in N. vectensis corresponded to the aroF-H genes of E. coli, which encode isoenzymes for 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase (DAHPS). However, BLAST searches showed the best hits (90% amino acid identity) to be the kdsA genes of Flavobacteria; these encode other isoenzymes of the DAHPS family that are involved in lipopolysaccharide synthesis.
The high similarity of the N. vectensis gene sequences to those of the bacterial shikimate pathway could be explained by either a recent HGT event or bacterial DNA contamination in the N. vectensis genome sequences. The codon usage was similar to Tenacibaculum rather than N. vectensis. Two sequences were identified that appeared to be significant fragments of bacterial 16S rRNA genes. One 16S rRNA sequence (985 bp; SI Dataset 8a) showed closest similarity to Pseudomonas sequences. However, as it did not belong to a scaffold containing other bacterial sequences and there were no other Pseudomonas-like genomic sequences detected, it is likely that it is derived from a sequencing contaminant. Because the original shotgun sequencing data were not available to us, we could not analyze the N. vectensis genome by using a recently released version of the Glimmer gene annotation tool (http://cbcb.umd.edu/software/glimmer; ref. 30), which would have been a useful way to quantify the percentage of the hologenome encoded on small scaffolds and likely, therefore, to be from living bacteria.
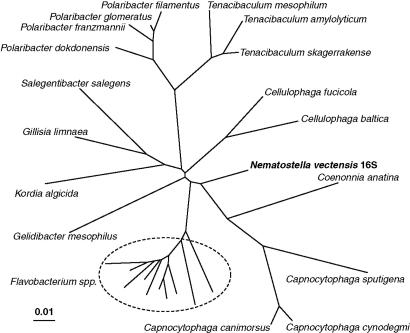
The other 16S rRNA sequence belonged to a scaffold, which also contained 23S rRNA sequences in an arrangement typical of rRNA operons (720 bp; SI Dataset 8b), and a phylogenetic tree of the 16S rRNA portion (Fig. 4) showed that it came from a flavobacterium, but it could not be assigned to a known genus. Phylogenetic trees were also constructed for the aroA, aroB, aroC, and aroE sequences, with similar results. A further consideration was that much of the genome of N. vectensis was organized into large scaffolds, whereas these 16S rRNA fragments were present in small scaffolds from which short contigs were sequenced, so that only incomplete gene sequences were revealed. This result gave the first indication that these 16S rRNA fragments might be from bacterial contamination rather than from genomic DNA of N. vectensis in the strict sense. The Tenacibaculum genome project has identified most of its genes, and the 2,679 predicted protein sequences from the genomic annotation were used for a BLAST search against translated N. vectensis DNA. When a stringent expected value of <10−30 was used, 509 of the Tenacibaculum sequences (19%) gave positive hits. However, a less stringent cutoff (10−10) gave 1,563 (58%) hits. In many of these cases, higher expected values were associated with partial sequences, as the hits were in smaller scaffolds having small contigs with many bases in the scaffolds not being determined. In fact, the aroE and kdsA genes were at the ends of contigs so that their sequences were truncated and lacked the last 40 or 25 aa, respectively. Although an accidental contamination of the original N. vectensis template cannot be ruled out, an exciting possibility is that the sequences come from a previously unsuspected flavobacterial associate similar to Tenacibaculum.
Fig. 4.
Phylogenetic tree showing the relationship of the 16S rRNA gene sequence found in the N. vectensis genome sequence (720-bp fragment in entry c429301624.Contig1 of StellaBase, http://evodevo.bu.edu/stellabase; SI Dataset 8a) to the sequences of the closest type strains in Ribosomal Data Base Project II (release 9.52; http://rdp.cme.msu.edu). Distances were calculated from a CLUSTAL W alignment using the F84 model, and the tree was constructed as in Fig. 3.
There is independent support for our contention that the foregoing sequences in the published genome for N. vectensis may come from bacteria associated with the early developmental stages of the anemone. The authors of the reported Nematostella vectensis genome (20), in their supporting online material (Supplement S2 in www.sciencemag.org/cgi/content/full/317/5834/86/DC1), did explicitly state that they prepared genomic DNA from larvae to avoid contamination by the commensals or symbionts that have been reported for the adults, although they gave no reference for the latter statement regarding such associates. Despite this precaution, there are separate findings that DNA isolates from embryos and early planula larvae of this sea anemone contain 16S rRNA sequences obtained from PCR amplicons attributed to bacteria, including those of the same groups (Flavobacteria and Pseudomonas) that we report here (H. Marlow and M. Q. Martindale, personal communication).
Bacterial associates of cnidarians have been known for at least 30 years (e.g., refs. 31 and 32), and most recently they have been visualized microscopically as epibionts and endosymbionts in two species of freshwater Hydra (33) and as envelope-wrapped aggregates in caverns between ectodermal cells of the nominally nonsymbiotic sea anemone Metridium senile (34). Such an intimate association with metazoan cells lacking an external physical barrier lends itself to direct host–microbe interactions manifested variously as pathogenicity in corals (35), the development of the immune response in Cnidaria (33), and a close symbiotic integration culminating in HGT from bacteria to host cnidarian as demonstrated here. Virtually nothing is known of the biosynthetic or other metabolic function of bacteria symbiotic with cnidarian hosts, a topic that, like so many others in modern marine microbiology, warrants investigation.
HGT between bacteria and certain metazoans (ecdysozoans, including insects and nematodes) was recently demonstrated by Baldo et al. (36) to be more widespread than suspected. They noted that bacterial sequences have been regarded previously as contamination and systematically excluded by eukaryotic genome sequencing projects, possibly masking the importance of such transfer in diverse invertebrates. Earlier, the genome sequence of the bacterial endosymbiont Carsonella ruddii found in aphids was made public (37, 38). Comparison of this genome sequence with that of another bacterial endosymbiont of aphids, Buchnera aphidicola, showed that both genomes had undergone considerable deletion, including loss of some genes encoding essential metabolic pathways. One such missing pathway leading to the formation of the aromatic amino acid tryptophan in C. ruddii caught our attention. According to dogma (10), precursors for this essential amino acid should be synthesized via the shikimic acid pathway in the commensal bacteria. Again, we searched global sequence alignments for genes encoding enzymes of the shikimic acid pathway in these bacterial genomes. We found one gene encoding a putative 5-enolpyruvylshikimate-3-phosphate phospholyase in C. ruddii (although whether this gene would transcribe a functional product is debatable owing to the large number of stop codons in the sequence), and only three (those encoding shikimate 5-dehydrogenase, 5-enolpyruvylshikimate-3-phosphate phospholyase, and 5-enolpyruvylshikimate-3-phosphate synthase) of the seven genes for the pathway were apparent in the B. aphidicola genome (SI Dataset 9). Taken together with our findings for the putative Tenacibaculum-like symbiont and its host N. vectensis, this evidence strongly suggests that the loss of essential metabolic function in the endosymbiont is an ongoing process of gene transfer and deletion in the evolution of symbioses that could ultimately lead to extinction of the symbiont by progressive assimilation of its genetic material into the host genome (37, 38).
The elucidation of “shared metabolic adaptations,” where the production of essential metabolites involves input by the partners of a symbiosis (even if one is degenerate), will require further genomic dissection of the unique organization and molecular functioning of invertebrate-microbial symbioses. This is highlighted by our finding that two of the genes for enzymes of the shikimic pathway, classically said to be absent from “animals,” are encoded in the metazoan host's genome. The extent to which such HGT, or the involvement of unsuspected bacterial consorts, may account for the apparent metabolic anomalies in cnidarians described in the Introduction, warrants further investigation. Understanding these processes may additionally provide critical insight into the cause of metabolic dysfunction evoked by climate change and environmental stress, particularly in the fragile symbioses of tropical corals and other marine cnidarians.
Materials and Methods
The DNA sequence of the N. vectensis genome was downloaded from StellaBase version 1.0 (http://evodevo.bu.edu/stellabase) and translated into all six reading frames by using Transeq (www.ebi.ac.uk/emboss/transeq). For profile analyses, HMMER version 2.3.2 (http://hmmer.janelia.org) and release 20 of the Pfam database (www.sanger.ac.uk/Software/Pfam) were used. Similarity searches used the BLAST service at NCBI (www.ncbi.nlm.nih.gov/BLAST).
Supplementary Material
ACKNOWLEDGMENTS.
We thank John R. Finnerty and James Sullivan of StellaBase (21) for useful comments and advice and Madeline van Oppen (Australian Institute of Marine Science) and Anthony Smith (University of London) for comment and advice in the preparation of this manuscript. Our insights regarding N. vectensis, and the revision of this manuscript, were greatly aided by the generous communication of information by Heather Marlow and Mark Q. Martindale (University of Hawaii). This work was supported by a cooperation grant from the German Academic Exchange Service and the Ministry of Science, Education and Sports, Republic of Croatia (to D.H. and J.C.), a stipendium from the German Academic Exchange Service (to A.S.), the School of Pharmacy, University of London (S.A. and P.F.L.), the Australian Institute of Marine Science (W.C.D.), and the University of Maine (J.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707388105/DC1.
References
- 1.Muscatine L. Coral reefs. In: Dubinsky Z, editor. Ecosystems of the World. Vol 35. Amsterdam: Elsevier; 1990. pp. 75–87. [Google Scholar]
- 2.Cook CB, D'Elia CF. Host feeding and nutrient sufficiency for zooxanthellae in the sea anemone Aiptasia pallida. Symbiosis. 1987;4:199–212. [Google Scholar]
- 3.Allemand D, Furla P, Benazet-Tambutté S. Mechanisms of carbon acquisition for endosymbiont photosynthesis in Anthozoa. Can J Bot. 1998;76:925–941. [Google Scholar]
- 4.Furla P, et al. The symbiotic Cnidarian: A physiological chimera between alga and animal. Integr Comp Biol. 2005;45:595–604. doi: 10.1093/icb/45.4.595. [DOI] [PubMed] [Google Scholar]
- 5.Shick JM, Dunlap WC. Mycosporine-like amino acids and related Gadusols: Biosynthesis, acumulation, and UV-protective functions in aquatic organisms. Annu Rev Physiol. 2002;64:223–262. doi: 10.1146/annurev.physiol.64.081501.155802. [DOI] [PubMed] [Google Scholar]
- 6.Favre-Bonvin J, Bernillon J, Salin N, Arpin N. Biosynthesis of mycosporines: mycosporine glutaminol in Trichothecium roseum. Phytochemistry. 1987;29:2509–2514. [Google Scholar]
- 7.Shick JM, Romaine-Lioud S, Ferrier-Pagès C, Gattuso J-P. Ultraviolet-B radiation stimulates shikimate pathway-dependent accumulation of mycosporine-like amino acids in the coral Stylophora pistillata despite decreases in its population of symbiotic dinoflagellates. Limnol Oceanogr. 1999;44:1667–1682. [Google Scholar]
- 8.Portwich A, Garcia-Pichel F. Biosynthetic pathway of mycosporines in the cyanobacterium Chlorogloeopsis PCC 6912. Phycologia. 2003;42:384–392. [Google Scholar]
- 9.Shick JM, Dunlap WC, Pearse JS, Pearse VB. Mycosporine-like amino acid content in four species of sea anemones in the genus Anthopleura reflects phylogenetic but not environmental or symbiotic relationships. Biol Bull. 2002;203:315–330. doi: 10.2307/1543574. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Mol Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald LM, Szmant AM. Biosynthesis of “essential” amino acids by scleractinian corals. Biochem J. 1997;322:213–221. doi: 10.1042/bj3220213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Douglas AE. Nitrogen recycling or nitrogen conservation in an alga-invertebrate symbiosis? J Exp Biol. 1998;201:2445–2453. doi: 10.1242/jeb.201.16.2445. [DOI] [PubMed] [Google Scholar]
- 13.Stochaj WR, Dunlap WC, Shick JM. Two new UV-absorbing mycosporine-like amino acids from the sea anemone Anthopleura elegantissima and the effects of zooxanthellae and spectral irradiance on chemical composition and content. Mar Biol. 1994;118:149–156. [Google Scholar]
- 14.Banaszak AT, Trench RK. Ultraviolet sunscreens in dinoflagellates. Protist. 2001;152:93–101. doi: 10.1078/1434-4610-00046. [DOI] [PubMed] [Google Scholar]
- 15.Banaszak AT, LaJeunesse TC, Trench RK. Synthesis of MAA by symbiotic dinoflagellates in culture. J Exp Mar Biol Ecol. 2000;249:219–233. doi: 10.1016/s0022-0981(00)00192-1. [DOI] [PubMed] [Google Scholar]
- 16.Banaszak AT, Santos MGB, LaJeunesse TC, Lesser MP. The distribution of mycosporine-like amino acids (MAAs) and the phylogenetic identity of symbiotic dinoflagellates in cnidarian hosts from the Mexican Caribbean. J Exp Mar Biol Ecol. 2006;337:131–146. [Google Scholar]
- 17.De la Cruz R, Davies J. Horizontal gene transfer and the origin of species: Lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 18.Steele RE, Hampson SE, Stover NA, Kibler DF, Bode HR. Probable horizontal transfer of a gene between a protist and a cnidarian. Curr Biol. 2004;14:R298–R299. doi: 10.1016/j.cub.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 19.Darling JA, et al. Rising starlet: The starlet sea anemone, Nematostella vectensis. BioEssays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- 20.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan JC, et al. StellaBase: The Nematostella vectensis Genomics Database. Nucleic Acids Res. 2006;34:D495–D499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards TA, et al. Evolutionary origins of the eukaryotic shikimate pathway: Gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot Cell. 2006;5:1517–1531. doi: 10.1128/EC.00106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eddy SR. Profile hidden Markov models. Bioinformatics. 1988;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 24.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waller RF, Slamovits CH, Keeling PJ. Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Protein Mol Biol Evol. 2006;23:1437–1443. doi: 10.1093/molbev/msl008. [DOI] [PubMed] [Google Scholar]
- 26.Technau U, et al. Maintenance of ancestral complexity and nonmetazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxylate cycle enzymes in Metazoa: Evidence of multiple horizontal transfer events and pseudogene formation. Biol Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habetha M, Bosch TC. Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. J Exp Biol. 2005;208:2157–2165. doi: 10.1242/jeb.01571. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Delcher AL, Bratke KS, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–697. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margulis L, Thorington G, Berger B, Stolz J. Endosymbiotic bacteria associated with the intracellular green algae of Hydra viridis. Curr Microbiol. 1978;1:229–232. [Google Scholar]
- 32.Palincsar EE, Jones WR, Palincsar JS, Glogowski MA, Mastro JL. Bacterial aggregates within the epidermis of the sea anemone Aiptasia pallida. Biol Bull. 1989;177:130–140. [Google Scholar]
- 33.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuett C, Doepke H, Grathoff A, Gedde M. Bacterial aggregates in the tentacles of the sea anemone Metridium senile. Helgol Mar Res. 2007;61:211–216. [Google Scholar]
- 35.Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin: Springer; 2004. [Google Scholar]
- 36.Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2007;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson SGE. Genetics: The bacterial world gets smaller. Science. 2006;314:259–260. doi: 10.1126/science.1133739. [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Brocal V, et al. A small microbial genome: The end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.