Abstract
Apoptosis is a highly regulated process of cell suicide that occurs during development, host defense, and pathophysiology. The transcription factor IFN regulatory factor 5 (IRF5), known to be involved in the activation of innate immune responses, recently has been shown to be critical for DNA damage-induced apoptosis and tumor suppression. Here, we report on a cell-type-specific role of IRF5 in promoting apoptosis upon signaling through the death receptor Fas (CD95/APO-1/TNFRSF6). In particular, we show that mice deficient in the Irf5 gene are resistant to hepatic apoptosis and lethality in response to the in vivo administration of a Fas-activating monoclonal antibody, and that IRF5 is involved in a stage of Fas signaling that precedes the activation of caspase 8 and c-Jun N-terminal kinase (JNK). In addition to hepatocytes, IRF5 is also required for apoptosis in dendritic cells activated by hypomethylated CpG but not in thymocytes and embryonic fibroblasts in vitro. Thus, these findings reveal a cell-type-specific function for IRF5 in the complex regulatory mechanism of death-receptor-induced apoptosis.
Keywords: death receptor, programmed cell death, transcription factor, liver, dendritic cell
IFN regulatory factor 5 (IRF5) plays a critical role in activating innate immune responses by transmitting pathogen-derived danger signals (i.e., pathogen-associated molecular patterns, or PAMPs) sensed by pattern recognition receptors to induce transcription of various cytokine genes (1–3). More recently, its tumor-suppressive function has been revealed by showing that IRF5 is required for DNA damage-induced apoptosis in c-Ha-Ras-expressing mouse embryonic fibroblasts (MEFs) and that MEFs undergo transformation by this single oncogene in the absence of IRF5 (3).
IRF5 is constitutively expressed in a variety of cell types and is further induced transcriptionally by treatment with type I IFNs or DNA damage (3). Upon stimulation with PAMPs or upon DNA damage, IRF5 is activated, presumably by phosphorylation, and then translocates from the cytoplasm to the nucleus where it promotes gene transcription by binding to target DNA sequences such as the IFN-stimulated response element in the cis-regulatory region of target genes (1, 3, 4). This finding is exemplified by the induction by IRF5 of the Il12b gene (1). DNA damage increases Irf5 transcript levels in a p53-dependent manner (3, 5). However, the induction of p53 target genes occurred normally in Irf5−/− cells (3), and overexpression of IRF5 enhanced the cellular susceptibility to anticancer drug-induced apoptosis even in p53-deficient cancer cell lines (6, 7), which suggests that IRF5 probably acts on a pathway that is distinct from that for p53. Nevertheless, how IRF5 participates in the apoptotic signaling pathway is not well understood.
Fas (CD95/APO-1/TNFRSF6) belongs to the death receptor family of the tumor necrosis factor (TNF) receptor superfamily, whose engagement induces apoptosis in a cell-type-specific and, typically, p53-independent manner (8). Ligation of Fas by its ligand (FasL) or agonistic antibodies allows the assembly of the death-inducing signaling complex (DISC) consisting of Fas, Fas-associated death domain-containing protein (FADD), cellular FLICE-like inhibitory protein (cFLIP), caspase 8, and, in human, caspase 10 (9, 10). The oligomerization and autoproteolytic cleavage of procaspase 8 to activated caspase 8 in turn leads to either the direct or indirect activation of caspase 3, depending on the cell type. The indirect activation of caspase 3, called the mitochondrial pathway, is mediated at least in part by the cleavage of the BH3-only protein Bid by caspase 8, leading to the release of cytochrome c, which then causes the oligomerization of Apaf-1 and subsequent activation of caspase 9. The contribution of the two Fas-mediated apoptosis pathways is cell-type-specific where, for example, Fas-stimulated hepatocytes require the mitochondrial apoptotic pathway, and thymocytes do not (11). The efficiency of the DISC formation depends on the cell type, which, it has been suggested, also would affect a requirement for the mitochondrial pathway (12). Indeed, the precise mechanism(s) underlying the cell-type-specific sensitivity of Fas-induced apoptosis is incompletely understood, and as-yet-unknown factor(s) also may participate.
In the present study, we address the issue of whether IRF5 plays a role in apoptosis induced by Fas activation. Using Irf5−/− mice, we show that hepatic apoptosis and mouse lethality upon administration of an agonistic anti-Fas monoclonal antibody (mAb) depends on IRF5. We also demonstrate that IRF5 is required for activation of c-Jun N-terminal kinase (JNK) and cleavage of caspase 8 in response to Fas stimulation. Interestingly, the contribution of IRF5 to Fas signaling is cell-type-specific because Fas-mediated apoptosis of thymocytes and MEFs does not require IRF5, whereas that of dendritic cells (DCs) prestimulated by a Toll-like receptor 9 (TLR9) ligand does. By revealing the involvement of IRF5 in death receptor signaling, our study contributes to our knowledge of the cell-type-specific process of death-receptor-induced apoptosis and exposes a unique function of the versatile biology of IRF5.
Results
Resistance to Fas-Induced Liver Damage and Lethality in Irf5−/− Mice.
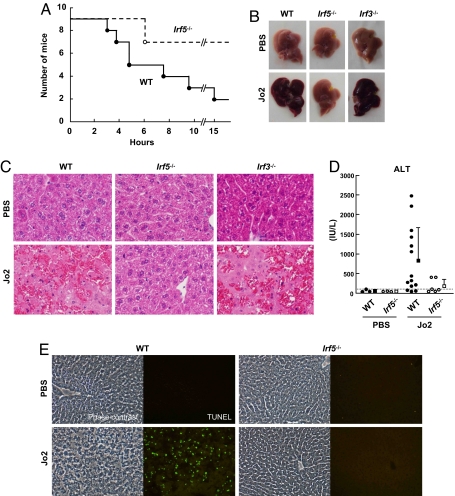
Acute fulminant hepatitis induced by the in vivo administration of agonistic Fas antibodies or FasL is characterized by extensive apoptosis of hepatocytes mediated by the Fas receptor on their cell surfaces, resulting in animal death within hours (8). To evaluate whether IRF5 plays a role in Fas-mediated apoptosis, we i.p. injected the Jo2 Fas-agonist mAb into wild-type (WT) and Irf5−/− mice. Although a majority of WT mice (78%) died within 15 h of treatment, only 22% of Irf5−/− mice succumbed to death, and the surviving Irf5−/− mice showed no signs of distress (Fig. 1A). Macroscopic examination 3 to 5 h after Fas mAb injection revealed that the livers from most Jo2-injected WT mice (7 of 9) turned dark red, which is indicative of widespread hemorrhage, whereas only 1 of 7 Irf5−/− mice showed a partial change in liver appearance (Fig. 1B). Furthermore, a histological examination revealed extensive hepatic apoptosis and hemorrhage in WT but not Irf5−/− livers (Fig. 1C). Consistent with these observations of liver destruction, the increase in serum alanine aminotransferase (ALT) levels was smaller in Irf5−/− mice than in WT mice injected with Jo2 antibodies (Fig. 1D), and a TUNEL assay confirmed the extensive hepatocyte apoptosis in WT but not Irf5−/− livers (Fig. 1E). We also examined whether IRF3, which also is implicated in p53-independent apoptosis (13), could contribute to Fas-induced liver damage and lethality. The survival rate of Irf3−/− mice was similar to that of WT mice (4 of 5 mice died in 15 h; data not shown). Moreover, macroscopic and microscopic examinations demonstrated a similar extent of liver damage in Irf3−/− mice to that observed in WT mice (Fig. 1 B and C). These results indicate that IRF5, but not IRF3, is required for hepatocyte apoptosis and lethality in response to in vivo administration of the Fas-agonist mAb.
Fig. 1.
Resistance to Fas-induced hepatic apoptosis and lethality in Irf5−/− mice. (A) Survival curves of WT and Irf5−/− mice i.p. injected with 15 μg of an agonistic Fas mAb (Jo2) or PBS. (B) Macroscopic appearance of livers 5 h after Jo2 injection. (C) Hematoxylin-eosin stains of livers 5 h after Jo2 injection. (Original magnification, ×200.) (D) Serum ALT levels 3 h after administration of Jo2 mAb. Circles indicate the ALT level of individual mouse. Squares and the error bars represent the average and SD of each group, respectively. (E) TUNEL assay of the livers 5 h after Jo2 mAb injection. (Original magnification, ×200.)
Differential Requirement for IRF5 in Fas-Induced Apoptosis.
To examine the extent to which IRF5 is involved in a general mechanism of Fas-induced apoptosis, we next tested Fas-mediated apoptosis of DCs, thymocytes, and MEFs in vitro. It is known that DCs prestimulated with TLR ligands become sensitive to Fas-mediated apoptosis (14, 15). As shown in Fig. 2, the Fas-induced apoptosis observed in WT DCs prestimulated by a TLR9 ligand hypomethylated DNA (CpG-B) (16) was strongly suppressed in prestimulated Irf5−/− DCs. Interestingly, however, when prestimulated with a TLR4 ligand lipopolysaccharide (LPS), both WT and Irf5−/− DCs underwent comparable rates of apoptosis (Fig. 2). These results suggest that there is a differential requirement for IRF5 in the sensitization of DCs to Fas by distinct TLR stimuli. It also is noteworthy that spontaneous cell death (without exogenous stimulation with Fas) also was substantially suppressed when Irf5−/− DCs were treated with CpG-B but not LPS. Significantly, no decrease in apoptosis was observed in Irf5−/− thymocytes or MEFs compared to WT cells (Fig. 2). These observations indicate that, depending on the type and state of a cell, IRF5 is differentially required for Fas-mediated apoptosis.
Fig. 2.
Cell-type-specific requirement of IRF5 for Fas-induced apoptosis. Bone-marrow-derived DCs were prestimulated with 0.35 μM CpG-B or 100 ng/ml LPS for 24 h and then treated with Jo2 mAb (100 ng/ml) and protein A (100 ng/ml) for 16 h. Thymocytes were incubated with or without Jo2 mAb plus protein A for 18 h. MEFs were incubated with or without Jo2 mAb plus protein A in the presence of 1 μg/ml cycloheximide for 18 h. Because cycloheximide was required to make MEFs sensitive to Fas, apoptosis occurred independently of de novo protein synthesis. The values represent the mean of three independent experiments ± SD.
Impaired Activation of Caspases in the Irf5−/− Liver.
In an attempt to understand the mechanism of action of IRF5, we next examined which step of the Fas signal transduction pathway is affected by the loss of IRF5 in the liver. We first analyzed the activation states of caspases by immunoblot analysis (Fig. 3A). Liver protein extracts from WT mice 3 h after Jo2 mAb administration showed clear signs of cleavage for caspases 8, 9, and 3. In contrast, liver extracts from Irf5−/− mice showed no or significantly reduced cleavage of all three caspases tested, including caspase 8, which is considered the initiator caspase in the Fas signaling pathway. In thymocytes, however, the extent of Fas-mediated cleavage of caspases 8, 9, and 3 was comparable between WT and Irf5−/− mice. Importantly, IRF5 protein was detectable in both liver and thymocyte extracts. These results indicate that the absence of IRF5 leads to a defect at or upstream of caspase 8 activation during Fas signaling in the liver but not in thymocytes.
Fig. 3.
Impaired activation of caspases and JNK in Irf5−/− livers. (A) Liver extracts were prepared from WT and Irf5−/− mice 3 h after i.p. injection of Jo2 mAb [indicated by (+); three mice were analyzed] or PBS [indicated by (−)]. Whole-cell extract from thymocytes were prepared 18 h after the addition of Jo2 mAb and protein A. Immunoblot analysis was performed by using specific antibodies as indicated. β-Actin was used as a loading control. (B) Immunoblot analysis for liver extracts was performed as in A using the indicated antibodies. Liver extracts from two mice injected with Jo2 mAb were analyzed.
Impaired cFLIP Down-Regulation and JNK Activation in the Absence of IRF5.
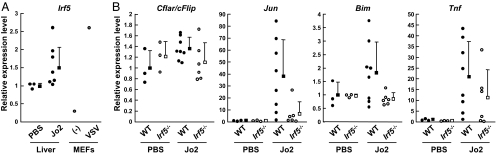
We next checked the protein expression levels of Fas, FADD, and cFLIPL, which are additional components of the DISC complex. No significant differences in the expression levels of Fas and FADD was observed between WT and Irf5−/− liver extracts (Fig. 3B). In WT livers, cFLIPL expression was, interestingly, down-regulated upon Fas engagement (Fig. 3B). Because mRNA transcript levels of the gene encoding cFLIP did not decrease after Fas activation (see Fig. 4B), the reduction of cFLIPL protein we observed is likely to be a posttranscriptional event, such as cleavage or proteasomal degradation. In contrast, such down-regulation was absent or small in Irf5−/− livers.
Fig. 4.
qRT-PCR analysis of WT and Irf5−/− livers. (A) qRT-PCR for Irf5 was performed by using RNA purified from WT mice 3 h after injection of Jo2 mAb or PBS. For comparison, RNA from WT MEFs before and 12 h after vesicular stomatitis virus (VSV) treatment also was analyzed. (B) qRT-PCR for the indicated transcripts was performed by using RNA purified from WT and Irf5−/− livers as in A. The Cflar primers detect the transcripts for both cFLIPL and cFLIPS, and the primers that specifically detect cFLIPL also gave a similar result (data not shown). The primers for Bim detect a common region for BimEL, BimL, and BimS.
It has been shown previously that cFLIPL, which is thought to be an inhibitor of caspase 8, is degraded via Itch, an E3 ubiquitin ligase that is activated by the JNK pathway downstream of TNFα signaling (17). Because JNK is also activated by Fas signaling, we next examined the activation status of this pathway in WT and Irf5−/− liver extracts. Strikingly, the phosphorylation of JNK upon Fas stimulation, observed to occur in liver extracts from WT mice, was completely absent in liver extracts from Irf5−/− mice (Fig. 3B). Thus, the defect in JNK activation in Irf5−/− livers may account for the failure of cFLIPL down-regulation, which could contribute to the impaired activation of caspase 8 (Fig. 3A). Interestingly, phosphorylation of p38 by Fas was observed in WT and Irf5−/− liver extracts (Fig. 3B). This demonstrates that some signal transduction events still occur upon Fas ligation in the absence of IRF5 and provides some insight into the step where IRF5 functions to support the Fas signaling in the liver (see Discussion).
Gene Expression Analysis in Irf5−/− Livers.
Next, we sought to find candidate genes whose expression is controlled by IRF5 and, therefore, could be important for the regulation of Fas-induced hepatic apoptosis. Thus, we performed a microarray analysis to measure the expression levels of ≈39,000 transcripts in RNA from WT and Irf5−/− mice 3 h after the administration of PBS or Jo2 mAb. It was revealed that among the ≈800 genes induced >4-fold by Jo2 injection in WT mice, ≈650 genes displayed only less than a 2-fold increase in Irf5−/− mice, suggesting that IRF5 is required for the activation of many genes in the liver upon Fas stimulation.
Consistent with the results of our immunoblot analysis, the expression of Irf5 mRNA was robustly detected in untreated WT liver cells and slightly induced by Fas activation [supporting information (SI) Table 1]. Subsequent quantitative reverse transcription PCR (qRT-PCR) analysis further showed that the level of IRF5 approached a magnitude similar to that observed in virus-infected MEFs (Fig. 4A), conditions in which IRF5 is critical for inducing apoptosis (3).
Importantly, our microarray data revealed that the expression of Jun and Bim mRNAs, both known to be regulated by the JNK-AP1 pathway (18–20), is induced by Fas activation in WT livers but severely impaired in Irf5−/− livers (SI Table 1). Analysis by qRT-PCR of samples obtained from 11 WT and 9 Irf5−/− mice revealed a trend that supports our microarray data (Fig. 4B). In addition, there was an, albeit slight, induction in Map3k5 (Ask1) and Apaf1 mRNA expression (1.8- and 1.9-fold, respectively) upon Jo2 mAb treatment that was absent in the liver of Irf5−/− mice (SI Table 1). Conversely, many other genes known to positively regulate Fas signaling were found to be expressed at comparable levels between WT and Irf5−/− mice before and after Jo2 mAb administration (SI Table 1 and SI Fig. 5). These genes include those encoding Bid, Bax, Bak, caspases 8, 9, and 3, DAXX, FLASH, RIP, FAF1, Dap3, JNK1, JNK2, TRAIL, and TNFα receptors. We also checked the expression of antiapoptotic genes such as those encoding Bcl-XL, Bcl-2, cIAP1, XIAP, and Mcl1 but found that most are induced more strongly in WT than in Irf5−/− samples (SI Table 1 and SI Fig. 5), which is rather counter to the observed phenotype.
Discussion
In this study, we have demonstrated that hepatic apoptosis in response to in vivo administration of the Jo2 Fas-agonist mAb requires IRF5. We have further shown that the activation of the JNK pathway is severely abolished, and that an early step at or upstream of caspase 8 is blocked in the livers of Irf5−/− mice. The former finding was shown by the absence of JNK phosphorylation and the impaired induction of the Jun gene; Jun is a target of AP1, the transcription factor complex activated by JNK, and c-Jun itself is a component of AP1 (18). The latter finding was demonstrated by the absence of caspase 8 cleavage and cleavage of the downstream caspases, caspase 9 and caspase 3. Although not examined in this study, we infer that a similar event may occur in DCs treated with CpG, which also required IRF5 to undergo Fas-mediated apoptosis (Fig. 2).
The role of JNK in Fas-mediated apoptosis and the upstream molecules that link Fas and JNK in the Fas signaling are controversial (21, 22). However, recent studies suggest that JNK may be a key link in two positive feedback loops involving Bim and cFLIP in the death receptor signaling pathway. Upon phosphorylation by JNK, the proapoptotic Bcl-2 family member BimEL (an isoform of Bim) translocates from the microtubules to the outer mitochondrial membrane to promote the release of cytochrome c, which ultimately activates caspase 9 and caspase 3 (23). Processed caspase 3 then cleaves phosphorylated BimEL, converting it to a more potent form and, thereby, generating an amplification loop (24). In fact, Bim−/− mice are resistant to Fas-induced hepatic apoptosis (25). Because Bim also is a target of AP1 (19, 20), the defects of JNK activation in Irf5−/− livers would result in both the failure to activate Bim protein and to induce transcription of the Bim gene, the latter of which we confirmed by gene expression analysis in this study (Fig. 4B).
Furthermore, JNK activation accelerates turnover of cFLIP, an inhibitor of caspase 8, via proteasomal degradation to generate a positive feedback loop in TNFα signaling (17). Our data reveal that in WT livers cFLIPL was down-regulated upon Fas activation, suggesting that a similar mechanism also, presumably, may operate in the Fas signaling pathway (Fig. 3). In Irf5−/− livers, however, the down-regulation of cFLIPL expression was attenuated concomitant with the absence of JNK activation. Thus, IRF5 seems to support JNK-mediated cFLIPL down-regulation, which would at least partly explain why caspase 8 activation is abolished in Irf5−/− livers.
Interestingly, mice deficient in TRAIL or receptors for TNFα are resistant to Jo2 mAb-induced hepatic apoptosis and lethality (25, 26). Fas, TRAIL, and TNFα all are notably capable of directly activating JNK, and TRAIL-deficient mice display an attenuation of JNK activation and Bim phosphorylation upon Jo2 mAb administration (25). We envisage that these ligands may cooperate to amplify an otherwise subtle, initial activation of the apoptotic signaling, ultimately leading to a massive and uncontrolled response. Given that the treatment of WT mice with a JNK inhibitor protects against Fas-induced liver damage (25), the regulation of JNK by IRF5 may represent a critical step for such cooperation, possibly by supporting the two positive feedback loops described above, i.e., phosphorylation and induction of Bim and degradation of cFLIP. Interestingly, the Fas-induced phosphorylation of JNK observed in WT livers was not detectable in thymocytes (K. Tamura, unpublished observations), congruent with the differential requirement of IRF5 and activation of JNK.
Exactly how IRF5 regulates Fas signaling remains to be clarified. However, our results provide some indication as to which step IRF5 is operating. In contrast to the defects in JNK and caspase 8 activation, p38 activation did occur in Irf5−/− mice (Fig. 3B), which indicates that the induction of the p38 pathway takes place independently of JNK and caspase 8, and that IRF5 may act at a bifurcation step for the activation of the two MAPK pathways, presumably by controlling the expression and/or the activity of an unidentified molecule(s) at the same or upstream level of caspase 8. In this context, it is noteworthy that the modest induction of Map3k (Ask1) mRNA by Fas stimulation was abolished in the Irf5−/− liver (SI Table 1) because ASK1 is known to activate the JNK pathway (27). However, it is not clear to what extent, if any, this defect accounts for the abrogation of JNK activation in Irf5−/− mice.
IRFs have been shown to function as DNA binding factors to regulate transcription of specific target genes. Therefore, it was of interest to examine whether IRF5 translocates from the cytoplasm to the nucleus during Fas stimulation. Unexpectedly, however, we did not detect translocation of a YFP-IRF5 chimera protein to the nucleus after Fas stimulation of sensitive cell lines, HepG2 and Jurkat (K.N. and T. Tamura, unpublished results). Taken at face value, we cannot exclude the possibility that IRF5 functions as a signal transducer in the cytoplasm; it is tempting to speculate that IRF5 may serve as an adaptor protein in the death receptor signaling pathway. If so, other IRF(s) may function independently or redundantly with IRF5 in this capacity in cells such as thymocytes and fibroblasts that do not require IRF5. Further studies will be required to clarify these issues.
However, because the failure of YFP-IRF5 nuclear translocation was observed by using cancer cell lines, these findings may not apply to the particular behavior of IRF5 in vivo. An alternative possibility is that the activation of IRF5 occurs when additional stimuli, other than Fas, are generated; systemic Fas activation appears to expose hepatocytes to many other stimuli, not only to Fas itself (see above). In support of this notion is the observation that pretreatment with CpG, a stimulus known to activate/translocate IRF5 via the TLR9-MyD88 pathway (1), makes DCs sensitive to Fas-induced apoptosis in an IRF5-dependent manner (Fig. 2). It will be interesting to address in the future the question of why IRF5 is required for CpG-treated, but not LPS-treated, DCs.
We must also consider that IRF5 may act in nonhepatocyte cell populations in the liver, such as Kupffer cells and sinusoidal epithelial cells, to induce the expression of a gene(s) that renders hepatocytes more sensitive to Fas-induced apoptosis. In this regard, it is interesting to note that Tnf induction by Fas, which is reported to occur in Kupffer cells (28), was reduced in some Irf5−/− mice (Fig. 4B). However, other Irf5−/− mice still showed a considerable induction of Tnf and yet still displayed no or only mild liver damage and diminished induction of Jun, suggesting that the reduced TNFα expression is not the main reason for the defective JNK activation and apoptosis in the liver. Nevertheless, the possibility that IRF5 plays a role in nonapoptotic functions of Fas, such as in the induction of proinflammatory cytokine genes, also is an interesting future issue to examine.
Our results indicate that IRF5 serves as a factor that sensitizes certain types of cells to Fas-induced apoptosis. Although the precise function of IRF5 still remains to be clarified, we infer that it contributes by effectively decreasing the threshold for Fas apoptotic signaling by vitalizing caspase 8 activation and the JNK-mediated positive feedback loops. Further investigation to clarify more precisely the details of the action of IRF5 will help to better understand the complex cell-type-specific mechanism(s) of Fas-induced apoptosis and the pathogenesis of liver diseases involving the Fas system. Finally, in view of the fact that Fas and other death receptors share common features in their signaling, it will be interesting to examine whether IRF5 is also involved in TRAIL- or TNFα-induced apoptosis, especially in the context of tumor suppression by death receptors.
Materials and Methods
Mice, Cells, and Measurement of ALT.
Six- to 10-week-old Irf5−/− and Irf3−/− mice in the C57BL/6 genetic background (1, 29) and their WT littermates or C57BL/6 mice were used. For Fas-induced liver injury studies, mice were i.p. injected with 15 μg of anti-Fas mAb (clone Jo2; BD PharMingen) diluted in 200 μl of PBS. Bone-marrow-derived DCs were generated in the presence of 20 ng/ml granulocyte–macrophage colony-stimulating factor (GM-CSF) as described in ref. 30. DCs were harvested on day 9 and prestimulated with CpG-B (0.35 μM) or LPS (100 ng/ml) in the presence of 5 ng/ml GM-CSF for 24 h. Serum ALT levels were measured by the UV method using a diagnostic kit (Nescauto VL ALT; Alfresa Pharma).
Histology and Detection of Apoptosis.
Liver tissues were fixed with 4% formaldehyde in PBS and were embedded in paraffin. Then 3- to 4-μm sections were stained with hematoxylin and eosin. For detection of apoptosis, liver was embedded and frozen in Tissue-Tek OCT Compound (Sakura Finetek), and 5-μm sections were subjected to TUNEL assay using the In Situ Cell Death Detection Kit (Roche). The percentage of apoptotic cells (sub-G1 cells) was determined by propidium-iodide staining of ethanol-fixed cells followed by flow cytometric DNA content analysis as described in ref. 31.
Immunoblot Analysis.
Immunoblot analysis was carried out by standard methods. The antibodies used were anti-Fas (M20) from Santa Cruz Biotechnology; anti-caspase 9 (5B4) and anti-FADD (1F7) from MBL; anti-caspase 8 (1G12) and anti-cFLIP (Dave-2) from Alexis; anti-IRF5, anti-caspase 3, anti-phospho-JNK, anti-JNK, anti-phospho-p38, and anti-p38 from Cell Signaling; and anti-β-actin from Sigma.
RNA Analysis.
Total RNA was prepared by using RNAiso reagent (Takara) followed by RNeasy (Qiagen) according to the manufacturer's instructions. Microarray analysis was performed by using Mouse Genome 430 2.0 Array (Affymetrix). qRT-PCR analysis was performed as described in ref. 1. Data were normalized against Gapdh expression levels of each sample. Primer sequences are available on request.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. T. W. Mak (University of Toronto, Toronto) for Irf5−/− mice, Drs. D. Savitsky and R. Eisenman for invaluable advice, and Mr. M. Shishido and Ms. R. Takeda for technical assistance. This work was supported by a Kakenhi Grant-in-Aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer” and “Dynamics of Extracellular Environments” and a Grant-in-Aid for Exploratory Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. A.C. and Z.W. are Japan Society for the Promotion of Science fellows.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712295105/DC1.
References
- 1.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 2.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 3.Yanai H, et al. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22:5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori T, Anazawa Y, Iiizumi M, Fukuda S, Nakamura Y, Arakawa H. Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene. 2002;21:2914–2918. doi: 10.1038/sj.onc.1205459. [DOI] [PubMed] [Google Scholar]
- 6.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63:6424–6431. [PubMed] [Google Scholar]
- 7.Hu G, Mancl ME, Barnes BJ. Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 2005;65:7403–7412. doi: 10.1158/0008-5472.CAN-05-0583. [DOI] [PubMed] [Google Scholar]
- 8.Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet. 1999;33:29–55. doi: 10.1146/annurev.genet.33.1.29. [DOI] [PubMed] [Google Scholar]
- 9.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 10.Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 11.Yin XM, et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 12.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 13.Weaver BK, Ando O, Kumar KP, Reich NC. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. 2001;15:501–515. doi: 10.1096/fj.00-0222com. [DOI] [PubMed] [Google Scholar]
- 14.Stranges PB, et al. Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity. 2007;26:629–641. doi: 10.1016/j.immuni.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Huang L, Wang J. Deficiency of Bim in dendritic cells contributes to overactivation of lymphocytes and autoimmunity. Blood. 2007;109:4360–4367. doi: 10.1182/blood-2006-11-056424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krieg AM, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 17.Chang L, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 19.Harris CA, Johnson EM., Jr BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem. 2001;276:37754–37760. doi: 10.1074/jbc.M104073200. [DOI] [PubMed] [Google Scholar]
- 20.Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 21.Park SM, Schickel R, Peter ME. Nonapoptotic functions of FADD-binding death receptors and their signaling molecules. Curr Opin Cell Biol. 2005;17:610–616. doi: 10.1016/j.ceb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Peter ME, et al. The CD95 receptor: Apoptosis revisited. Cell. 2007;129:447–450. doi: 10.1016/j.cell.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Putcha GV, et al. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Zhou Q. Caspase cleavage of BimEL triggers a positive feedback amplification of apoptotic signaling. Proc Natl Acad Sci USA. 2004;101:1235–1240. doi: 10.1073/pnas.0308050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corazza N, et al. TRAIL receptor-mediated JNK activation and Bim phosphorylation critically regulate Fas-mediated liver damage and lethality. J Clin Invest. 2006;116:2493–2499. doi: 10.1172/JCI27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costelli P, et al. Mice lacking TNFα receptors 1 and 2 are resistant to death and fulminant liver injury induced by agonistic anti-Fas antibody. Cell Death Differ. 2003;10:997–1004. doi: 10.1038/sj.cdd.4401281. [DOI] [PubMed] [Google Scholar]
- 27.Ichijo H, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki Y, et al. Soluble Fas gene therapy protects against Fas-mediated apoptosis of hepatocytes but not the lethal effects of Fas-induced TNF-α production by Kupffer cells. Cell Death Differ. 2002;9:626–635. doi: 10.1038/sj.cdd.4401016. [DOI] [PubMed] [Google Scholar]
- 29.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 30.Lutz MB, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 31.Hueber AO, Bernard AM, Herincs Z, Couzinet A, He HT. An essential role for membrane rafts in the initiation of Fas/CD95-triggered cell death in mouse thymocytes. EMBO Rep. 2002;3:190–196. doi: 10.1093/embo-reports/kvf022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.