Abstract
There has been progressively heightened interest in the development of targeted nanoparticles (NPs) for differential delivery and controlled release of drugs. Despite nearly three decades of research, approaches to reproducibly formulate targeted NPs with the optimal biophysicochemical properties have remained elusive. A central challenge has been defining the optimal interplay of parameters that confer molecular targeting, immune evasion, and drug release to overcome the physiological barriers in vivo. Here, we report a strategy for narrowly changing the biophysicochemical properties of NPs in a reproducible manner, thereby enabling systematic screening of optimally formulated drug-encapsulated targeted NPs. NPs were formulated by the self-assembly of an amphiphilic triblock copolymer composed of end-to-end linkage of poly(lactic-co-glycolic-acid) (PLGA), polyethyleneglycol (PEG), and the A10 aptamer (Apt), which binds to the prostate-specific membrane antigen (PSMA) on the surface of prostate cancer (PCa) cells, enabling, respectively, controlled drug release, “stealth” properties for immune evasion, and cell-specific targeting. Fine-tuning of NP size and drug release kinetics was further accomplished by controlling the copolymer composition. By using distinct ratios of PLGA-b-PEG-b-Apt triblock copolymer with PLGA-b-PEG diblock copolymer lacking the A10 Apt, we developed a series of targeted NPs with increasing Apt densities that inversely affected the amount of PEG exposure on NP surface and identified the narrow range of Apt density when the NPs were maximally targeted and maximally stealth, resulting in most efficient PCa cell uptake in vitro and in vivo. This approach may contribute to further development of targeted NPs as highly selective and effective therapeutic modalities.
Keywords: aptamer, controlled release, chemotherapy, combinatorial screening
The development of polymeric nanoparticles (NPs) for targeted delivery and controlled drug release may improve the therapeutic index of drugs. Such improvement is particularly important when administering chemotherapeutics that have toxicities that often limit their dose, resulting in suboptimal efficacy (1–4). The potential of drug targeting was first suggested by the visionary Paul Ehrlich in 1906 and despite significant work over many years (1, 3, 5–8), targeted NPs have yet to make an impact on human health. A central challenge has been defining the optimal interplay of biophysicochemical parameters that confer molecular targeting, immune evasion, and drug release to overcome the physiological barriers in vivo. The challenge is confounded by the fact that the parameters are interrelated and improving one may adversely impact another, and thus the narrow window where each of these are in the optimal range and working in concert has been difficult to achieve. The conventional methods of synthesizing targeted NPs involves serial chemical processing of particles during the formulation process, whereby drug-encapsulated NPs are first formed, followed by surface functionalization, which may include the conjugation of biomaterials to render the NPs “stealth” by decreasing their nonspecific immune clearance and biofouling by plasma proteins, and the attachments of targeting ligands to direct the cell- or tissue-specific delivery of NPs. In some cases the chemical processing of NPs has been done with functionalized biomaterials that simultaneously render the NPs both stealth and targeted (9–11), or alternatively self-assembled stealth NPs have been conjugated with a targeting ligands to achieve targeted NPs (12, 13). Regardless of the strategies taken, the methods for postsynthesis NP surface modification often require the addition of an excess amount of reactants to drive the chemical reaction (14), thus making it difficult to adjust the properties of NP surface in a meaningfully reproducible manner. Consequently, the postsynthesis particle-processing methods offer limited ability to precisely engineer the NP surface properties, and the targeted NPs produced by such methods may have significant batch-to-batch variations in their biophysicochemical properties (2, 4, 14). The development of prefunctionalized biomaterials that have all of the desired NP components present, and engineering these biomaterials to self-assemble into a targeted NP would eliminate the need of postparticle modification. Furthermore, the use of prefunctionalized biomaterials for the self-assembly of NPs could result in precisely engineered NPs, whereas simpler conjugation and purification procedures are amenable to scale-up with little batch-to-batch variability (15–17). In addition, the ability to precisely engineer targeted NPs may enable the formulation of distinct NPs that narrowly vary from each other, making it possible to optimize their biophysicochemical properties.
In this study, we used the A10 2′-fluoropyrimidine RNA aptamer (Apt) (18), which binds to the prostate-specific membrane antigen (PSMA) on the surface of prostate cancer (PCa) cells as a model hydrophilic targeting molecule; the poly(d,l-lactide-co-glycolide) (PLGA) as a model controlled-release polymer system; and polyethylene glycol (PEG) as a model hydrophilic polymer with antibiofouling properties, to develop a proof-of-concept biointegrated amphiphilic triblock copolymer (TCP) composed of PLGA-b-PEG-b-Apt that forms targeted NPs by macromolecular self-assembly in one single step. Using distinct ratios of PLGA-b-PEG-b-Apt TCP with PLGA-b-PEG diblock copolymer (DCP) lacking the A10 PSMA Apt, we developed NPs with varying Apt surface density and identified the narrow range of Apt density for maximum PCa cell uptake in vitro and in vivo. This narrow range was determined experimentally to be the minimum amount of Apt on the NP surface that conferred the maximal targeted cellular uptake. By avoiding the unnecessary masking of PEG on NP surface by excess Apt, which did not confer additional targeting benefit, we were able to maximize the exposure of PEG and its antibiofouling properties, resulting in maximally stealth and maximally targeted NPs. The delicate balance between each distinct biophysicochemical property of NPs needs to be experimentally determined and precisely and reproducibly engineered for ultimate in vivo success.
Results and Discussion
Development of PLGA-b-PEG-b-Apt TCP for Self-Assembly of Targeted NPs.
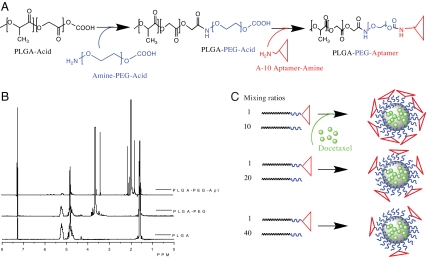
To engineer biomaterials that can self-assemble into a targeted NP, we developed a biointegrated block copolymer consisting of the following three polymer blocks: PLGA, PEG, and the A10 2′-fluoropyrimidine RNA Apt (18). The PLGA polymer block provides a biodegradable matrix for the encapsulation and sustained release of therapeutic agents and the PEG polymer acts as the antibiofouling coating on the NP surface. Both PLGA and PEG have been approved by the Food and Drug Administration for a number of clinically used products. The A10 RNA Apt (18) that binds to the extracellular domain of the PSMA (18–22) directs the delivery and uptake of NPs in a cell-specific manner. We postulated that a nucleic acid composition of Apt would be uniquely suited for the development to our TCP to be used for NP self-assembly, because it would remain stable in organic solvents during polymer synthesis and NP formulation. The PLGA-b-PEG-b-Apt TCP was synthesized by using a two-step reaction (Fig. 1A): first, the carboxyl-capped PLGA was conjugated to the amine terminal of heterobifunctional PEG (amine-PEG-carboxyl), forming the PLGA-b-PEG DCP; second, the carboxyl end of PEG-b-PLGA DCP was conjugated to the amine functional group of the A10 PSMA Apt, forming the PLGA-b-PEG-b-Apt TCP. The NMR characterization of PLGA, PLGA-PEG DCP, and PLGA-b-PEG-b-Apt TCP is shown in Fig. 1B. By precipitating the PLGA-b-PEG-b-Apt TCP in water, the polymer self-assembles to form PSMA-targeted polymeric NPs without postparticle conjugation steps: the hydrophobic PLGA blocks form a core to minimize their exposure to aqueous surroundings, the hydrophilic PEG blocks form a corona-like shell to stabilize the core, and the hydrophilic Apt blocks thrust into aqueous solution on NP surface as the targeting moieties (Fig. 1C).
Fig. 1.
Development of PSMA-targeted NPs by using PLGA-b-PEG-b-Apt TCP. (A) The PLGA-b-PEG-b-Apt-biointegrated TCP was synthesized in two steps: (i) synthesis of PLGA-b-PEG by conjugating carboxyl-capped PLGA (PLGA-acid) to the amine terminals of heterobifunctional PEG (amine-PEG-acid) and (ii) formation of PLGA-b-PEG-b-Apt by conjugating the carboxyl ends of PLGA-b-PEG-acid to the amine ends of A10 PSMA Apt. (B) 1H NMR characterization of PLGA-b-PEG and PLGA-b-PEG-b-Apt. For the synthesis of PLGA-b-PEG, the yield of PLGA and PEG conjugation was 73–91% and the purified PLGA-b-PEG DCP was used for the subsequent conjugation to Apt. The presence of Apt on the PLGA-b-PEG-b-Apt TCP was visualized by the peaks between 1.8 and 2.2 ppm. The Apt conjugation efficiency of the PLGA-b-PEG DCP for seven independent reactions was 13–21%. (C) By titration in water, the PLGA-b-PEG-aptamer TCPs self-assemble and form PSMA-targeted NP-Apt bioconjugates. By using distinct ratios of PLGA-b-PEG-b-Apt TCP with PLGA-b-PEG DCP lacking the A10 Apt during NP formulation, the Apt surface density can be precisely and reproducibly changed.
Effects of Copolymer Composition on NP Biophysicochemical Properties.
We next demonstrated that by combinatorially varying the individual components of the TCP we can systematically change of the following NP biophysicochemical properties: (i) NP size, (ii) drug release kinetics, and (iii) differential targeting.
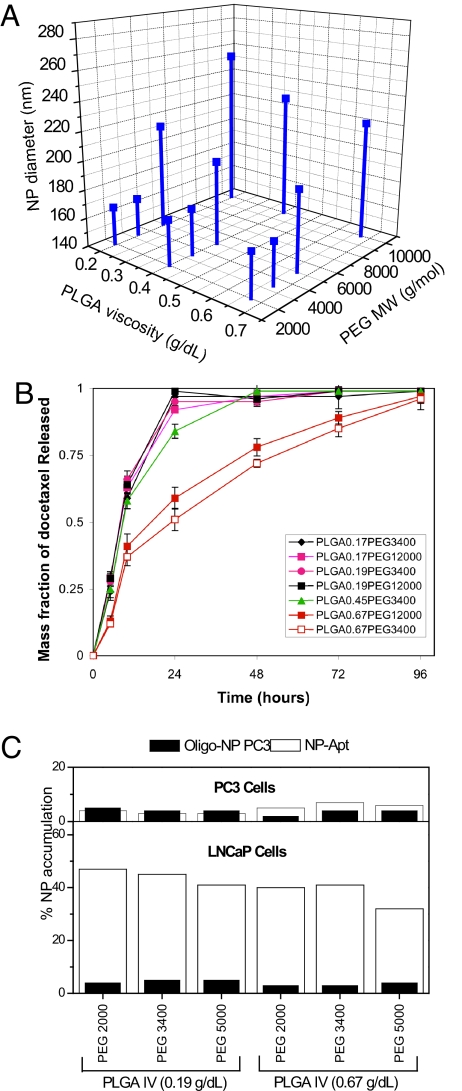
The optimization of the NP size was performed by varying the composition of the TCP. A series of targeted PLGA-b-PEG-b-Apt NPs were formulated by using combinations of various molecular masses of PLGA and PEG (Fig. 2A). Previous work on PLGA-PEG DCP showed that NP size depended on size of the PEG polymer (23, 24). We sought to determine whether varying the length of the PEG segment in the TCP could control the NP-Apt bioconjugate size. We found that, indeed, the molecular mass of PEG but not PLGA was a key factor in controlling the NP size for the triblock system. By shortening the PEG segment on the TCP from 10,000 Da to 5,000 Da, the diameter of NP reduced from 291 ± 5.2 nm to 160 ± 3.7 nm (Fig. 2A).
Fig. 2.
Fine-tuning the nanoparticle physiochemical properties by using PLGA-b-PEG-b-Apt TCPs. (A) Effect of TCP composition on NP size. Synthesis of TCP containing different molecular mass segments of PLGA and PEG. Targeted NPs were formulated by TCP self-assembly. The size of each NP formulation was measured by dynamic light scattering. Each data point represents the average of four experiments (n = 4). The standard deviation varied between 1 and 6%. The range of nanoparticle size polydispersity index (PDI) was between 0.05 and 0.10 and that the of zeta potential of various nanoparticles formulations was between −20 and −29. (B) NP drug release properties as a function of PLGA-b-PEG-b-Apt TCP composition. Dtxl was encapsulated into various NPs at a mass loading of 1.5 wt/wt%. Each data point represents the average of four experiments (n = 4). (C) Quantification on the percentage of NP accumulation in LNCaP and PC3 cells. NP-Apt bioconjugates were formulated by self-assembly of the PLGA-b-PEG-b-Apt TCP containing different lengths of PLGA and PEG segments. LNCaP and PC3 cells were incubated with 50 μg of 3H-labeled PLGA-b-PEG-b-Apt NPs for 30 min. The amount of NPs endocytosed was determined by 3H radiation counts collected in the cells. Each data point represents the average of four experiments (n = 4).
Using docetaxel (Dtxl) as a model drug, we varied the intrinsic PLGA viscosity to control drug release kinetics. We determined that increasing the length of the PLGA segment of the PLGA-b-PEG-b-Apt TCP prolonged the rate of drug release in vitro. The length of PEG chain had a minimal effect on the drug release properties of NP-Apt bioconjugates. By using PLGA with an intrinsic viscosity of >0.67 g/dl in hexafluoroisopropanol (≈100,000 molecular mass), Dtxl was released at a sustained rate for 3 days (Fig. 2B).
We next evaluated the bioactivities of the PSMA Apt expressed on the NP surface. After the TCP self-assembles into targeted NPs, the Apt on the particle surface can be heat denatured and slowly cooled to allow proper Apt tertiary conformation necessary for PSMA binding and differential targeting. We aimed to confirm that the Apt on NP surface maintain their bioactivity after the chemical steps of triblock formation and Dtxl encapsulation, by quantifying the differential accumulation of Dtxl-encapsulated NP-Apt bioconjugates within PCa cells that express the PSMA (LNCaP) and control cells lacking PSMA (PC3). Tritium-labeled NPs were formulated by mixing PLGA-b-PEG-b-Apt with a trace amount of tritium-labeled PLGA (3H-PLGA). The percentage of NP-Apt bioconjugates accumulated in LNCaP and PC3 cells was determined by quantifying the 3H detected in the cells. NPs were formulated by using two types of TCPs: (i) PLGA-b-PEG-b-Apt by using PSMA Apt (NP-Apt) and (ii) PLGA-b-PEG-b-MutApt by using a scrambled nonfunctional aptamer (NP-MutApt). LNCaP cells retained >30% of NPs at 30 min irrespective of the PLGA and PEG composition (Fig. 2C). The NP-MutApt incubated with LNCaP cells and NP-Apt incubated with PC3 cells demonstrated minimal NP accumulation. These results suggested that the Apt expressed on the Dtxl-encapsulated NP-Apt bioconjugates were bioactive and were capable of differentially guiding the NPs to accumulate in PSMA+ LNCaP cells.
Cell-Specific Endocytosis and Endosomal Colocalization of Targeted NPs.
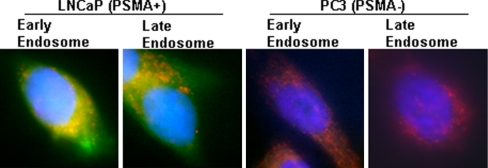
Our data suggested that PLGA0.67-b-PEG3400-b-Apt TCP had the most desirable combination of small particle size, sustained drug release kinetics, and differential accumulation in LNCaP cells. Next, LNCaP and PC3 cells were used to examine whether the self-assembled NP-Apt bioconjugates were differentially endocytosed by LNCaP cells. NP-Apt bioconjugates were visualized by encapsulating a green fluorescent dye, (22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol) cholesterol (NBD-cholesterol). By using z-axis scanning fluorescence microscopy and colocalization staining, both early and late endosomal markers were colocalized with NPs in LNCaP cells, confirming a relatively rapid endocytosis of NP-Apt bioconjugates by LNCaP cells (Fig. 3). In comparison, the NP-Apt bioconjugates seeded onto PC3 cells showed minimal cellular uptake consistent with the lack of PSMA expression in these cells.
Fig. 3.
NP-Apt cellular uptake. The NBD dyes (green) were formulated into PLGA-PEG-aptamer triblock nanoparticles by nanoprecipitation. LNCaP (PSMA+) and PC3 (PSMA−) cells were incubated with 50 μg of NBD-encapsulated PLGA-PEG-aptamer nanoparticles for 30 min. The early and late endosomal markers were visualized in red. The cell nuclei were stained by DAPI (blue).
Precise Control of Apt Density on NP Surface.
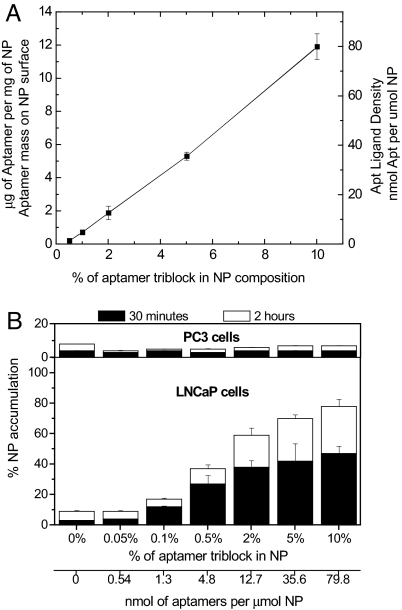
Once we confirmed that NP-Apt bioconjugates resulting from the PLGA0.67-b-PEG3400-b-Apt TCP were effectively endocytosed by PSMA+ LNCaP cells, we proceeded to optimize the Apt density on the NP surface. We postulated that the Apt density on the NP surface can be narrowly controlled by mixing known quantities of Apt in the form of PLGA0.67-b-PEG3400-b-Apt TCPs with various molar ratios of PLGA0.67-b-PEG3400 DCPs lacking the A10 Apt (Fig. 1C). The Apt densities on the NP surface were determined by Apt cleavage assay and nucleic acid quantification after hydrolyzing the Apt from the NP surface. Because PLGA is also prone to alkaline hydrolysis, the NPs were constantly monitored to ensure that they remained stable during the Apt cleavage assay. We determined that the Apt density can be precisely fine-tuned by controlling the mixing ratios of PLGA-b-PEG-b-Apt TCP to PLGA-b-PEG DCPs lacking the A10 Apt (Fig. 4A). For example, by achieving a molar ratio of 5:95 of TCP/DCP (NP-Apt 5%) in the formulation process, we can reproducibly achieve an Apt surface density of 5.31 ± 0.233 μg of Apt per mg of NP. By changing the triblock/diblock ratio from 5:95 (NP-Apt 5%) to 1:99 (NP-Apt 1%), the Apt density on NP surface is reproducibly and linearly reduced to 0.225 ± 0.0691 μg of Apt per mg of NPs. Because the average particle size was 160 nm, and the density of PLGA-PEG NP core was estimated at 1.27 g/ml (25) we estimated the Apt ligand density for NP-Apt 0.5% formulation to be 2.59 ± 0.79 Apt per NP. By increasing the TCP composition to 2%, 5%, and 10%, the Apt ligand density increased to 21.7 ± 4.69, 61.0 ± 2.67, and 139.4 ± 8.41, Apt per NPs, respectively (Fig. 4A).
Fig. 4.
Fine-tuning the Apt ligand density for targeted cell uptake in vitro. (A) Estimation of Apt ligand density on NP surface. The aptamer density can be precisely controlled in a linear manner by varying the ratio of PLGA0.67-b-PEG3400-b-Apt and PLGA0.67-b-PEG3400. (B) NPs containing 0.05–10% of PLGA0.67-PEG3400-Apt were synthesized by mixing with different amounts of PLGA0.67-PEG3400. LNCaP and PC3 cells were incubated with 3H-labeled PLGA-PEG-Apt NPs for 30 min (black) and 2 h (white). Data represent the mean and the standard error of mean of triplicates per group. Data points for NP-Apt 0.1–10% were statistically significant compared with NP-Apt 0% by t test at 95% confidence interval. One-way ANOVA with Fisher's least significant difference (LSD) post hoc comparisons at 95% confidence interval was used for statistical comparisons between groups.
Determination of Optimal Apt Density on NP Surface in Vitro and in Vivo.
We next determined the optimal Apt density on NP surface in vitro. The optimal ligand density was defined as the minimum amount of Apt on the NP surface to confer maximal targeted cellular uptake. We believed that by avoiding the unnecessary masking of PEG on NP surface by excess Apt that did not confer additional targeting benefit, we could maximize the exposure of PEG to retain its antibiofouling properties, resulting in maximally stealth and maximally targeted NPs. Our group and others had shown that PEG can increase the antibiofouling properties of NPs in vivo resulting in long circulating NPs for systemic delivery (24, 26, 27). NPs containing different Apt ligand densities were synthesized by controlling the mixing ratios of PLGA0.67-b-PEG3400-b-Apt to PLGA0.67-PEG3400. The experimental control groups were: nontargeted NPs made of PLGA0.67-PEG3400 DCPs lacking the A10 Apt (NP), and NPs made of mutated Apt TCPs (NP-MutApt). NPs were radiolabeled with 3H-PLGA, and the amount of NPs endocytosed by LNCaP and PC3 cells was quantified by measuring the 3H in the cells. All NP-Apt bioconjugates regardless of the Apt surface densities consistently demonstrated minimal uptake by the PC3 cells that do not express PSMA (Fig. 4B). In contrast, the amount of NPs endocytosed by LNCaP cells that express the PSMA can be precisely controlled by adjusting the Apt ligand densities. NPs containing <0.05% of Apt TCPs (NP-Apt 0.05%) had virtually no uptake by the LNCaP cells (P < 0.05). There was a significant increase in NP-Apt bioconjugate uptake by the LNCaP cells when the NPs contain >0.1% PLGA-b-PEG-b-Apt TCP (P < 0.05). By increasing the PLGA-b-PEG-b-Apt TCP in the formulation to 5%, there was a 5-fold increase in the amount of NP uptake by the LNCaP cells compared with the NP-Apt 0.05% formulation. Any further increase in Apt triblock proportion in the NP resulted in a modest increase in NP uptake (Fig. 4B). Thus, we estimated the optimum ligand density for PSMA-specific endocytosis in vitro was in the range of 10 to 80 nmol of Apt per μmol of NPs.
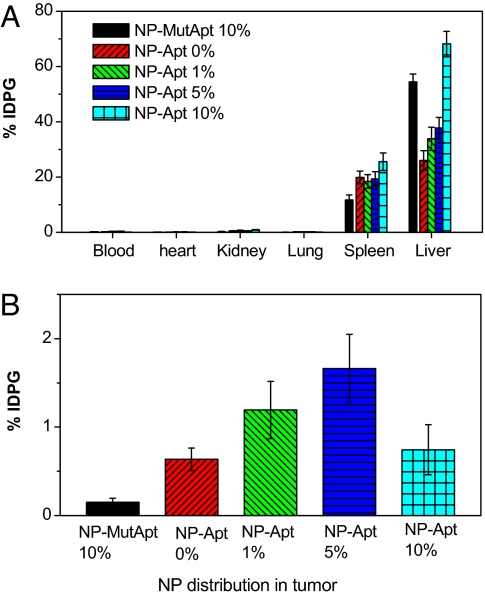
After determining the optimal Apt density for targeted cell uptake in vitro, we correlated this finding to tumor targeting in vivo by using a LNCaP s.c. xenograft mouse models of PCa to confirm whether a narrow window exists for when a PCa-targeted NP is both maximally targeted and maximally stealth. Comparative biodistribution studies were performed by using similar 3H-labeled NP-Apt bioconjugates that were used in the in vitro studies: NP-Apt 10%, NP-Apt 5%, NP-Apt 1%, NP-Apt 0%, and NP-MutApt 10% (Fig. 5 A and B). Systemic administration of targeted NPs showed that increasing the Apt surface density from 0% (no Apt) to 1% significantly increased NP retention in tumors from 0.576 ± 0.116% of injected dose per g (IDPG) of tissue to 1.09 ± 0.293% of IDPG (P < 0.05). The maximal tumor targeting was achieved by using the NP-Apt 5% formulation at 1.82 ± 0.421% of IDPG (P < 0.05). As the Apt triblock formulation in the NPs reached >10%, there was a decrease in NP accumulation in the tumors. The decrease in tumor accumulation for the NP-Apt 10% (0.743 ± 0.282% of IDPG) is possibly caused by diminishing NP stealth properties by the increased Apt surface density, resulting in its rapid clearance by the liver. Interestingly, the NP-MutApt 10% control group accumulated less in the tumor compared with the NP-Apt 0% control group, consistent with the hypothesis that former particles are less stealth and nontargeted, whereas the NP-Apt 0% particles are maximally stealth and nontargeted NPs, and thus can reach the tumor by passive targeting alone.
Fig. 5.
The effect of Apt surface density on NP biodistribution in vivo. The comparative biodistribution study of a single systemic administration of NPs containing different 1% (green), 5% (blue), and 10% (cyan) of NP-Apt were administered by retro-orbital injection into LNCaP tumor-bearing mice. The control groups were NPs without aptamers (NP-Apt 0%) (red), and NPs with scrambled nonfunctional Apt (NP-MutApt 10%) (black). Data represent the mean and the standard error of the mean of five mice per group. Data points for NPs-Apt 1% and 5% were statistically significant compared with NP-Apt 0% and NP-MutApt 10% by t test at 95% confidence interval. One-way ANOVA with Fisher's LSD post hoc comparisons at a 95% confidence interval were used for statistical comparisons between groups.
In addition to affecting the particle accumulation in the tumor tissues, the ligand surface density was directly proportional to the rate of NP accumulation in the liver. Both NP-Apt 10% (68.1 ± 4.56% of IDPG) and NP-MutApt 10% (73.0 ± 3.89% of IDPG) formulations showed significantly higher particle accumulation in the liver compared with NPs with a lower Apt surface density (P < 0.05). By decreasing the TCP to 5% (NP-Apt 5%), 37.7 ± 3.85% of IDPG was retained in the liver. The NP-Apt 1% formulation had 33.9 ± 4.21% of IDPG accumulations in the liver. NPs without Apt ligands (NP) showed that 26.1 ± 7.9% of the injected NPs was retained in the liver. These data suggest that a narrow window exists where the ligand density of targeted NPs is in the optimal range for maximal targeting and maximal stealth properties, and this range needs to be determined experimentally by using precisely engineered targeted NPs. A similar approach can be used to optimize other important NP biophysicochemical parameters to achieve the combination of parameters that result in maximal targeting in vivo.
Summary.
We demonstrated that a biointegrated TCP consisting of PLGA, PEG, and the A10 Apt could be used for the self-assembly of targeted NPs for PCa targeting. NPs with distinct biophysicochemical properties can be generated by mixing PLGA-b-PEG-b-Apt TCPs with a desired amount of PLGA-b-PEG DCPs lacking the A10 Apt and having the desired PLGA and PEG characteristics. Increasing the A10 PSMA Apt density on NP surface can increase the rate of NP uptake by LNCaP cells in vitro. However, the presence of high Apt surface density also leads to an increase in NP accumulation in the liver and spleen. This is likely due to Apt masking the PEG layer on the NP surface, and compromising the NP antibiofouling properties in vivo. Thus, in engineering targeted NPs, one must balance the tumor-targeting ligand surface density and the antibiofouling surface properties. The use of a PLGA-b-PEG-b-Apt triblock copolymer approach demonstrated the ability to systematically fine-tune Apt ligand density to maintain tumor targeting while minimizing the amount of NP accumulation in the liver. The use of this triblock copolymer approach can be used as a technology platform for developing large-scale synthesis processes of prefunctionalized targeted NPs while minimizing production times and NP batch-to-batch variations. This technology may have significance in designing delivery vehicles that target cancer cells and may contribute to the development of the next generation of nano-scale diagnostic and therapeutic modalities.
Methods
Synthesis of PLGA-b-PEG-b-Apt TCP.
The PLGA-b-PEG-b-Apt TCP was synthesized by conjugating a modified A10 2′-fluoropyrimidine RNA Apt to the PEG terminal of PEG-PLGA DCP. All materials were purchased from Sigma Aldrich. All reagents were tissue culture grade unless otherwise noted. The RNA Apt (sequence: 5′-NH2-spacer GGG/AGG/ACG/AUG/CGG/AUC/AGC/CAU/GUU/UAC/GUC/ACU/CCU/UGU/CAA/UCC/UCA/UCG/GCiT-3′ with 2′-fluoro pyrimidines, a 5′-amino group attached by a hexaethyleneglycol spacer and a 3′-inverted T cap) was custom synthesized by RNA-TEC. Carboxylate-functionalized copolymer PLGA-b-PEG was synthesized by the conjugation of COOH-PEG-NH2 (Nektar Therapeutics) to PLGA-COOH (Lactel) in methylene chloride as described by Farokhzad et al. (2). To synthesize PLGA-b-PEG-b-Apt TCP, PLGA-b-PEG DCP was dissolved in methylene chloride and activated with 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) in the procedure described earlier. The resulting PLGA-b-PEG-NHS was washed in cold methanol, dried in vacuum, and resolubilized in acetonitrile. The RNA Apt were solubilized in a mixture of formamide/acetonitrile (vol/vol 50:50), and then reacted with PLGA-b-PEG-NHS at room temperature for 24 h. The resulting PLGA-b-PEG-b-Apt TCPs were dialyzed in cold methanol to remove formamide from the reaction mixture. The resulting PLGA-b-PEG-b-Apt TCP was dried under vacuum and used for NP preparation without further treatment. The presence of PLGA and PEG were visualized by using a 400-MHz 1H NMR (Bruker) at δ 1.6, 3.6, 4.8, and 5.2. The protons from the Apt on the TCP were visualized at δ 1.8–2.2 and 3.78. The Apt conjugation efficiency to the PLGA-b-PEG was determined by degrading the PLGA segment of the TCP in 0.5 M Tris buffer at pH 9 at 40°C for 3 h, and the mass of Apt recovered from the TCP was measured by measuring UV absorbance at 260 nm (Varian).
NP Preparation and Characterization:
NPs were prepared by using a nanoprecipitation method. In brief, PLGA-b-PEG-b-Apt (50 mg/ml) and Dtxl (0.5 mg/ml) were dissolved in acetonitrile/formamide (60:40 vol/vol) and together mixed dropwise into water, giving a final NP concentration of 5 mg/ml. The NPs were stirred for 3 h, and the remaining organic solvent was removed by washing the NPs three times by using an Amicon centrifugation filtration membrane with a molecular mass cutoff of 10 kDa. The Apt ligand surface density was determined by hydrolyzing the Apts from the NP surface in 0.01 M Tris buffer at pH 9.5 at 40°C. Apts were separated from the remaining NP supernatant by centrifugation by 12,000 × g for 30 min. After centrifugation, nanoparticle debris formed a pellet, whereas the free aptamers remained suspended in the supernatant. The mass of Apt recovered in the supernatant was determined spectrophotometrically. The NP size was measured by quasi-elastic laser light scattering (QELS) by using a ZetaPALS dynamic light-scattering detector (15 mW laser, incident beam = 676 nm, Brookhaven Instruments).
Dtxl Release Study.
NPs encapsulating Dtxl were aliquoted into a series of semipermeable dialysis, minidialysis tubes (100 μl in volume, molecular mass cutoff 10,000 Da, Pierce) and were then placed in a large 40-liter PBS reservoir to mimic the infinite sink condition. At a predetermined time, a fraction of the NPs sampled were collected from the dialysis tubes and were dissolved in acetonitrile. The amount of Dtxl that remained in the NPs at each release time point was measured by HPLC in triplicate as described by Cheng et al. (14).
NP Endocytosis in Vitro.
For the fluorescent imaging study, LNCaP (PSMA+), PC3 (PSMA−) were seeded at 30,000 cells per well in a eight-well chamber slide and incubated in humidified incubators with 5% CO2 at 37°C overnight. Green fluorescent dyes, NBD-cholesterol (Invitrogen), were mixed in various formulations of PLGA-b-PEG-b-Apt TCP in organic solutions and were coprecipitated in sterile PBS. NBD-encapsulated NPs were washed and concentrated at 5 mg/ml in PBS. LNCaP and PC3 cells were washed with prewarmed PBS and incubated with 50 μg per well of prewarmed NP formulations for 30 min. Endosomal markers were visualized according to manufacturer-recommended procedures. In brief, LNCaP and PC3 cells were washed five times to remove the unbound NPs, and fixed with 4% paraformaldehyde for 20 min. Cells were then washed five times, and permeabilized and blocked in PBS containing 1% BSA and 0.1% Trition X-100, respectively. Early endosomal markers, mouse monoclonal EEA-1, and late endosomal markers, mouse monoclonal Mannose 6 Phosphate Receptor (Abcam), were incubated with LNCaP and PC3 cells for 1 h at room temperature. These endosomal markers were visualized by using Cy5 goat anti-mouse antibodies. The cell nuclei were stained with DAPI. Cells were imaged under a DeltaVision fluorescent microscope. To quantify the amount of NPs endocytosed by LNCaP and PC3 cells, NPs were radiolabeled with 3H by coprecipitating various PLGA-PEG-Apt formulations with 3H-PLGA (custom synthesized by Perkin–Elmer). LNCaP (PSMA+) and PC3 (PSMA−) were seeded in six-well plates and incubated in humidified incubators with 5% CO2 at 37°C overnight. On the day of the experiments, 50 μg of 3H-abeled NPs were incubated with LNCaP and PC3 cells for 30 min and 2 h, respectively. The cells were washed five times to remove the unbound NPs and subsequently lysed in 100–200 μl of lysis buffer (PBS with 0.1% Triton X-100). The NP-cell mixtures were then collected and solubilized in Solvable (Perkin–Elmer), and then mixed with Hionic-Fluor scintillation mixture (Perkin–Elmer). The 3H content in the cells was assayed in a Packard Tri-Carb Scintillation Analyzer (Perkin–Elmer).
Tumor Targeting in Vivo.
All animal studies were carried out under the supervision of the Division of Comparative Medicine, Massachusetts Institute of Technology, and in compliance with the Principles of Laboratory Animal Care of the National Institutes of Health. Human xenograft prostate cancer tumors were induced in 8-week-old balb/c nude mice (Charles River Laboratories). Mice were injected s.c. in the right flank with 4 × 106 LNCaP cells suspended in a 1:1 mixture of media and matrigel (BD Biosciences). Tumor-targeting studies were carried out after the mice developed ≈300-mm3 tumors. Mice were randomly divided to minimize tumor size variations between groups. Mice were anesthetized by i.p. injection of avertin (200 mg/kg body weight), and dosed with NP formulations by retro-orbital injection. NP biodistributions were traced by encapsulating 3H-PLGA into the NPs before administration. Mice were killed at 24 h after NP injections, and 200 μl of blood was drawn by cardiac puncture from each mouse. The tumor, heart, lungs, liver, spleen, and kidneys were harvested from each animal as described by Cheng et al. (14). The 3H content in the tissues was assayed in a Packard Tri-Carb Scintillation Analyzer. To determine 100% dose, vials of the formulated NPs were counted along with the tissues.
ACKNOWLEDGMENTS.
We thank Drs. Philip Kantoff and Neil Bander for helpful discussions throughout this study. This work was supported by National Institutes of Health Grants CA119349 and EB003647 and Koch–Prostate Cancer Foundation Award in Nanotherapeutics. F.G. was supported by a Postdoctoral Fellowship from the Canadian Natural Sciences and Engineering Research Council.
Footnotes
Conflict of interest statement: O.C.F. and R.L. have financial interest in BIND Biosciences. The rest of the authors declare no conflict of interest.
References
- 1.Langer R. Drug delivery and targeting. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- 2.Farokhzad OC, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari M. Cancer nanotechnology: Opportunities and challenges. Nature Rev. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 4.Gu F, et al. Targeted nanoparticles for cancer therapy. Nano Today. 2007;2:14–21. [Google Scholar]
- 5.Farokhzad OC, Langer R. Nanomedicine: Developing smarter therapeutic and diagnostic modalities. Adv Drug Delivery Rev. 2006;58:1456–1459. doi: 10.1016/j.addr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Heath TD, Fraley RT, Papahdjopoulos D. Antibody targeting of liposomes: Cell specificity obtained by conjugation of F (ab′)2 to vesicle surface. Science. 1980;210:539–541. doi: 10.1126/science.7423203. [DOI] [PubMed] [Google Scholar]
- 7.Leserman LD, Barbet J, Kourilsky F, Weinstein JN. Targeting to cells of fluorescent liposomes covalently coupled with monoclonal antibody or protein A. Nature. 1980;288:602–604. doi: 10.1038/288602a0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, et al. Nanoparticles in medicine: Therapeutic applications and developments. Clin Pharmacol Ther. 2007 doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 9.Leamon CP, Cooper SR, Hardee GE. Folate-liposome-mediated antisense oligodeoxynucleotide targeting to cancer cells: evaluation in vitro and in vivo. Bioconjugate Chem. 2003;14:738–747. doi: 10.1021/bc020089t. [DOI] [PubMed] [Google Scholar]
- 10.Popielarski SR, Pun SH, Davis ME. A nanoparticle-based model delivery system to guide the rational design of gene delivery to the liver. 1. Synthesis and characterization. Bioconjugate Chem. 2005;16:1063–1070. doi: 10.1021/bc050113d. [DOI] [PubMed] [Google Scholar]
- 11.Chiu SJ, Liu S, Perrotti D, Marcucci G, Lee RJ. Efficient delivery of a Bcl-2-specific antisense oligodeoxyribonucleotide (G3139) via transferrin receptor-targeted liposomes. J Controlled Release. 2006;112:199–207. doi: 10.1016/j.jconrel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Farokhzad OC, et al. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 13.Sun B, Ranganathan B, Feng SS. Multifunctional poly(d,l-lactide-co-glycolide)/montmorillonite (PLGA/MMT) nanoparticles decorated by Trastuzumab for targeted chemotherapy of breast cancer. Biomaterials. 2008;29:475–486. doi: 10.1016/j.biomaterials.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, et al. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28:869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubowicz S, et al. Multicompartment micelles formed by self-assembly of linear ABC triblock copolymers in aqueous medium. Angew Chem. 2005;44:5262–5265. doi: 10.1002/anie.200500584. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Shi L, Ma R, An Y, Huang N. Formation of complex micelles with double-responsive channels from self-assembly of two diblock copolymers. Angew Chem. 2006;45:4959–4962. doi: 10.1002/anie.200600172. [DOI] [PubMed] [Google Scholar]
- 17.Reynhout IC, Cornelissen JJ, Nolte RJ. Self-assembled architectures from biohybrid triblock copolymers. J Am Chem Soc. 2007;129:2327–2332. doi: 10.1021/ja066790f. [DOI] [PubMed] [Google Scholar]
- 18.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. Invitro selection of Rna molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.McNamara JO, II, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 21.Chu TC, et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 22.Chu TC, et al. Labeling tumor cells with fluorescent nanocrystal-aptamer bioconjugates. Biosens Bioelectron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Xie J, Tong YW, Wang CH. Effect of PEG conformation and particle size on the cellular uptake efficiency of nanoparticles with the HepG2 cells. J Controlled Release. 2007;118:7–17. doi: 10.1016/j.jconrel.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Avgoustakis K, et al. Effect of copolymer composition on the physicochemical characteristics, in vitro stability, and biodistribution of PLGA-mPEG nanoparticles. Int J Pharm. 2003;259:115–127. doi: 10.1016/s0378-5173(03)00224-2. [DOI] [PubMed] [Google Scholar]
- 25.Vauthier C, Schmidt C, Couvreur P. Measurement of the density of polymeric nanoparticulate drug carriers by isopycnic centrifugation. J Nanoparticle Res. 1999;1:411–418. [Google Scholar]
- 26.Mosqueira VC, et al. Biodistribution of long-circulating PEG-grafted nanocapsules in mice: effects of PEG chain length and density. Pharm Res. 2001;18:1411–1419. doi: 10.1023/a:1012248721523. [DOI] [PubMed] [Google Scholar]
- 27.Gref R, et al. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]