Abstract
Severe skeletal muscle wasting is the most debilitating symptom experienced by individuals with myotonic dystrophy type 1 (DM1). We present a DM1 mouse model with inducible and skeletal muscle-specific expression of large tracts of CTG repeats in the context of DMPK exon 15. These mice recapitulate many findings associated with DM1 skeletal muscle, such as CUG RNA foci with Muscleblind-like 1 (MBNL1) protein colocalization, misregulation of developmentally regulated alternative splicing events, myotonia, characteristic histological abnormalities, and increased CUGBP1 protein levels. Importantly, this DM1 mouse model recapitulates severe muscle wasting, which has not been reported in models in which depletion of MBNL1 is the main feature. Using these mice, we discovered previously undescribed alternative splicing events that are responsive to CUGBP1 and not MBNL, and these events were found to be misregulated in individuals with DM1. Our results indicate that increased CUGBP1 protein levels are associated with DMPK-CUG RNA expression, suggesting a role for CUGBP1-specific splicing or cytoplasmic functions in muscle wasting.
Keywords: alternative splicing, CUG-binding protein 1, Muscleblind-like 1, muscle atrophy, microsatellite expansion
Myotonic dystrophy type 1 (DM1) is a multisystemic, autosomal dominant disease caused by a CTG repeat expansion in the 3′ untranslated region (UTR) of the DMPK gene (exon 15). Individuals with the disease have expansions ranging from 50 to >2,000 repeats. Onset and severity of disease correlate with repeat expansion size. The dominant organ system affected is skeletal muscle, which exhibits myotonia and degeneration. Severe skeletal muscle wasting is the primary cause of morbidity and mortality (1).
After transcription of the expanded allele, CUG repeats accumulate within the nucleus in discrete RNA foci. Pathology in DM1 is a result of toxic RNA expression and not alteration of DMPK gene expression. This model was solidified by work from Mankodi et al. (2) using HSALR transgenic mice that expressed 250 CUG repeats in the human skeletal actin 3′ UTR specifically in skeletal muscle. HSALR mice develop DM1-characteristic RNA foci, misregulated alternative splicing, myotonia, and histological abnormalities (2). Additionally, a second form of DM (DM2) is caused by expanded CCTG repeats in intron 1 of an unrelated gene, ZNF9, and yet has similar symptoms to those seen in DM1 (3, 4). Accumulation of toxic CUG or CCUG repeats leads to a transdominant misregulation of RNA homeostasis. In particular, developmentally regulated alternative splicing is affected such that there is a failure to express adult isoforms. Thus, pathology results at least in part from inappropriate expression of embryonic splicing patterns in adult tissues (5).
Two proteins identified as interacting with CUG RNA repeats, Muscleblind-like 1 (MBNL1) and CUG-binding protein 1 (CUGBP1), play important roles in DM1 pathogenesis. Both proteins normally regulate alternative splicing of exons that are misregulated in DM1 (7, 24). Interestingly, they are antagonistic regulators of developmentally regulated splicing events. The predominant model for DM1 pathogenesis is MBNL1 sequestration on repeat RNA foci (8, 9). MBNL1 colocalizes with the repeat RNA foci leading to its nuclear depletion and reversion to the embryonic splicing pattern of MBNL1 target exons both in DM1 tissues and HSALR mice (10–14). Support for this model also comes from mice in which MBNL1 exon 3 has been deleted, designated MbnlΔ3/Δ3 resulting in loss of the predominant CUG-binding isoforms. These mice display myotonia, cataracts and splicing changes characteristic of DM1 and similar to HSALR mice (15). Delivery of MBNL1 to HSALR skeletal muscle using AAV vectors reverses abnormal splicing patterns (16). However, whereas HSALR and MbnlΔ3/Δ3 mice reproduce loss of functional MBNL1 and have splicing misregulation, they fail to exhibit the increased CUGBP1 protein levels and overt muscle wasting that is seen in skeletal muscle from individuals with DM1 (10, 17, 18).
In this article, we describe a DM1 mouse model that incorporates key features necessary to recapitulate the severe and progressive muscle wasting that arises in individuals with DM1. Expression of 960 CTG repeats in the context of DMPK exon 15 is tissue specific and inducible so that onset and progression of pathology can be studied specifically in adult skeletal muscle after induction. These mice are a robust model of skeletal muscle pathology in DM1 and display established molecular features of the disease including increased CUGBP1 protein levels. Additionally, we discovered previously undescribed DM1 misregulated alternative splicing events that are specific to this model, and that may contribute to muscle wasting.
Results
Inducible and Skeletal Muscle-Specific Expression of the DMPK 3′ UTR Containing 960 CUG Repeats.
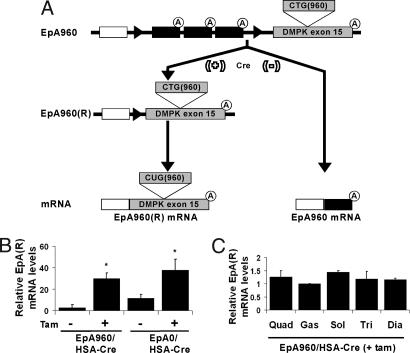
We have previously described transgenic mice in which expression of DMPK exon 15 containing either 960 interrupted CTG repeats (EpA960) or no repeats (EpA0) is induced by Cre-mediated recombination in cardiomyocytes (19). Ubiquitous transcription of the nonrecombined construct is driven by the pCAGG CMV enhancer/chicken β-actin promoter and a floxed concatemer of three SV40 polyadenylation sites prevents expression of DMPK exon 15, which is located downstream of the polyadenylation cassette (Fig. 1A). In this study we crossed the EpA lines with HSA-Cre-ERT2 mice, expressing the tamoxifen-dependent Cre-ERT2 recombinase selectively in skeletal muscle tissue (20). To induce recombination and expression of mRNA containing the expanded CTG repeats [EpA960(R)], EpA960/HSA-Cre-ERT2 bitransgenic mice 3 to 4 months of age were injected daily for 5 days with 1 mg of tamoxifen (20). EpA0/HSA-Cre-ERT2 bitransgenic mice were also treated with tamoxifen to induce expression of the identical mRNA containing human DMPK exon 15, but lacking CTG repeats [EpA0(R)]. The levels of expression of repeat containing mRNA from the recombined EpA960 and EpA0 alleles were quantified by real time RT-PCR in gastrocnemius muscle. EpA960/HSA-Cre-ERT2 and EpA0/HSA-Cre-ERT2 animals exhibited a basal level of expression of the EpA recombined alleles in the absence of tamoxifen. However, tamoxifen administration resulted in a 10-fold induction of EpA960(R) and three-fold induction of EpA0(R) mRNA (Fig. 1B). In addition, tamoxifen-treated EpA0/HSA-Cre-ERT2 mice expressed levels of EpA0(R) mRNA that were consistently equivalent to or higher than EpA960(R) mRNA from EpA960/HSA-Cre-ERT2 mice; thus EpA0/HSA-Cre-ERT2 mice serve as a proper control (Fig. 1B). There was comparable expression across proximal and distal, as well as predominantly fast twitch and slow twitch muscle groups (Fig. 1C). RT-PCR analysis demonstrated that expression of EpA960(R) mRNA is limited to skeletal muscle tissue and was not detected in the heart, kidney, liver, brain, or testes (data not shown). We also determined that the level of EpA960(R) mRNA in skeletal muscle is consistently lower than CUG repeat mRNA levels found in skeletal muscle of two other DM1 mouse models: HSALR and (CTG)5 5-336 [supporting information (SI) Fig. 5].
Fig. 1.
Bitransgenic animals expressing either 0 or 960 CUG repeats within exon 15 of the DMPK gene. (A) Transgene driven by the CMV enhancer was created with a floxed concatemer of three polyadenylation sites followed by exon 15 of the DMPK gene with either 0 (designated EpA0) or 960 (EpA960) CTG repeats (19). EpA lines transcribe RNAs that terminate at one of the three polyadenylation sites. Cre-mediated recombination induces expression of mRNA containing DMPK exon 15 with [EpA960(R)] or without [EpA0(R)] CUG repeats. (B) Quantitative RT-PCR performed on RNA extracted from gastrocnemius muscle in EpA960/HSA-Cre-ERT2 mice (n = 5 for each group) and EpA0/HSA-Cre-ERT2 mice (n = 3 for each group) before and 4 weeks after tamoxifen administration. GAPDH was used as a standard. Differences in mRNA levels from the recombined allele [EpA0(R) or EpA960(R)] between pre- and posttamoxifen in EpA960/HSA-Cre-ERT2 and EpA0/HSA-Cre-ERT2 mice were statistically significant (*) as assessed by Student's t test, P < 0.05. (C) Expression of mRNA from the recombined allele in different muscle groups (Quad, quadriceps; Gas, gastrocnemius; Sol, soleus; Tri, triceps; Dia, diaphragm) in EpA960/HSA-Cre-ERT2 mice 4 weeks after tamoxifen. There is no statistically significant difference between the five muscle groups using one-way ANOVA, n = 3 for each group.
EpA960/HSA-Cre-ERT2 (+ tam) Mice Display DM1-Like Features.
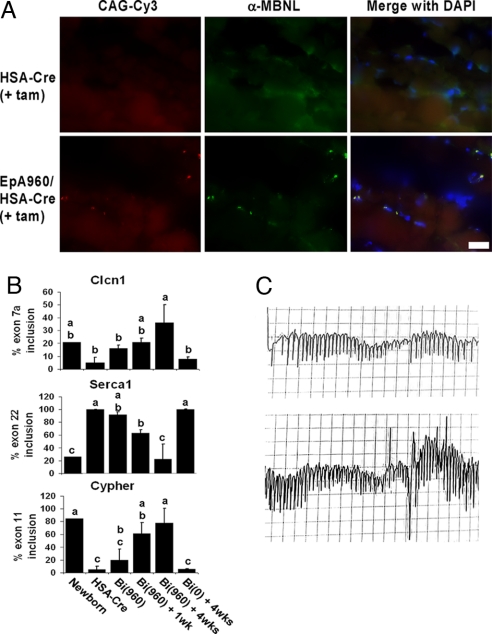
Intranuclear CUG RNA foci, which colocalize with MBNL1 are seen in individuals with DM1 and other DM1 mouse models that express expanded CUG repeats (2, 13). EpA960/HSA-Cre-ERT2 (+ tam) mice display CUG RNA foci with MBNL1 colocalization (Fig. 2A). Foci formation and MBNL1 colocalization is observed before tamoxifen administration as well in EpA960/HSA-Cre-ERT2 (− tam) mice (data not shown), consistent with the basal leakage of EpA960(R) mRNA expression (Fig. 1B).
Fig. 2.
EpA960/HSA-Cre-ERT2 (+ tam) mice reproduce the molecular features of DM1. (A) Intranuclear CUG repeat RNA foci with MBNL1 colocalization is not detected in triceps muscle isolated from HSA-Cre-ERT2 (Upper) and is present in EpA960/HSA-Cre-ERT2 (Lower) 4 weeks after tamoxifen administration. All images acquired at ×63 and with the same exposure times for the red and green filters. (Scale bar: 20 μm.) (B) EpA960/HSA-Cre-ERT2 (+ tam) mice display misregulation of developmentally regulated alternative splicing events as seen in DM1. RT-PCR was performed by using primers that flank the alternative exon (SI Table 1). The percentage inclusion of alternative exons from three genes is graphically represented. Lanes are as follows: Newborn, WT day 1 newborn limb; HSA-Cre, HSA-Cre-ERT2 no tam; Bi(960), EpA960/HSA-Cre-ERT2 no tam; Bi(960) + 1 wk, EpA960/HSA-Cre-ERT2 1 week posttamoxifen (post tam); Bi (960) + 4 wks, EpA960/HSA-Cre-ERT2 4 weeks post tam; Bi (0) + 4wks, EpA0/HSA-Cre-ERT2 4 weeks post tam. Each bar represents the mean of the three biological replicates with standard deviation, except for the Newborn sample, which represents a pool of >12 animals. One-way ANOVA followed by Tukey–Kramer analysis was used to assign the samples into groups (a, b, or c) for each graph. Animals assigned to one group but not another are statistically different. Gel images are in SI Fig. 6. (C) Electromyogram traces from the gastrocnemius muscle of an EpA960/HSA-Cre-ERT2 (+ tam) mouse, showing portions of two distinct, sustained myotonic runs with characteristically audible waxing and waning frequency. Vertical scale, 0.2 mV per division; horizontal scales, 25 ms per division (Upper) and 50 ms per division (Lower).
We investigated three well established misregulated alternative splicing events that occur in DM1 skeletal muscle tissue (Clcn1, Serca1, and Cypher). Clcn1 is a skeletal muscle-specific chloride channel responsible for repolarizing the skeletal muscle cell sarcoplasmic membrane after contraction. Adult DM1 individuals have increased inclusion of the fetal exon 7a, which contains a premature stop codon, resulting in reduced expression of functional Clcn1 because of nonsense mediated decay, ultimately causing myotonia (21, 22). Serca1 (SR/endoplasmic reticulum Ca2+ ATPase) regulates Ca2+ homeostasis and adults with DM1 have increased exclusion of exon 22, similar to that found in fetal tissue (23). Cypher localizes to the Z line and exhibits aberrant inclusion of exon 11 in DM1 (16). In EpA960/HSA-Cre-ERT2 mice there was a significant and progressive misregulation of these alternative splicing events after tamoxifen administration, which was not seen in EpA0/HSA-Cre-ERT2 (+ tam) mice (Fig. 2B). Splicing misregulation initiates in EpA960/HSA-Cre-ERT2 (− tam) mice; because of basal leakiness of the transgene; however, the progressive degree of worsening splicing misregulation after tamoxifen administration was statistically significant (Fig. 2B).
EpA960/HSA-Cre-ERT2 (+ tam) mice have sustained myotonia, with waxing and waning myotonic runs noted on electromyogram (Fig. 2C). EpA960/HSA-Cre-ERT2 (− tam) mice develop myotonia as well, which appears indistinguishable from the myotonia seen in EpA960/HSA-Cre-ERT2 (+ tam), indicating that the basal expression of the recombined allele with subsequent Clcn1 alternative splicing misregulation is sufficient to induce myotonia. Whereas this level of basal expression in (− tam) mice leads to foci formation, alternative splicing misregulation and myotonia, it is not sufficient to cause overt skeletal muscle wasting, histological abnormalities or muscle dysfunction (see below).
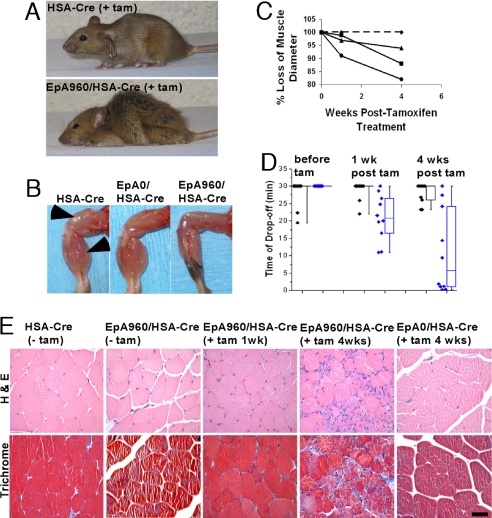
EpA960/HSA-Cre-ERT2 (+ tam) Mice Exhibit Severe and Progressive Skeletal Muscle Wasting and Degeneration with Marked Loss of Muscle Function.
EpA960/HSA-Cre-ERT2 (+ tam) mice exhibit a strong DM1–like muscle phenotype, whereas neither EpA960/HSA-Cre-ERT2 (− tam) or EpA0/HSA-Cre-ERT2 (+ tam) mice exhibit muscle degeneration. By 4 weeks after tamoxifen administration, 70% of EpA960/HSA-Cre-ERT2 mice display significant muscle wasting, leading to reduced mobility, abnormal gait and kyphosis (Fig. 3A and SI Movie 1) that is unresponsive to daily gavage feeding (data not shown). Four weeks after tamoxifen administration there is a severe reduction in muscle size of EpA960/HSA-Cre-ERT2 (+ tam) mice compared with HSA-Cre-ERT2 (+ tam) littermates and age/sex-matched EpA0/HSA-Cre-ERT2 (+ tam) mice (Fig. 3B). MRI analysis of the paraspinous muscle (SI Fig. 7) demonstrates that the muscle wasting progressively worsens with a ≈15% reduction in diameter by 4 weeks after tamoxifen administration (Fig. 3C). Although muscle wasting in DM1 predominates in distal compared with proximal muscle groups, there was no obvious difference in muscle wasting between different muscle groups in EpA960/HSA-Cre-ERT2 (+ tam) mice (data not shown). It may be that the characteristic muscle wasting pattern seen in individuals with DM1 results from higher expression of DMPK in the more affected muscle groups. Expression of EpA960(R) mRNA remains relatively constant across different muscle groups (Fig. 1C), possibly explaining the lack of preference for distal over proximal muscle group wasting.
Fig. 3.
EpA960/HSA-Cre-ERT2 mice exhibit severe and progressive skeletal muscle wasting and dysfunction after tamoxifen administration. (A) HSA-Cre-ERT2 (Upper) and EpA960/HSA-Cre-ERT2 (Lower) littermates 4 weeks after tamoxifen administration. (B) From left to right, the hind limbs of age- and sex-matched HSA-Cre-ERT2, EpA0/HSA-Cre-ERT2, and EpA960/HSA-Cre-ERT2 mice 4 weeks after tamoxifen administration. Arrows indicate quadriceps and gastrocnemius muscle bodies. (C) Wasting of the paraspinous muscle determined by MRI from one HSA-Cre-ERT2 (dashed line) and three EpA960/HSA-Cre-ERT2 (solid lines) mice during a 4-week time course after tamoxifen administration. Measurements were normalized to the kidney diameter. (D) Progressive loss of muscle function in EpA960/HSA-Cre-ERT2 mice after tamoxifen administration. All mice were assayed before tamoxifen administration and then at 1 and 4 weeks after tamoxifen by using a graded treadmill protocol (see Materials and Methods). The box plot represents 10 HSA-Cre-ERT2 mice (black diamonds) and 10 EpA960/HSA-Cre-ERT2 mice (blue diamonds). Data points between the upper 75th and lower 25th quartiles are boxed; the horizontal line within the box is the median value. Using the Student t test, there is no statistically significant difference in HSA-Cre-ERT2 mice between the three time points, but there is a statistically significant difference in EpA960/HSA-Cre-ERT2 mice when the pretamoxifen time point is compared with the 1-week- and 4-weeks-after-tamoxifen time points, P < 0.05. (E) EpA960/HSA-Cre-ERT2 (+ tam) mice show a progressive muscle dystrophy by histology. Note the increasing numbers of central nuclei, small basophilic fibers, fiber diameter variation, and fibrosis seen in EpA960/HSA-Cre-ERT2 mice after tamoxifen administration. All images are at ×40 magnification. (Scale Bar: 50 μm.)
To determine whether the degree of the skeletal muscle wasting in EpA960/HSA-Cre-ERT2 (+ tam) mice was sufficient to lead to impaired muscle function, a graded treadmill protocol was used. Ten EpA960/HSA-Cre-ERT2 mice and 10 HSA-Cre-ERT2 littermates were tested before and then at 1 and 4 weeks after tamoxifen administration. There was a significant decrease in the time of drop off for EpA960/HSA-Cre-ERT2 (+ tam) mice compared with HSA-Cre-ERT2 (+ tam) littermates, and this difference worsened from 1 week to 4 weeks after tamoxifen (Fig. 3D and SI Fig. 8).
Consistent with gross examination, MRI analysis and the treadmill test, pathological changes were seen by histology as early as 1 week after tamoxifen administration, and significantly worsened by 4 weeks (Fig. 3E). These changes ranged from mildly increased numbers of smaller myofibers and scattered basophilic fibers to acute myofiber degeneration, with significant numbers of necrotic fibers and many fibers with basophilic cytoplasm and central nuclei consistent with regeneration. Trichrome staining revealed focal regions of increased fibrosis, consistent with dystrophy. Normal mouse quadriceps tissue is primarily composed of fibers expressing the fast twitch myosin isoform, with focal areas containing a mixture of fibers expressing either fast or slow twitch myosin isoforms in a mutually exclusive distribution. Four weeks after tamoxifen administration EpA960/HSA-Cre-ERT2 mice show a relative loss in fibers expressing solely slow twitch myosin and an increase in the number of fibers showing simultaneous expression of slow and fast twitch myosin isoforms (SI Fig. 9).
Elevated CUGBP1 Protein Levels and Previously Undescribed Misregulated Splicing Events in EpA960/HSA-Cre-ERT2 (+ tam) Mice.
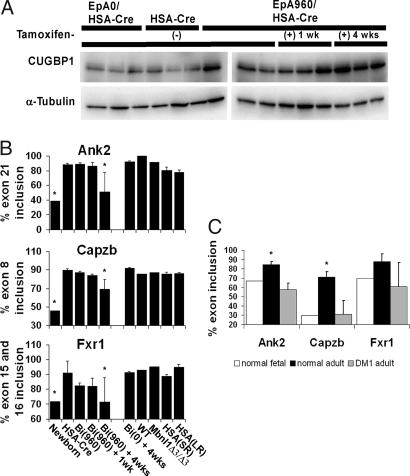
CUGBP1 is a splicing factor that has been shown to regulate alternative splicing events that are misregulated in DM1 (17, 24, 25). Individuals with DM1 show increased CUGBP1 protein steady state levels (10, 17, 26). EpA960/HSA-Cre-ERT2 (+ tam) mice express higher protein levels of CUGBP1, compared with HSA-Cre-ERT2 (+ tam) littermates and age/sex-matched EpA0/HSA-Cre-ERT2 (+ tam) mice (Fig. 4A). CUGBP1 levels are elevated in EpA960/HSA-Cre-ERT2 (− tam) mice compared with EpA0/HSA-Cre-ERT2 (+ tam) and HSA-Cre-ERT2 (+ tam) littermates, consistent with leaky expression of EpA960(R) mRNA. Importantly, we observe a small but consistent increase of CUGBP1 levels between 1 and 4 weeks after tamoxifen administration, suggesting that there might be a critical threshold CUGBP1 level correlating with muscle wasting (Fig. 4A and SI Fig. 10).
Fig. 4.
Increased CUGBP1 expression is a potential mechanism for muscle wasting in EpA960/HSA-Cre-ERT2 mice. (A) EpA960/HSA-Cre-ERT2 mice show increased steady state levels of CUGBP1 protein by Western blot analysis. (B) Percentage inclusion of alternative exons in Ank2, Capzb, and Fxr1 in skeletal muscle tissues across several time points after tamoxifen in EpA960/HSA-Cre-ERT2 mice and from two other DM1 mouse models. Each bar is a mean of biologic replicates with standard deviation when n is >1. Lanes: Newborn, day 1 newborn limb (n = 1, sample is pool of 12 animals); HSA-Cre, HSA-Cre-ERT2 no tam (n = 3); Bi (960), EpA960/HSA-Cre-ERT2 no tam (n = 3); Bi (960) + 1 wk, EpA960/HSA-Cre-ERT2 1 week after tam (n = 3); Bi (960) + 4 wks, EpA960/HSA-Cre-ERT2 4 weeks after tam (n = 3); Bi (0) + 4wks, EpA0/HSA-Cre-ERT2 4 weeks after tam (n = 3); WT, wild-type control for Mbnl1Δ3/Δ3 (n = 1); Mbnl1Δ3/Δ3, Mbnl1Δ3/Δ3 (n = 1); HSA(SR), HSASR, similar to HSALR but contains 5 CTG repeats (n = 3); HSA(LR), HSALR (n = 3). Each animal group was compared with HSA-Cre, and any statistically significant differences were determined by Student's t test, P < 0.05 (*). (C) Percentage inclusion of alternative exons in Ank2, Capzb, and Fxr1 in human skeletal muscle samples from normal fetal (white n = 1) and adult (black n = 3) tissue as well as adult DM1 tissue (gray n = 5 or 6). *, The normal adult skeletal muscle sample has a statistically significant difference from both the normal fetal and adult DM1 tissue samples, determined with the Student's t test, P < 0.05. Gel images for B and C are in SI Figs. 11 and 12, respectively; information on human samples is located in SI Table 2.
Other DM1 mouse models that do not exhibit overt skeletal muscle wasting also do not exhibit increased CUGBP1 protein levels (2, 15); therefore, alternative splicing events regulated specifically by CUGBP1 could be relevant to muscle wasting. Our lab has identified several alternative splicing events regulated during heart development that are CUGBP1 but not MBNL1 responsive (data not shown). From this list, we identified alternative splicing events in three genes that are misregulated in skeletal muscle from EpA960/HSA-Cre-ERT2 (+ tam) mice and are not misregulated in skeletal muscle from two MBNL1 depletion models (HSALR and MbnlΔ3/Δ3 mice). These events were found to be developmentally regulated in skeletal muscle, and adult EpA960/HSA-Cre-ERT2 (+ tam) mice aberrantly produce the newborn isoforms. Ankyrin-2 (Ank2) belongs to the ankyrin protein family, which is involved in linking integral membrane proteins to the cytoskeleton (27). F actin capping protein beta subunit (Capzb) binds to the fast growing barbed filament ends of actin and contributes to filament growth (28). Fragile X mental retardation-related protein 1 (Fxr1) is an RNA binding protein thought to transport mRNAs and mice expressing lower levels of Fxr1 display reduced limb musculature (29).
For all three genes the amount of exon inclusion in adult EpA960/HSA-Cre-ERT2 mice 4 weeks after tamoxifen administration was similar to that seen in wild-type newborn skeletal muscle tissue and was statistically different from that seen in adult HSA-Cre-ERT2 mice (Fig. 4B). Misregulation of these exons in EpA960/HSA-Cre-ERT2 mice appears specifically at 4 weeks after tamoxifen administration (Fig. 4B). In contrast, exon inclusion in skeletal muscle tissue samples from adult EpA0/HSA-Cre-ERT2 (+ tam), MbnlΔ3/Δ3, HSASR, and HSALR mice was not statistically different from HSA-Cre-ERT2 mice for all three genes (Fig. 4B). Therefore, the splicing events correlate with the unique feature of muscle wasting in this mouse model.
To assess whether these splicing events are relevant to DM1 pathogenesis it was critical to test these events in human tissue samples. Indeed, regulation of exon inclusion for all three genes is developmentally regulated and misregulated in adult DM1 patients in that they express the fetal splicing pattern; this difference was statistically significant for both Ank2 and Capzb (Fig. 4C). We conclude that these mice, which express expanded repeats in the context of DMPK exon 15 exhibit splicing abnormalities unique to those observed in MBNL1 depletion models.
Discussion
Understanding the mechanism underlying skeletal muscle wasting in DM1 is important because it contributes to the majority of morbidity and mortality. We have produced a DM1 mouse model that exhibits severe and progressive muscle wasting by expressing high levels of 960 CUG repeats within the natural context of DMPK exon 15. Other tri-nucleotide repeat mouse models often require high expression and large tracts of repeats to achieve a phenotype similar to human disease (30). We chose to incorporate the repeats within their natural context of exon 15 of the DMPK gene because cell based studies in a mouse myoblast cell line (C2C12) have shown that this combination is critical for inhibiting myoblast differentiation (31). Our results emphasize the critical role of expressing expanded CUG repeats within their natural context to reproduce a muscle wasting phenotype.
Histological examination of EpA960/HSA-Cre-ERT2 (+ tam) skeletal muscle depicts a dystrophic scenario in which there is an overall decrease in myofiber size, increase in the variation of myofiber diameters, necrotic fibers, small basophilic fibers with central nuclei (presumed to be regenerating fibers), and increased fibrosis. Importantly, the muscle wasting is functionally debilitating as demonstrated by treadmill tests. Two pieces of evidence argue that the muscle wasting seen is a result of pathogenesis specific to DM1 and not secondary to high expression of an exogenous RNA. First, EpA0/HSA-Cre-ERT2 (+ tam) mice, express comparable or higher levels of an identical RNA lacking CUG repeats without effects. In addition, mice from two other DM1 models, HSALR and (CTG)5 5-336, express higher levels of CUG repeat-containing mRNA than EpA960/HSA-Cre-ERT2 (+ tam) mice and yet do not display muscle wasting. Second, EpA960/HSA-Cre-ERT2 (+ tam) mice develop well characterized skeletal muscle pathologies seen in individuals with DM1 such as: intranuclear CUG RNA foci with MBNL1 colocalization, DM1-like misregulated alternative splicing, myotonia, and increased expression of CUGBP1.
The role of MBNL1 sequestration on misregulation of alternative splicing in DM1 is well established. MBNL1 controls multiple postnatal splicing transitions in skeletal muscle and studies using HSALR and MbnlΔ3/Δ3 mice demonstrate that MBNL1 is required for these transitions (7). MBNL1 depletion models suggest a predominant role for MBNL1 in many misregulated splicing events in DM1. However, the observations that MBNL1 depletion models do not exhibit overt muscle wasting and expression of MBNL1 in HSALR skeletal muscle reverses splicing but not histologic abnormalities, suggest that other defects contribute to muscle pathology (2, 15, 16). Evidence also suggests a role for increased CUGBP1 activity in DM1 pathogenesis. CUGBP1 protein levels normally decrease during postnatal heart and skeletal muscle development in mice, and this drop also correlates with alternative splicing transitions (7, 32–35). In adult DM1 skeletal muscle tissue, CUGBP1 protein levels are abnormally elevated and these same splicing events revert to the fetal splicing pattern (10, 17, 26). Increased levels of CUGBP1, by transient transfection or transgenic overexpressing mice, lead to a switch in the splicing pattern to that observed in the embryonic state and in DM1 (6, 17, 24, 25). Mice overexpressing CUGBP1 are neonatal lethal and display a developmental delay of muscle tissue, dystrophic muscle, and a myofiber-type switch all characteristic of DM1 skeletal muscle tissue (18, 24). Therefore, we hypothesize that increased CUGBP1 contributes to the skeletal muscle wasting phenotype in DM1, and that expression of repeats within the DMPK 3′ UTR is necessary for increased CUGBP1 levels. This hypothesis may explain why individuals with DM2, who do not display increased CUGBP1 protein levels, have mild skeletal muscle wasting compared with individuals with DM1 (1, 36). DM1 mouse models, which do not express CUG repeats within the DMPK 3′ UTR (HSALR and MbnlΔ3/Δ3) do not display overt skeletal muscle wasting and do not have increased CUGBP1 protein levels (2, 15). For other DM1 mouse models that do express repeats in their natural context the story is a bit more complex. Mice expressing the entire DMPK gene with tracts of >300 pure repeats develop abnormal skeletal muscle histology without overt muscle atrophy (37). It has recently been demonstrated that these mice developed large intergenerational expansions of >1,000 repeats. In the homozygous state these mice display significant growth retardation; however, it remains unclear whether there is an overt skeletal muscle wasting phenotype and whether CUGBP1 levels are altered (38). Mice with robust expression of five CUG repeats in the DMPK 3′ UTR exhibit increased CUGBP1 levels (39). However, these mice are not reported to develop overt skeletal muscle atrophy. This lack of reported muscle wasting may be due to small repeat size (five CUG repeats) or premature death from severe cardiac pathology. Our results from the EpA0/HSA-Cre-ERT2 (+ tam) mice illustrate that expression of exon 15 of DMPK alone is not sufficient to cause muscle pathology.
CUGBP1 could play a role in skeletal muscle wasting through altered RNA processing events that are regulated by CUGBP1 but not MBNL proteins. In this study we examined three such events involving alternative splicing: exon 21 in Ank2, exon 8 in Capzb and exon 15 and 16 in Fxr1. These events were found to be developmentally regulated in both mouse and human skeletal muscle. Furthermore, these events are misregulated in EpA960/HSA-Cre-ERT2 (+ tam) mice (increased CUGBP1), but not HSALR and MbnlΔ3/Δ3 mice (unchanged CUGBP1). Additionally, unpublished data in our lab indicates that transgenic mice overexpressing CUGBP1 in cardiac tissue exhibit the embryonic splicing pattern for these three alternative splicing events. These events are misregulated in DM1 skeletal muscle tissue strengthening the significance of these findings. Further investigation into whether these misregulated isoforms mechanistically contribute to muscle wasting and identification of additional CUGBP1-specific events is of interest. Elevated CUGBP1 levels may also contribute to muscle wasting through its cytoplasmic activities such as translation regulation, RNA stability, or deadenylation (18, 40, 41).
Our recent studies have shown that phosphorylation of CUGBP1 increases its stability and steady state levels (42). Additionally, expression of CUG repeats within the DMPK 3′ UTR increases phosphorylation of CUGBP1 by a mechanism that requires protein kinase C (PKC) activation (42). Although strongly suggestive from the available data, the role of increased CUGBP1 levels in skeletal muscle atrophy still remains correlative. EpA960/HSA-Cre-ERT2 mice are a robust model for DM1 skeletal muscle pathology, and can be used to study the mechanism for skeletal muscle wasting as well as used to develop therapeutic approaches.
Materials and Methods
RT-PCR.
cDNA was generated from 4 μg of RNA by using oligo dT following standard procedures. One TaqMan primer/probe mix for the EpA 960 and 0 repeat recombined allele mRNA products [EpA(R)] and a second for the human DMPK 3′ UTR were obtained from Applied Biosystems. Q-PCR was performed on the 7500 Fast Real-Time PCR System (Applied Biosystems). All samples were normalized to GAPDH (Applied Biosystems part no. 4352339E). For assaying alternative splicing events flanking primer pairs were designed for Clcn1, Seca1, Cypher, Ank2, Capzb, and Fxr1 genes. See SI Table 1 for oligo sequences. Band products were quantified by using Kodak Gel Logic 2200 with Molecular Imaging Software. Normal human RNA samples were purchased from Invitrogen and Clontech and obtained from the University of Miami Tissue Bank.
Slot Blot.
RNA was loaded onto a nylon membrane (Bio-Rad) using a slot blot apparatus (Minifold II; Schleicher & Schuell) under vacuum. The membrane was air dried and then cross linked with UV light before incubating with a 20mer CAG 2′-O-methyl Digoxygenin (Dig)-labeled RNA probe at 45°C overnight (2× SSC, 0.1% N-lauroyl sarcosine, 0.02% SDS, 1% Roche blocking solution, 30% formamide, 2 mM vanadyl complex, DMSO). The membrane was washed twice with 2× SSC, 0.1% SDS for 5 min each at room temperature, and then twice in 0.1% SSC, 0.1% SDS for 15 min each at 65°C. The Dig-labeled probe was detected by using anti-Dig HRP-conjugated antibody (Roche) followed by exposure with the Kodak Gel Logic 2200 system.
MRI.
Mice were anesthetized with 2–4% isoflurane gas with an oxygen flow rate of 2.5 liters per minute, placed in a Bruker Pharmascan 7.0T spectrometer, 16-cm bore horizontal imaging system (Bruker Biospin, Billerica, MA) and maintained on 2–2.5% isoflurane gas anesthesia with an oxygen flow rate of 1 liter per minute. We used FLASH (Fast Low Angle Shot) sequence to acquire images, with respiratory gating (SA Instruments, Stony Brook, NY). Coronal slices were captured every 1 mm, with a matrix size 256 × 256, and a FOV of 44.3 × 44.3 mm. Data were analyzed by using Amira software version 4.1. The diameter of the paraspinous muscle group was measured at the second vertebral body counting up from the tip of the iliac crest. Muscle diameters were normalized against the diameter of the kidney at the point of renal vein exit.
Histology.
Tissues were fixed in 10% formalin overnight, followed by dehydration in 70% ethanol overnight. Samples were then embedded in paraffin, sectioned, and stained. Slow and fast twitch myosin heavy chain isoforms were stained with monoclonal anti-Myosin Slow clone NOQ7.5.4D (1:100) and monoclonal anti-Myosin Fast clone MY-32 (1:1000) antibodies respectively (Sigma). Incubation for 20 min in a pressure cooker with citric acid pH 6.0 was used for antigen retrieval.
Treadmill.
Mice were placed on a horizontal two-channel AccuPacer Treadmill with rear electrical shock grid (AccuScan Instruments Inc.) at a starting speed of 10 m/min. Every two minutes, the speed was increased by 2 m/min for 30 min or until the mouse could no longer run.
Electromyogram.
Mice were lightly anesthetized with 0.1 ml of ketamine 37.6 mg/ml, xylazine 1.92 mg/ml, and acepromazine 0.38 mg/ml. An Xltek EMG system was used to acquire electrical signals from a monopolar electrode inserted in the left gastrocnemius muscle, at filter settings of 5 Hz and 5 kHz.
In Situ Hybridization and Immunofluorescence.
Skeletal muscle was frozen in OTC media immediately after dissection. Five-micrometer-thick sections were placed on superfrost plus slides (VWR cat. no. 48311-703) and fixed with 2% paraformaldehyde for 30 min on ice. In situ hybridization protocol (2) was performed with a CAG6-LNA-Cy3-labeled oligo probe. Sections were incubated in blocking solution (5% dehydrated milk, 0.05% Tween 20, 1× PBS) for 1 h at room temperature. Incubation with anti-MBNL1 polyclonal (19) 1:1,000 in blocking solution incubated overnight at 4°C was followed by incubation with anti-rabbit Alexa Fluor 488 (Molecular Probes) 1:1,000 for 1 h at room temperature. Sections were washed in PBS, and then DAPI stained and mounted by using Vectashield hard set mounting media with DAPI (Vector).
Western Blot Analysis.
Protein extracts from skeletal muscle tissue were resolved with SDS/PAGE, followed by transfer to PVDF membrane and antibody incubation. CUGBP1 clone 3B1 monoclonal antibody (Biotechnology Program, University of Florida) was HRP conjugated and used at 1:500 (34). α-Tubulin monoclonal antibody (Sigma) was used at 1:10,000 followed by goat anti-mouse light chain (Jackson ImmunoResearch) 1:10,000. Bands were quantified by using Kodak Gel Logic 2200 with Molecular Imaging Software.
Statistical Analysis.
All statistical analysis was performed with JMP IN software version 5.1 (SAS Institute). For datasets where three or more groups were analyzed simultaneously, one way ANOVA was used to determine whether there was a statistically significant difference between groups, and only datasets with an F value of <0.05 were further analyzed with Tukey–Kramer HSD. For datasets where two groups were compared, Student's t test was used, and only P values <0.05 were labeled (*) as having a statistically significant difference.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Donnie Bundman (Baylor College of Medicine) for generating the EpA mouse lines, Ankur Nagaraja for real time RT-PCR advice, Xander Wehrens for advice with the treadmill apparatus, BCM Phenotyping Core and Robia Pautler for help with MRI, Srinivas Rao for interpretation advice, Najeeba Ali and Penny Gregg for assistance with EMG recordings, Charles Thornton (University of Rochester, Rochester, NY) and Hannes Vogel (Stanford University, Stanford, CA) for DM1 tissue samples, Charles Thornton, Maurice Swanson (University of Florida, Gainesville, FL), and Mani Mahadevan (University of Virginia, Charlottesville, VA) for tissue samples from their mouse models, and Charles Thornton and Ami Mankodi for the slot blot protocol. Support for this work was provided by National Institutes of Health Grant R01AR45653, by the Muscular Dystrophy Association, and by the Department of Veterans Affairs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708519105/DC1.
References
- 1.Harper PS. Myotonic Dystrophy. London: W. B. Saunders; 2001. [Google Scholar]
- 2.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 3.Liquori CL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 4.Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am J Hum Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orengo JP, Cooper TA. Alternative Splicing in the Postgenomic Era. Austin, TX: Landes Bioscience; 2007. [Google Scholar]
- 6.Ho TH, et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 8.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 9.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 10.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 11.Fardaei M, et al. Three proteins, MBNL, MBLL and MBXL, colocalize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 13.Mankodi A, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 14.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 16.Kanadia RN, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 18.Timchenko NA, et al. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 19.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41:165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 21.Lueck JD, et al. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am J Physiol Cell Physiol. 2007;292:C1291–1297. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 22.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 23.Kimura T, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 24.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 25.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 26.Timchenko NA, et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 27.Sorrentino V. Molecular determinants of the structural and functional organization of the sarcoplasmic reticulum. Biochim Biophys Acta. 2004;1742:113–118. doi: 10.1016/j.bbamcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Littlefield R, Fowler VM. Defining actin filament length in striated muscle: Rulers and caps or dynamic stability? Annu Rev Cell Dev Biol. 1998;14:487–525. doi: 10.1146/annurev.cellbio.14.1.487. [DOI] [PubMed] [Google Scholar]
- 29.Mientjes EJ, et al. Fxr1 knockout mice show a striated muscle phenotype: Implications for Fxr1p function in vivo. Hum Mol Genet. 2004;13:1291–1302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 30.Watase K, Zoghbi HY. Modelling brain diseases in mice: The challenges of design and analysis. Nat Rev Genet. 2003;4:296–307. doi: 10.1038/nrg1045. [DOI] [PubMed] [Google Scholar]
- 31.Amack JD, Mahadevan MS. Myogenic defects in myotonic dystrophy. Dev Biol. 2004;265:294–301. doi: 10.1016/j.ydbio.2003.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Charlet BN, et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 33.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 35.Leroy O, et al. Brain-specific change in alternative splicing of Tau exon 6 in myotonic dystrophy type 1. Biochim Biophys Acta. 2006;1762:460–467. doi: 10.1016/j.bbadis.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Mankodi A, et al. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 37.Seznec H, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 38.Gomes-Pereira M, et al. CTG trinucleotide repeat “big jumps”: Large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahadevan MS, et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paillard L, et al. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.