Abstract
The phosphatidylinositol 3-kinase (PI3K) signaling pathway is up-regulated in cancer. PIK3CA, the gene coding for the catalytic subunit p110α of PI3K, is mutated in ≈30% of tumors of the prostate, breast, cervix, and endometrium. The most prominent of these mutants, represented by single amino acid substitutions in the helical or kinase domain, show a gain of enzymatic function, activate AKT signaling, and induce oncogenic transformation. We have carried out a genetic and biochemical analysis of these hot-spot mutations in PIK3CA. The results of this study suggest that the helical and kinase domain mutations trigger gain of function through different mechanisms. They show different requirements for interaction with the PI3K regulatory subunit p85 and with RAS-GTP. The gain of function induced by helical domain mutations is independent of binding to p85 but requires interaction with RAS-GTP. In contrast, the kinase domain mutation is active in the absence of RAS-GTP binding but is highly dependent on the interaction with p85. We speculate that the contrasting roles of p85 and RAS-GTP in helical and kinase domain mutations reflect two distinct states of mutated p110α. These two states differ in mutation-induced surface charges and also may differ in conformational properties that are controlled by interactions with p85 and RAS-GTP. The two states do not appear mutually exclusive because the helical and kinase domain mutations act synergistically when present in the same p110α molecule. This synergism also supports the conclusion that the helical and kinase domain mutations operate by two different and independent mechanisms.
Keywords: cancer, molecular mechanisms, p85, RAS, AKT
Phosphatidylinositol 3-kinases (PI3Ks) phosphorylate phosphatidylinositols at the 3′ position of the inositol ring, generating second messengers that control cellular activities and properties, including proliferation, survival, motility, and cell shape. Mutations that block PI3K function disrupt these processes (1–12). PI3Ks form a family that is divided into three classes, differing in structure, substrate preference, tissue distribution, mechanism of activation, and, ultimately, function (12–16). For the regulation of cell proliferation and in tumorigenesis, the most important PI3K proteins are those of class IA, notably the catalytic subunit p110α and its associated regulatory subunits (p85α, p55α, p50α, p85β, and p55γ).
p110α contains an N-terminal p85-binding domain (p85BD), a Ras-binding domain (RBD), a protein-kinase-C homology-2 (C2) domain, a helical domain, and a C-terminal kinase domain (17). p110α is constitutively associated with regulatory subunits, of which p85α is the best studied (herein referred to as p85). p110α and p85 exist in the cell as a heterodimeric complex. In quiescent cells, p85 stabilizes p110α and inactivates PI3K activity (18). Upon growth factor stimulation, receptor tyrosine kinases (RTKs) undergo autophosphorylation, creating binding sites for Src homology 2 (SH2) domain-containing proteins. The SH2 domains of p85 bind to phospho-YxxM motifs in RTKs (19). This binding relieves the inhibition of p110α and mediates the recruitment of PI3K to the plasma membrane. Direct interaction between GTP-bound Ras and p110α through the RBD further augments the activity of p110α (20–22).
p110α shows mutations in ≈30% of tumors of the prostate, breast, cervix, and endometrium (Catalog of Somatic Mutations in Cancer, www.sanger.ac.uk/genetics/CGP/cosmic). Genetic, biochemical, and cell-based analyses have shown that such mutated p110α functions as an oncoprotein playing an important role in tumorigenesis (1, 10, 23, 24). The most frequently occurring mutations, E542K, E545K, and H1047R, increase enzymatic activity, stimulate AKT (protein kinase B) signaling, induce growth factor and anchorage independent growth in culture, and cause tumors in vivo (10, 25–29).
In this article, we examine the biochemical and cell-transforming activities of several mutant forms of p110α in chicken embryonic fibroblasts (CEFs). We demonstrate that helical and kinase domain mutations operate by two different and independent gain-of-function mechanisms. Our results show that (i) gain of function by E542K and E545K is highly dependent on RAS-GTP binding, (ii) gain of function by H1047R is likely dependent on allosteric change mediated by p85, and (iii) this allosteric change triggered by H1047R may mimic RAS-GTP binding, making this mutant independent of an interaction with RAS-GTP.
Results
Helical Domain and Kinase Domain Mutations Act Synergically in Cell Transformation.
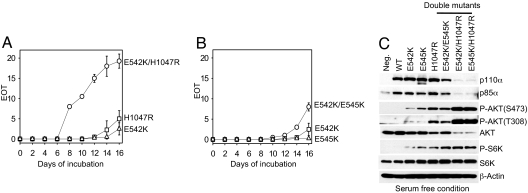
The modeled structure of p110α suggests that the helical domain mutations and kinase domain mutation map to the surface of the protein and affect protein–protein or domain–domain interactions (24, 30). We hypothesized that the spatially separated kinase domain and helical domain mutations may trigger gains of function through different molecular mechanisms. To test this hypothesis, we examined the properties of the double mutants E542K/E545K, E542K/H1047R, and E545K/H1047R (Fig. 1 A and B). These double mutants transformed CEFs more efficiently, inducing the foci of transformed cells earlier and in greater number than single hot-spot mutants. The increase in activity was additive when both mutations were in the helical domain. However, combining helical and kinase domain mutations within the same p110α molecule had a clear synergistic effect and generated a potently transforming protein. The product of PI3K, PIP3, mediates the activation of several signaling pathways, including that of AKT (31). We tested the cellular phospho-AKT level in CEFs expressing the p110α double mutants under the condition of serum starvation. Although the total AKT levels were down-regulated in CEF-expressing double mutants E542K/H1047R or E545K/H1047R, the phospho-AKT levels were ≈1,000-fold higher than in CEFs expressing the single mutants E542K, E545K, or H1047R (normalized to the protein levels of p110α). The highly up-regulated AKT signals detected in CEFs expressing E542K/H1047R or E545K/H1047R correspond to the strong cell-transforming activity of these mutants (Fig. 1C). In contrast, a combination of the two helical domain mutations E542K and E545K conferred a moderate increase in the transforming activity of the protein and enhanced AKT signaling slightly, compared with E542K and E545K alone.
Fig. 1.
Helical domain and kinase domain mutations act synergically in cell transformation. (A and B) Focus growth curves for CEFs transfected with RCAS vectors encoding the p110α mutants. The focus growth curve of E545K/H1047R is similar to that of E542K/H1047R and is therefore not shown. EOT, number of foci per nanogram of DNA. The means for two experiments are shown. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT and S6K. Cells were starved in basal medium, and lysates were prepared as described in Materials and Methods.
Binding to p85 Is Essential for H1047R-Induced Cell Transformation.
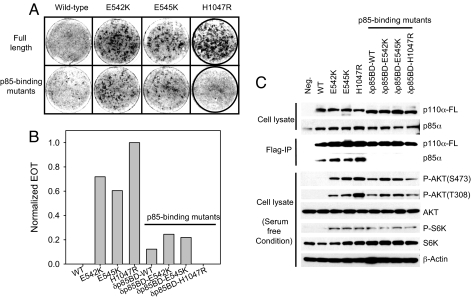
Biochemical and structural studies support the proposal that the helical domain mutations E542K and E545K relieve an inhibitory interaction between the N-terminal SH2 domain of p85 and the helical domain of p110α (32, 33). To examine the effect of p85 binding on mutant activity, we eliminated the binding of p110α to p85 by deleting the 72 N-terminal amino acids from all p110α constructs. The truncated p110α proteins no longer interacted with p85, and truncated and full-length p110α proteins were expressed at approximately equal levels (Fig. 2C). The elimination of p85 binding had a minor effect on the transforming activity of the helical domain mutations, but completely abolished transformation induced by the H1047R kinase domain mutation (Fig. 2 A and B). In the absence of p85 binding, WT p110α gained weak transforming activity (Fig. 2 A and B). This observation is in agreement with previous studies (28) and probably reflects the removal of a p85-mediated inhibitory effect.
Fig. 2.
Binding to p85 is essential for H1047R-induced cell transformation. (A and B) Cell transformation (focus formation) induced by full-length or p85-binding domain deletion mutants of p110α (δp85BD-p110α). EOT by H1047R is normalized to one. (C) Western blots comparing the p110α expression levels and the phosphorylation levels of AKT and S6K. δp85BD-p110α constructs do not coimmunoprecipitate with endogenous p85α. The assays were carried out as described in Materials and Methods.
Cells expressing mutant constructs that fail to interact with p85, including truncated E542K, E545K, and H1047R, show constitutive phosphorylation of AKT and S6K, albeit at lower levels than cells expressing the full-length proteins (Fig. 2C). Because truncated H1047R fails to transform cells, an up-regulated AKT pathway alone appears to be insufficient for PI3K-induced oncogenic transformation. Our results suggest that an unknown process dependent on p85 binding plays a critical role in H1047R oncogenicity.
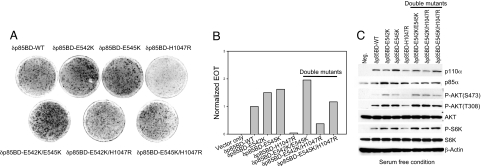
We also examined the effect of the N-terminal truncation on the double mutants E542K/H1047R and E545K/H1047R. These constructs show transforming activity with the same potency as that of the single helical domain mutants E542K and E545K (Fig. 3 A and B). Activation of AKT also corresponds to the levels induced by single helical domain mutants (Fig. 3C). The synergistic effect that is seen in the full-length double mutant is lost in the deletions, further suggesting that p85 binding is essential for the gain of function associated with H1047R.
Fig. 3.
Helical domain mutations are incapable of rescuing the oncogenic activity of δp85BD-p110α H1047R. (A and B) Cell transformation induced by δp85BD-p110α constructs. EOT by δp85BD-p110α is normalized to one. (C) Western blots comparing the p110α expression levels and the phosphorylation levels of AKT and S6K. The assays were carried out as described in Materials and Methods.
RAS Binding Is Essential for Transformation by Helical Domain Mutants of p110α.
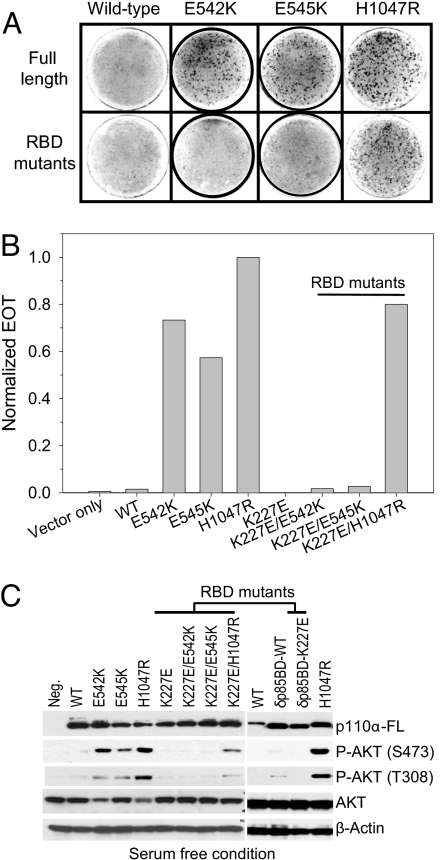
Previous studies have shown that a point mutation in the RBD of p110α, K227E, blocks the interaction of p110α and RAS and abrogates the ability of RAS to activate PI3K (21). We constructed a series of RAS-binding mutants of p110α proteins changing lysine 227 to glutamate. We then determined the transforming activities of these mutants in focus assays (Fig. 4 A and B). The K227E mutation does not affect oncogenic transformation induced by the kinase domain mutant H1047R, but completely abolishes the transforming activities of the helical domain mutants E542K and E545K. The mutant constructs were expressed at approximately equal levels (Fig. 4C). These differences in transforming potency are in agreement with the effects on AKT signaling. K227E/H1047R transforms cells and leads to constitutive activation of AKT signaling, albeit at a lower level than H1047R. In contrast, K227E/E542K and K227E/E545K fail to transform cells and do not induce constitutive activation of AKT signaling.
Fig. 4.
Oncogenic transformation by the helical domain mutation depends on binding to RAS. (A and B) Cell transformation induced by full-length or RAS-binding mutants of p110α. EOT by H1047R is normalized to one. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT. The assays were carried out as described in Materials and Methods.
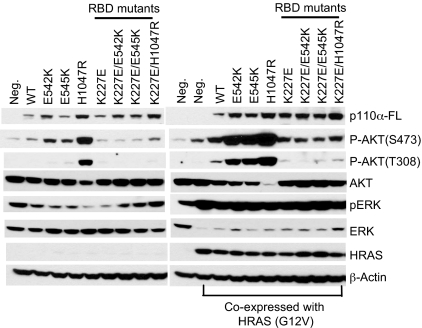
To confirm the effect of K227E on RAS-mediated activation, we tested the AKT signaling in CEFs transfected with a p110α expression vector only and CEFs cotransfected with p110α and HRAS (G12V) expression vectors (Fig. 5). Our results show that the WT and the hot-spot mutants of p110α, when coexpressed with HRAS (G12V), lead to activation of both ERK and AKT. In contrast, CEFs expressing the RBD mutants of p110α failed to activate AKT, although ERK activation was detected. The basal level of AKT phosphorylation in these cells is probably due to endogenous PI3K activity. Because of the short exposure time, the level of AKT phosphorylation at T308 in cells expressing E542K, E545K, and K227E/H1047R is not shown in the Western blot of Fig. 5.
Fig. 5.
RAS-binding mutation of p110α abolishes the activation by HRAS (G12V). CEFs were transfected with the p110α expression vector only or cotransfected with p110α and HRAS (G12V) expression vectors. Cells were maintained in a nutrient medium containing 3% FBS and 1% chicken serum and were then harvested and probed with the indicated antibodies by Western blotting.
In the previous section, we showed that WT p110α with a deletion in the p85-binding domain gained transforming activity and constitutive lipid kinase activity possibly by the removal of an inhibitory interaction between p85 and p110α. To test whether the gain of function by this truncated WT p110α depends on RAS binding, we introduced the K227E mutation in this construct. CEFs expressing this double mutant did not undergo transformation (data not shown) and did not show the up-regulation of AKT signaling seen with this mutant in the presence of Ras binding (Figs. 2C and 4C). These results suggest that RAS-mediated activation is indispensable for p110α activation.
The H1047R Mutation Can Substitute for RAS Binding.
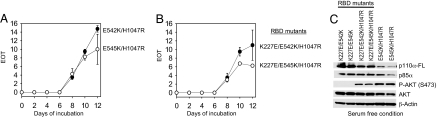
Because the cell-transforming activity of H1047R is independent of RAS binding, we speculated that an active conformation triggered by the H1047R mutation might mimic the activation mediated by binding to RAS. We therefore constructed the triple mutants K227E/E542K/H1047R and K227E/E545K/H1047R. Expression of these two mutant forms of p110α in CEFs resulted in potent and efficient transformation (Fig. 6B). The quantitative aspects of this transformation as measured in focus assays are similar to those determined with E542K/H1047R or E545K/H1047R, leading to focus formation within 7 days (Fig. 6A). In agreement with this transforming activity, AKT signaling in these cells is constitutively up-regulated, similar to the levels seen in CEFs expressing E542K/H1047R or E545K/H1047R (Fig. 6C).
Fig. 6.
The H1047R mutation rescues the cell-transformation phenotypes of the p110α mutants K227E/E542K and K227E/E545K from RAS-binding mutation. (A and B) Focus growth curves for CEFs transfected with RCAS vectors encoding the p110α mutants. EOT, number of foci per nanogram of DNA. The means for two experiments are shown. (C) Western blots comparing the protein expression levels of p110α and the phosphorylation levels of AKT. The assays were carried out as described in Materials and Methods.
Discussion
Gain-of-function and oncogenic activity make the cancer-specific mutants of p110α ideal therapeutic targets. However, to take advantage of this unique situation and design mutant-specific interventions, it is necessary to ascertain the molecular mechanisms responsible for the gain of function in the p110α mutants. We used genetic and functional analyses to gain insight into mutant mechanisms. As detailed below, our conclusions are in accord with recent structural data on p110α (33, 34).
The three hot-spot mutations in PIK3CA account for ≈80% of the mutated p110α proteins found in diverse cancers (Catalog of Somatic Mutations in Cancer, www.sanger.ac.uk/genetics/CGP/cosmic). These mutations map to two functionally and structurally distinct domains of p110α. E542K and E545K are localized to the helical domain, and H1047R resides in the kinase domain. Mapping of these mutations in two distinct functional domains suggests that helical and kinase domain mutations operate by different molecular mechanisms. A simple test that could support the hypothesis of two molecular mechanisms is to introduce two mutations concurrently in the same protein. The tests of such double mutants revealed a strong synergism of the helical kinase domain combination of mutations, but only simple additivity for the helical domain double mutant. These data are best explained by the two-mechanism proposal, but they do not rule out the possibility of a single mechanism that is greatly enhanced by a combination of mutations in different domains. The E545K/H1047R double mutant has been found to be naturally occurring in human cancer (6). This natural occurrence may reflect the strong selective growth advantage conferred on cells by this mutant combination.
The properties of mutant p110α that lack either p85 or RAS binding provide more compelling evidence for two different mechanisms triggered by helical and kinase domain mutations, respectively. The p110α protein occurs in the cell as a heterodimer bound to p85. The p85 subunit stabilizes p110α but also inhibits the lipid kinase activity of p110α. We generated p110α constructs that are defective in p85 binding by deleting the 72 N-terminal amino acids of p110α. These constructs are stable, are efficiently expressed, and fail to interact with p85 as determined by coimmunoprecipitation. The defect in p85 binding has contrasting consequences in helical and kinase domain mutants, leaving the former active, but completely abolishing the transforming activity of the latter. Phosphorylation of AKT was, however, not completely inhibited in cells expressing H1047R that lacked p85 binding. WT p110α with the 72 N-terminal amino acid truncation showed modest gain of function and became weakly oncogenic.
Previous biochemical and structure modeling studies have provided evidence for an interaction between the N-terminal SH2 domain of p85 (N-SH2) and the helical domain of p110α (32, 33). This interaction is responsible for the p85-induced inhibition of p110α. The helical domain mutations may interfere with this p85–p110α interaction and thus remove the inhibition. A weakened interaction between N-SH2 and p110α by the helical domain mutations E542K and E545K also is suggested by the failure of the lipid kinase activity of these mutants to become activated by the phosphorylated insulin receptor substrate, in contrast to WT and H1047R p110α (35). Despite the disconnection from upstream input, the N-terminally truncated E542K and E545K induced constitutive activation of AKT. The N-terminally truncated helical domain mutations show significantly higher oncogenic potency than the truncated WT p110α. Because both have lost p85 binding, the difference in oncogenic potency then suggests an effect of the helical domain mutations that goes beyond interference with the p85 interaction.
PI3K is an important RAS effector and has a role in mediating the proliferative and survival functions of RAS (20, 21). Direct interaction between GTP-bound RAS and p110α mediated by the RBD augments the activity of p110α (22) possibly by inducing a conformational change at the substrate-binding site (36). Disrupting the interaction between p110α and RAS inhibits the in vitro transformation of mouse embryo fibroblasts by oncogenic KRAS (G12V) (37). Because the product of PI3K, PIP3, can activate RAS by stimulating the GEF activity of SOS (38), we postulated that PI3K-RAS binding also may play a critical role in oncogenic transformation by PI3K. We disabled RAS binding by introducing a point mutation K227E and found again contrasting effects on helical and kinase domain mutations, but opposite to the effects seen with deleted p85 binding. In the case of disabled RAS binding, transforming ability and AKT activation by the helical domain mutations were abolished, whereas those by the kinase domain mutation H1047R remained unaffected. The H1047R mutation could even rescue the helical domain mutants that are unable to bind RAS and could restore their activity to the same levels as detected in the synergistic double mutants E542K/H1047R and E545K/H1047R. The gain of function induced by H1047R is independent of RAS binding, whereas that of E542K and E545K requires RAS interaction. We hypothesize that H1047R induces an allosteric change of p110α that mimics RAS-GTP binding. A schematic summary of mutant properties is presented in Fig. 7.
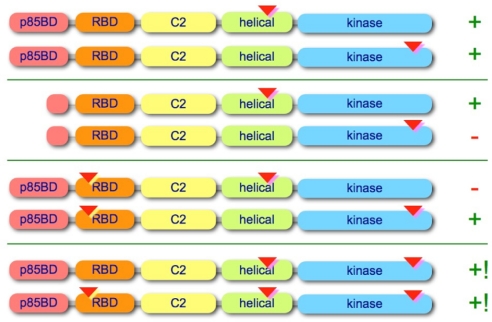
Fig. 7.
A schematic summary of mutant properties. p85BD, p85-binding domain; RBD, Ras-binding domain; C2, C2 domain. The approximate locations of the point mutations are indicated by inverted triangles, and the deletion in the p85BD is marked by a truncated alias of the p85BD. Cell-transforming activity is qualitatively denoted by + and −, and synergistic activity is denoted by +!.
Our data and conclusions are in broad agreement with the recently published crystal structure of the p110α–p85 complex (34). In this publication, the interaction between the N-SH2 domain of p85 and the helical domain is modeled, and the model allows for the possibility that this interaction could be weakened by the helical domain mutations of p110α. However, the model does not definitely exclude a possible effect of the helical domain mutations on the interaction with another p110α-binding partner. The kinase domain mutation H1047R maps close to the activation loop and may affect the conformation of the loop, altering the interaction with the substrate. Previous structural studies on the p110γ–RAS complex also have demonstrated a change in the conformation of the substrate-binding site as a result of RAS binding (36). These data are consistent with our hypothesis that H1047R activates p110α by inducing a conformational shift that is similar to that caused by RAS. The crystal structure of the p110α–p85 complex also reveals an unexpected interaction between the p85-binding domain (adaptor-binding domain) and the kinase domain. This interaction might be important for the active conformation induced by H1047R and could explain the sensitivity of H1047R to a loss of p85 binding.
Our studies strongly suggest that helical and kinase domain mutations trigger a gain of p110α function by different molecular mechanisms. Helical domain mutations function independently of p85, but require RAS. The kinase domain mutation H1047R requires p85, but not RAS (Fig. 7). The two gain-of-function states represented by helical and kinase domain mutations differ in surface charge and most likely reflect different conformational changes mediated by p85 or RAS, respectively. These conformational changes may make it possible to identify mutant-specific small-molecule inhibitors of p110α. Such inhibitors will not affect the WT enzyme and, therefore, will have superior therapeutic properties as anticancer drugs.
Materials and Methods
Plasmid Construction.
The construction of the pBSFI vector KOZ-cp3k encoding WT, E542K, E545K, and H1047R chicken p110α has been described previously (27, 39). The double mutants were constructed by using the QuikChange site-directed mutagenesis kit (Stratagene) with E542K as the template to generate E542K/H1047R and with E545K as the template to generate E545K/H1047R. The primers used were described previously (27). To generate E542K/E545K, E542K was used as the template, and the primer used was: E542K/E545K, 5′-CGA GAT CCT TTG TCT AAA ATC ACT AAG CAA GAG-3′ (only the forward primer is listed). The construction of pBSFI-Δ72-cp3k (encoding p110α with a deletion of the first 72 amino acids) has been described previously (39). The mutant constructs containing an E542K, E545K, or H1047R substitution were generated by using the QuikChange site-directed mutagenesis kit (Stratagene). To generate the K227E mutant constructs, the pBSFI vector KOZ-cp3k encoding WT, E542K, E545K, H1047R, E542K/E545K, E542K/H1047R, and E545K/H1047R was used as a template, and the primer used was: K227E, 5′-GCT GAA GCA ATT AGG GAG AAA ACA CGA AGT ATG-3′ (only the forward primer is listed). The mutated genes were subsequently cloned into the avian retrovirus vector RCAS.Sfi (40). All mutations were confirmed by sequencing.
Cell Culture and Transfection.
Fertilized chicken eggs (white Leghorn) were obtained from Charles River Breeding Laboratories. Preparation and cultivation of primary CEFs have been described previously (41). CEFs were maintained in Ham's F-10 medium (Sigma–Aldrich) supplemented with 7% FBS (Sigma–Aldrich), 5% chicken serum (Sigma–Aldrich), 1x MEM vitamin solution (Sigma–Aldrich), 8 mg/liter folic acid, and 1% l-glutamine–penicillin–streptomycin solution (Sigma–Aldrich) at 37°C in 5% CO2. Cells were plated at 80% confluence in 100-mm tissue culture and were then transfected with relevant RCAS vectors by using the dimethyl sulfoxide/Polybrene method (42). CEFs were passaged three times to ensure the high infection rate by RCAS viruses. After three passages, the cells were harvested for further analysis. For serum starvation, CEFs were first maintained in Ham's F-10 medium with 0.25% FBS and 0.05% chicken serum for 40–44 h, followed by an additional 2 h in Ham's F-10 medium. They were then harvested for protein analyses.
Transformation Assays.
DNA was transfected into CEFs by using either the Lipofectamine reagent (Invitrogen) or the dimethyl sulfoxide/Polybrene method (42). Transfected cells were fed every other day with nutrient agar and then stained with crystal violet. After incubation of cells under nutrient overlay, the foci of transformed cells were counted ≈2 weeks after transfection. For focus growth curves, the foci of transformed cells were counted every other day.
Immunoprecipitation and Western Blotting.
Lysis of cells for Western blotting was performed in Nonidet P-40 buffer [20 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P-40, and 10% glycerol] and supplemented with 1 mM DTT, 1 mM PMSF, 50 mM β-glycerophosphate, 50 mM NaF, and 1x protease Inhibitor mixture (Roche). Lysates were clarified by centrifugation at 16,000 × g for 15 min at 4°C, and the protein concentration was determined. Whole-cell lysates were incubated with mouse monoclonal anti-FLAG M2 affinity gel (Sigma–Aldrich) at 4°C overnight. The immunocomplexes were pelleted and washed three times with cold lysis buffer in the presence of protease and phosphatase inhibitor. For Western blot analysis, the bound proteins were eluted by boiling in SDS-loading buffer, resolved on SDS/PAGE gel, and then transferred to Immobilon P membranes (Millipore). After the membranes were blocked with 5% BSA in Tris-buffered saline with 0.05% Tween-20 (TBS-T) for 1 h at room temperature, they were incubated overnight at 4°C with primary antibodies. Anti-FLAG, anti-p85α, anti-AKT, anti-phospho-AKT (Ser-473), anti-phospho-AKT (Thr-308), anti-S6K, anti-phospho-S6K (Thr-389), anti-ERK (p44/42), anti-phospho-ERK (p44/42, Thr-202/Tyr-204), and anti-β-tubulin were purchased from Cell Signaling Technology. Membranes were washed three times in TBS-T and incubated in a 1:10,000 dilution of peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (H+L) (Pierce) in 5% nonfat dry milk in TBS-T for 1 h at room temperature. The reactive bands were visualized by chemiluminescence (Pierce) according to the manufacturer's protocol.
ACKNOWLEDGMENTS.
We thank Lynn Hamaguchi-Ueno for expert and skilled technical assistance and Drs. Sohye Kang and Lingyun Zhao for helpful discussion of the manuscript. This work was supported by grants from the National Cancer Institute, The Stein Foundation, and National Cancer Institute Postdoctoral Fellowship F32CA130304 (to L.Z.). This article is manuscript no. 19271-MEM from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 2.Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 3.Broderick DK, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 4.Campbell IG, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann C, et al. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol. 2005;109:639–642. doi: 10.1007/s00401-005-1000-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee JW, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 7.Levine DA, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 8.Li VS, et al. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer. 2005;5:29. doi: 10.1186/1471-2407-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saal LH, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 10.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, et al. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25:322. doi: 10.1002/humu.9316. [DOI] [PubMed] [Google Scholar]
- 12.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 13.Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 14.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 15.Wymann MP, Zvelebil M, Laffargue M. Phosphoinositide 3-kinase signalling–which way to target? Trends Pharmacol Sci. 2003;24:366–376. doi: 10.1016/S0165-6147(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 16.Deane JA, Fruman DA. Phosphoinositide 3-kinase: Diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 17.Walker EH, et al. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: Stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skolnik EY, et al. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Viciana P, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Viciana P, et al. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 22.Chan TO, et al. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- 23.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 24.Kang S, Bader AG, Zhao L, Vogt PK. Mutated PI3-kinases: Cancer targets on a silver platter. Cell Cycle. 2005;4:578–581. doi: 10.4161/cc.4.4.1586. [DOI] [PubMed] [Google Scholar]
- 25.Ikenoue T, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 26.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 27.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corvera S, Czech MP. Direct targets of phosphoinositide 3-kinase products in membrane traffic and signal transduction. Trends Cell Biol. 1998;8:442–446. doi: 10.1016/s0962-8924(98)01366-x. [DOI] [PubMed] [Google Scholar]
- 32.Shekar SC, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 33.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 34.Huang CH, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 35.Carson JD, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphatidylinositol 3-kinase. Biochem J. 2008;409:519–524. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 36.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 38.Nimnual AS, Yatsula BA, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 39.Aoki M, et al. The catalytic subunit of phosphoinositide 3-kinase: Requirements for oncogenicity. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 40.Aoki M, et al. The akt kinase: Molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogt PK. In: Fundamental Techniques in Virology. Habel K, Salzman NP, editors. New York: Academic; 1969. pp. 198–211. [Google Scholar]
- 42.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]