Abstract
Mutations in the leucine-rich repeat kinase (LRRK2) gene cause late-onset autosomal dominant Parkinson's disease (PD) with pleiomorphic pathology. Previously, we and others found that expression of mutant LRRK2 causes neuronal degeneration in cell culture. Here we used the GAL4/UAS system to generate transgenic Drosophila expressing either wild-type human LRRK2 or LRRK2-G2019S, the most common mutation associated with PD. Expression of either wild-type human LRRK2 or LRRK2-G2019S in the photoreceptor cells caused retinal degeneration. Expression of LRRK2 or LRRK2-G2019S in neurons produced adult-onset selective loss of dopaminergic neurons, locomotor dysfunction, and early mortality. Expression of mutant G2019S-LRRK2 caused a more severe parkinsonism-like phenotype than expression of equivalent levels of wild-type LRRK2. Treatment with l-DOPA improved mutant LRRK2-induced locomotor impairment but did not prevent the loss of tyrosine hydroxylase-positive neurons. To our knowledge, this is the first in vivo“gain-of-function” model which recapitulates several key features of LRRK2-linked human parkinsonism. These flies may provide a useful model for studying LRRK2-linked pathogenesis and for future therapeutic screens for PD intervention.
Keywords: dopaminergic neuron, Parkinson's disease
Mutations in the leucine-rich repeat kinase (LRRK2) gene cause late-onset autosomal dominant Parkinson's disease (PD) with pleiomorphic pathology, including nigral degeneration, Lewy bodies, and neurofibrillary tau-positive tangles (1–4). The LRRK2 gene spans a 144-kb genomic region, with 51 exons encoding 2,527 aa. The gene is expressed in all tissues examined, although at low levels. LRRK2 contains multiple conserved domains including MAP kinase kinase kinase (MAPKKK), leucine-rich repeat (LRR), GTPase (ROC and COR), and WD40 domain (1, 2). The normal biological function of LRRK2 is unclear, although suppression of LRRK2 with siRNAs or a dominant inhibitory allele leads to increased neurite length and complexity (5). The discovery of PD-linked point mutations in almost all of the predicted domains of LRRK2, the absence of deletions or truncations, and the dominant inheritance of the disease suggest a “gain-of-function” mechanism for LRRK2-linked PD.
The LRRK2 MAPKKK domain contains sequence homology to both serine/threonine and tyrosine kinases. Several pathogenic mutations of LRRK2 in PD have been found within the protein kinase domain active segment (e.g., G2019S), suggesting that these mutations may cause pathology through altering the enzymatic activity of LRRK2 (1, 2, 6). G2019S is the most common mutation in LRRK2-associated PD (7–9) and is believed to increase LRRK2 kinase activity in assays to measure autophosphorylation or phosphorylation of generic substrates (5, 10–14). Controversy exists regarding whether the other PD mutations alter LRRK2 kinase activity (14, 15). Abolishing LRRK2 kinase activity diminishes the toxicity of all PD mutants tested in cell culture (11, 12), suggesting that LRRK2 protein kinase activity may play an important role in PD pathogenesis (6).
Increasing evidence suggests that Drosophila melanogaster is an excellent model organism for studying neuronal degenerative diseases (16, 17) such as PD, Alzheimer's disease, tauopathies, amyotrophic lateral sclerosis, hereditary spastic paraplegia and polyglutamine diseases, and spinocerebellar ataxia (17, 18). The LRRK2 gene is highly conserved across species. Drosophila has a single orthologue of the human LRRK2 (CG5483), and a loss of function in this gene has been described (19). However, the loss-of-function mutation of Drosophila CG5483 does not constitute an adequate model for the most common forms of LRRK2-linked PD, which appear to be gain-of-function mutations (like G2019S). To create a pathogenic gain-of-function model for LRRK2-linked disease, we generated transgenic flies expressing full-length human wild-type LRRK2 and mutant LRRK2-G2019S. We found that overexpression of LRRK2 or LRRK2-G2019S led to retinal degeneration, selective loss of dopaminergic (DA) neurons, decreased climbing activity, and early mortality. Expression of mutant LRRK2-G2019S caused a more severe phenotype than wild-type LRRK2. Thus, the LRRK2 transgenic flies recapitulated several key features of human parkinsonism, indicating that overexpression of LRRK2 in flies may provide a model for the human disease.
Results
LRRK2 Induces Retinal Degeneration.
To address whether overexpression of wild-type human LRRK2 and the mutant LRRK2-G2019S phenocopy the human disease in flies, we introduced these proteins in specific subsets of cells using the GAL4/UAS system (20). This system takes advantage of the yeast GAL4 transcription factor, which binds specifically to the upstream activation sequence (UAS). Thus, UAS-linked transgenes can be expressed in specific cell types under the control of a given promoter (promoter-GAL4). To determine whether introduction of LRRK2 and LRRK2-G2019S causes a parkinsonism-like phenotype, we first assayed for retinal degeneration, because photoreceptor cell death has been used to assay neurodegeneration in other fly models of PD (18, 21). Therefore, we expressed two lines of UAS-LRRK2 (1 and 4) and two lines of UAS-LRRK2-G2019S (2 and 3) in photoreceptor cells, under the control of the glass multiple reporter (GMR)-GAL4. Using antibodies directed against the N-terminal Flag tags, we found that the wild-type and mutant proteins were stably expressed (Fig. 1A).
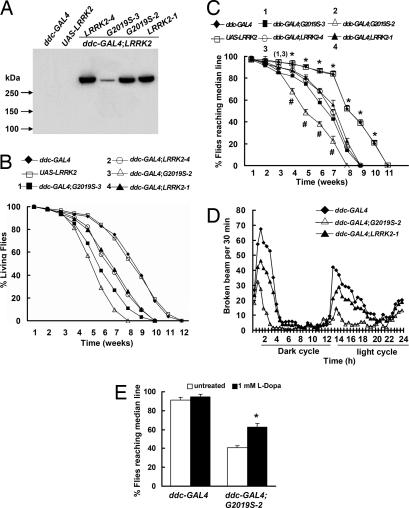
Fig. 1.
LRRK2 induced retinal degeneration. (A) Expression of Flag-LRRK2 and Flag-LRRK2-G2019S in photoreceptor cells. Fly head extracts prepared from the indicated transgenic flies were fractionated and subjected to Western blot analysis using anti-Flag antibodies. (B) Time course of photoreceptor degeneration determined by the optical neutralization technique. Each data point was based on examination of ≥90 ommatidia from at least six flies. Statistically significant differences between control and LRRK2 or LRRK2-G2019S transgenic flies are indicated: *, P < 0.05 by ANOVA. (C) Ommatidia of 5-week-old flies examined by transmission electron microscopy.
The fly compound eye comprises ≈800 repeat units, ommatidia, each including seven photoreceptor cells in any given plane of section. Each photoreceptor cell has a microvillar structure, the rhabdomere, which is the site of photoreception and is the invertebrate equivalent of the rod and cone outer segment. To examine the kinetics of retinal degeneration, we used the optical neutralization technique. As shown, retinal degeneration was detectable by 3 weeks after eclosion in LRRK2 transgenic flies and increased markedly in older flies in comparison with GMR-GAL4 or UAS only control flies (Fig. 1 B and C).
Expression of LRRK2 by ddc-GAL4 Causes Early Mortality and Locomotion Impairment.
To express LRRK2 in DA neurons, we combined the UAS-WT-LRRK2 and UAS-G2019S-LRRK2 transgenes with the dopa decarboxylase (ddc)-GAL4 driver. Using an anti-Flag antibody, both wild-type and mutant LRRK2 were readily detected in fly head homogenates (Fig. 2A). Survival curves used to examine whether expression of either LRRK2 or LRRK2-G2019S in DA neurons affected fly viability revealed that expression of either LRRK2 or LRRK2-G2019S caused premature mortality (Fig. 2B), although expression of mutant LRRK2-G2019S caused more severe mortality at equivalent expression level (Fig. 2 A and B). The ages at which 50% of the LRRK2-1 and G2019S-2 transgenic flies survived were 48 and 38 days, respectively. The G2019S-3 line had much lower expression than wild-type LRRK2-1 but had a faster rate of mortality (Fig. 2B).
Fig. 2.
Expression of LRRK2 protein by ddc-GAL4 driver caused locomotor dysfunction. (A) Expression of LRRK2 proteins in flies containing the ddc-GAL4 in combination with the UAS-LRRK2 transgenes. Head extracts from the indicated fly stocks were subjected to Western blot analysis by using anti-Flag antibodies [20 μg of protein per lane, except for the ddc-GAL4;LRRK2–4 (10 μg)]. (B) Survival curves of flies expressing either LRRK2 or LRRK2-G2019S (n = 50). (C) Cohorts of 60 flies from each genotype were subjected to climbing assays weekly. Statistically significant differences between the control and LRRK2 transgenic lines (except the 4 week data) are indicated: *, P < 0.05 by ANOVA. Statistically significant differences between LRRK2-1 and G2019S-2 flies are indicated: #, P < 0.05 by ANOVA. (D) Cohorts of 20 flies from each genotype at 5 weeks of age were subjected to the actometer to measure locomotor activity. Shown are representative data from three separate experiments. (E) Flies at 5 weeks of age were untreated or were treated with 1 mM l-DOPA for 10 days, then subjected to climbing assays. Statistically significant differences between untreated ddc-GAL4;G2019S-2 flies are indicated: *, P < 0.05 by ANOVA.
To measure the behavioral differences resulting from expression of LRRK2 in DA neurons, we used a climbing assay (negative geotaxis test). When tapping the flies to the bottom of the vial, nearly all control flies (ddc-GAL4 or the UAS-LRRK2) that were <7 weeks old climbed rapidly to the top of the vial (Fig. 2C). As the control flies aged, they were no longer able to climb to the top of the vial but instead made short abortive climbs and fell back to the bottom of the vial. We found that young (≤7 days old) flies expressing either wild-type or mutant LRRK2-G2019S climbed as well as nontransgenic control flies. However, over time, their performance declined more rapidly than that of the control flies, revealing a locomotor dysfunction of the LRRK2 transgenic flies (Fig. 2C). Moreover, G2019S-2-expressing flies displayed more severe impairment than flies expressing wild-type LRRK2-1 at comparable levels (Fig. 2C).
To further assess the deficits in locomotor activity, we used an actometer to assess the locomotor activity in 5-week-old flies during a 12-h dark/12-h light cycle. The control flies displayed two peaks of activity, and expression of either wild-type or LRRK2-G2019S did not affect the times at which peak activity occurred. By contrast, expression of the LRRK2 transgenes in DA neurons decreased the frequency of locomotor activity (Fig. 2D). Consistent with the climbing assay, the activity of the flies expressing LRRK2-G2019S was more severely impaired than that of flies expressing wild-type LRRK2 at equivalent levels (Fig. 2D). Finally, treatment of ddc-GAL4;G2019S-2 flies (5 weeks of age) with 1 mM l-DOPA for 10 days significantly improved the locomotor activity of ddc-GAL4;G2019S-2 flies (Fig. 2E).
LRRK2 Induces DA Neuronal Degeneration.
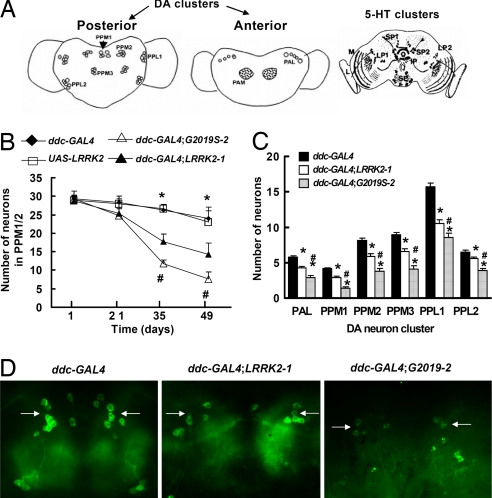
Six neuronal DA clusters are normally present in each Drosophila adult brain hemisphere (22, 23). These neurons express tyrosine hydroxylase (TH), which is an enzyme required for the biosynthesis of dopamine. Flag-LRRK2-linked immunofluorescence was evident in neurons of all DA neuron clusters and colocalized with anti-TH immunostaining (data not shown). To assess whether expression of LRRK2 resulted in degeneration of DA neurons, brains from transgenic flies at 1, 21, 35, and 49 days after eclosion were dissected and immunostained with anti-TH antibodies. In control flies (ddc-GAL4 or UAS-LRRK2 flies), the DA clusters did not change significantly in number or morphology during aging, as monitored by anti-TH staining (Fig. 3B). At 1 day after eclosion, there were no differences in anti-TH-positive staining between the control and LRRK2 or LRRK2-G2019S flies (Fig. 3B). However, at 5 weeks of age, anti-TH staining decreased significantly in flies expressing either wild-type or mutant LRRK2-G2019S (Fig. 3 B–D). We counted the TH-positive cells in all clusters except in the paired anterolateral medial (PAM) cluster, because the density of the neurons in PAM was too high to allow precise quantification. We found statistically significant TH-positive neuronal loss in all of the DA clusters examined (Fig. 3C). In addition, mutant LRRK2-G2019S caused more TH-positive neuronal loss than wild-type LRRK2 at equivalent expression levels (Fig. 3 B–D). Although l-DOPA improved the mutant LRRK2-induced locomotor impairment, it did not prevent the loss of TH-positive neurons (data not shown).
Fig. 3.
Expression of LRRK2 protein by ddc-GAL4 driver induced loss of TH-positive DA neurons. (A) Diagram of DA and 5-HT neuron clusters in the medial and lateral areas of the adult fly brain as in previous publications (35, 36). (Left) Five clusters: PPM1 (unpaired), PPM2 (paired), PPM3 (paired; protocerebral posterior medial), and PPL1 and PPL2 (paired; protocerebral posterolateral) on the posterior side. (Center) Two DA clusters: PAL (protocerebral anterolateral) and PAM (paired anterolateral medial) on the anterior side. (Right) Five distinct 5-HT neuronal clusters (SP1, SP2, LP1, LP2, and IP) in the two brain hemispheres. (B–D) Dissected whole brains were subjected to anti-TH immunofluorescent staining. (B) Quantitation of TH-positive neurons in PPM1/2 clusters in transgenic flies of the indicated ages. (C) Average numbers of TH-positive neurons per DA cluster in 5-week-old flies of the indicated genotypes. (D) Representative images of anti-TH staining in PPM1 and PPM2 clusters from 5-week-old flies of the indicated genotypes. Statistically significant differences between the control and all LRRK2 transgenic lines are indicated: *, P < 0.05 by ANOVA. Statistically significant differences between LRRK2-1 and G2019S-2 flies are indicated: #, P < 0.05 by ANOVA.
Because ddc-GAL4 can also lead to LRRK2 transgenes expressing in serotonin (5-HT) neurons, we also examined whether LRRK2 protein affected 5-HT neurons using anti-5-HT whole-mount brain immunohistochemical analysis. We found that the brains of flies expressing either wild-type or mutant LRRK2-G2019S at 5 and 7 weeks after eclosion displayed 5-HT immunoreactivity (data not shown) similar to that of the control flies.
Expression of LRRK2 in All Neurons Causes Late-Onset Locomotion Impairment and Selective Loss of TH-Positive Neurons.
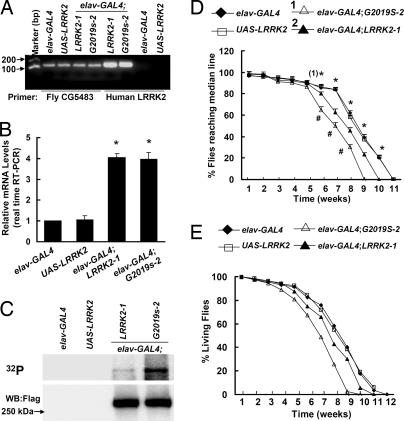
To further determine the effect of LRRK2, we expressed LRRK2 protein using the panneuronal driver, the embryonic lethal abnormal visual system gene (elav)-GAL4, achieving comparable levels of expression in elav-GAL4;LRRK-1 and elav-GAL4;G2019S-2 fly brains by RT-PCR (Fig. 4 A and B) and anti-Flag Western blot (Fig. 4C). The human LRRK2 transgene transcripts were more abundant than CG5483 (Fig. 4 A and B). Importantly, we found that the protein kinase activity in homogenates from elav-GAL4;G2019S-2 fly heads was ≈2.8-fold higher than that measured in elav-GAL4;LRRK2-1 (Fig. 4C). At 1 week after eclosion, flies expressing either wild-type or mutant LRRK2 under the control of elav-GAL4 displayed normal locomotor activity (Fig. 4D). However, by 6 weeks, the climbing assay showed significant motor impairment in elav-GAL4;G2019S-2 flies and a slight deficit in elav-GAL4;LRRK2-1 flies compared with control flies (Fig. 4D). The climbing performance of flies expressing either LRRK2-1 or LRRK2-G2019S-2 declined more rapidly than that of control flies. Moreover, the mutant LRRK2-G2019S flies exhibited more severe impairment than did the wild-type flies 6 week after eclosion (Fig. 4D). We also found that expression of LRRK2-1 and LRRK2-G2019S-2 shortened the lifespan compared with control flies (Fig. 4E). The ages at which 50% of the LRRK2-1 and G2019S-2 transgenic flies survived were 49 and 55 days, respectively.
Fig. 4.
Expression of LRRK2 by elav-GAL4 driver caused late-onset locomotor impairment. (A and B) Expression of CG5483 and human LRRK2 in various types of fly brain tissues. Total RNA was prepared from fly brain tissues, and cDNA was generated. Semiquantitative RT-PCR was performed by using primers for CG5483 and human LRRK2 to assess mRNA levels. (A) Representative image of RT-PCR products. (B) Quantitative analysis of relative mRNA levels of CG5483 and human LRRK2. *, P < 0.05 versus UAS-LRRK2 by ANOVA. (C) LRRK2 autophosphorylation (kinase) analysis of various fly head homogenates. Anti-Flag-LRRK2 immunoprecipitated samples from fly head homogenates were incubated with [γ-32P]ATP, subjected to SDS/PAGE, and blotted onto PVDF membranes. The samples were then imaged by using a phosphoimaging system. The incorporation of [γ-32P]ATP into LRRK2 protein increased by ≈2.8-fold in elav-GAL4;G2019S-2 flies compared with elav-GAL4;LRRK2-1 flies. Shown are representative images from three independent experiments. (D) Cohorts of 60 flies from each genotype were subjected to climbing assays weekly. Statistically significant differences between the control and all LRRK2 transgenic lines are indicated: *, P < 0.05 by ANOVA. Statistically significant differences between LRRK2-1 and G2019S-2 flies are indicated: #, P < 0.05 by ANOVA. (E) Survival curves of flies expressing either LRRK2 or LRRK2-G2019S (n = 50).
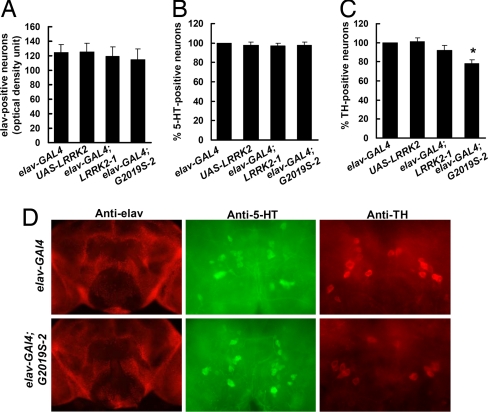
Analysis of brains of elav-GAL4;G2019S-2 flies at 5 weeks of age revealed reductions in TH immunoreactivity up to 22% in DA clusters whereas the elav-GAL4;LRRK2-1 flies showed only a slight decrease (Fig. 5 C and D). At 7 weeks of age, flies expressing LRRK2-1 or LRRK2-G2019S-2 showed significant decreases in anti-TH staining up to 28% and 50%, respectively. In contrast, there were no significant changes in anti-elav and anti-5-HT immunostaining at 5 weeks (Fig. 5) and 7 weeks (data not shown) of age compared with nontransgenic control flies. These results indicate that expression of LRRK2 proteins was selectively toxic for DA neurons and that LRRK2-G2019S was more toxic than wild-type LRRK2.
Fig. 5.
Expression of LRRK2 in all neurons caused selective loss of anti-TH-positive neurons. (A) Anti-elav whole-mount brain immunofluorescence of flies at 5 weeks after eclosion showed that expression of either LRRK2-1 or G2019S-2 did not significantly change the density of immunofluorescence. There was a slight but not significant decrease in anti-elav staining in LRRK2 transgenic flies. This could be due to loss of anti-TH-positive neurons. (B) Anti-5-HT whole-mount brain immunofluorescence of flies at 5 week after eclosion showed that expression of either LRRK2-1 or G2019S-2 did not cause loss of 5-HT-positive neurons. (C) Anti-TH whole-mount brain immunofluorescence of flies at 5 weeks after eclosion showed that expression of G2019S induced loss of TH-positive neurons. (D) Representative images of whole-mount brain sections of flies 5 weeks after eclosion. (Left) Anti-elav staining brain section. (Center) SP1, SP2, and IP 5-HT neuronal clusters with anti-5-HT staining. (Right) PPM1 and PPM2 TH-stained DA neuronal clusters.
Discussion
Mutations in the LRRK2 gene represent the most common known cause of PD (1, 2, 24). Unlike mutations in other PD-linked genes, LRRK2-linked disease has a clinical progression and neurochemical phenotype similar to that of typical late-onset disease, but little is known about LRRK2-linked molecular pathogenesis. Here we have created gain-of-function LRRK2 Drosophila models by overexpressing the human wild-type LRRK2 and the mutant form LRRK2-G2019S. Expression of both forms of LRRK2 led to retinal degeneration, selective loss of DA neurons in the brain, early mortality, and locomotor impairment. Moreover, LRRK2-G2019S caused a more severe parkinsonism-like phenotype than wild-type LRRK2. Treatment with l-DOPA improved the mutant LRRK2-induced locomotor impairment but did not prevent the loss of TH-positive neurons, similar to LRRK2-linked human PD.
Expression of LRRK2 in all neurons under the control of the elav-GAL4 caused a less severe phonotype in flies than specific expression of LRRK2 in DA neurons under the control of the ddc-GAL4, although the expression levels of proteins in fly head homogenates was higher in elav-GAL4;LRRK2 flies. This paradox may be explained by the lower expression of LRRK2 proteins in DA neurons by elav-GAL4. LRRK2 triggered the loss of anti-TH immunostaining but not the significant loss of anti-elav or anti-5-HT immunostaining, indicating that LRRK2-induced toxicity is preferentially localized to DA neurons in the brain, which is reminiscent of human PD. The manifestation of symptoms in PD patients is associated with the loss of 50–60% of DA neurons (25–27). We found that 5-week-old ddc-GAL4;G2019S-2 flies and 7-week-old elav-GAL4;G2019S-2 flies had an ≈50% reduction of TH-positive neurons. These results indicated that LRRK2 induced the loss of DA neurons or TH expression. In either case there would be loss of DA function. Moreover, the LRRK2 flies displayed parallel kinetics in the loss of TH-positive neurons and locomotor dysfunction, suggesting that these abnormalities may be causally related.
Expression of wild-type LRRK2 protein was toxic, albeit less so than expression of LRRK2-G2019S. These results raise the possibility that an elevated concentration of wild-type LRRK2 protein under some circumstances, such as genetic variation or cellular stress, may lead to DA neuronal degeneration and locomotor impairment and subsequently may contribute to some cases of human PD. As a related example, genetic duplication or triplication at the α-synuclein locus leading to overexpression of wild-type α-synuclein caused PD (28), although there are as yet no similar reports that elevated expression of wild-type LRRK2 links to human PD. A recent report shows that the disease phenotype and mortality of patients with heterozygous versus homozygous G2019S mutations are similar (29). However, there was an expression-level-dependent effect in phenotype between different G2019S fly lines. G2019S-2, which had a higher expression level than G2019S-3, has a faster rate of mortality and locomotor impairment than G2019S-3. At comparable expression levels, the mutant G2019S-LRRK2 had a more severe phenotype than wild-type LRRK2. The G2019S-3 line had much lower expression than wild-type LRRK2-1 but had a faster rate of mortality. We further found that mutant G2019S-2 had higher autophosphorylation activity than wild-type LRRK2-1. These results are consistent with in vitro findings that mutant LRRK2-G2019S has higher protein kinase activity and causes greater toxicity than wild-type LRRK2 (5, 10–14). A recent report shows that the G2019S mutation in LRRK2 appears to increase autophosphorylation through a process that seems to involve reorganization of the kinase activation segment and suggests a molecular explanation for how the G2019S mutation enhances the catalytic activity of LRRK2, thereby leading to pathogenicity (13). Another possibility is that LRRK2 may act as a scaffold protein to alter other signaling molecules leading to pathogenicity through its specific protein–protein interaction domains (LRR and WD40). Clarifying the effects of PD-associated mutation on kinase activity and PD pathogenesis awaits the identification of true LRRK2 substrates and interaction partners. LRRK2 Drosophila may be a potential useful in vivo system to identify these LRRK2 interactors or substrates.
Drosophila CG5483 (the fly homolog of LRRK2) is expressed in all tissues examined and may be enriched in brain and thoracicoabdominal ganglion according to the FlyBase database (http://flybase.bio.indiana.edu/reports/FBgn0038816.html) by cDNA array. The loss-of-function mutant studies indicate that the Drosophila CG5483 protein is critical for the integrity of fly DA neurons (16). Transgenic expression of Drosophila wild-type CG5483 and a mutation (R1069C) corresponding to the human R1441C mutation does not show any significant defects (16). This mutation in the context of Drosophila CG5483 may not be as pathogenic as the same R1441C change in the context of the human LRRK2 patients. Alternatively, the expression level of this mutant allele may not reach the pathology threshold in the fly. A recent report shows that overexpression of mouse LRRK2 with a mutation corresponding to the R1441C mutation in human LRRK2 results in biochemical features similar to those of human LRRK2-R1441C; whether these mice display motor dysfunction and DA neuronal loss has not been reported (30). Introduction of the human LRRK2 protein kinase domain fragment into adult rat substantia nigra, via adenoassociated virus-2-mediated gene transduction, has been reported to cause of degeneration of DA neurons (5).
To our knowledge, our study is the first report of an animal gain-of-function model expressing full-length human LRRK2. The limitations of our study are that we have used only one mutation (albeit the most common) and that LRRK2 proteins may be overexpressed relative to the endogenous fly LRRK2 homolog. However, flies with lower expression had a phenotype similar to those with higher expression, just less severe. The findings in this study need to be extended to vertebrate animals and compared with human patients. It is noteworthy that the phenotype of the LRRK2 flies recapitulates several key features of the human disorder and can be improved by l-DOPA, thereby representing a valuable genetic model for pathogenesis study of LRRK2-linked parkinsonism. This model may be useful to screen for LRRK2 interactors and to search for LRRK2 substrates. No effective treatments are as yet available to prevent the progressive death of DA neurons in PD. The Drosophila model of mutant α-synuclein has unveiled a gain-of-function mechanism for mutant α-synuclein-linked PD and provided a model for testing neuroprotective strategies for α-synuclein-mediated toxicity (18, 31). Similarly, the LRRK2 flies may provide a useful in vivo model for therapeutic screens to prevent neuronal loss and to rescue locomotor dysfunction in PD.
Methods
Generation of Human LRRK2 Transgenic Flies.
To generated UAS-LRRK2 and UAS-LRRK2-G2019S transgenic flies, the genes encoding these human proteins with N-terminal Flag tags were excised from pcDNA3.1 vectors, cloned between the XhoI site of pUAST vector (20), and verified by sequencing. The resulting constructs were microinjected in w1118 fly embryos (Rainbow Transgenic Flies). We obtained two transgenic lines each of UAS-LRRK2 and UAS-LRRK2-G2019S. The LRRK2 expression levels were examined by anti-Flag Western blot analysis. We used elav-GAL4, ddc-GAL4 (32), and GMR-GAL4 (33) to express UAS-LRRK2 and UAS-LRRK2-G2019S in all neurons, DA neurons, and photoreceptor cells, respectively. We selected two representative wild-type (LRRK2-1 and LRRK2–4) and mutant (G2019S-2 and G2019S-3) lines to conduct the phenotype characterization. Drosophila were grown on standard cornmeal medium at 25°C.
Western Blot Analysis.
Adult fly heads were homogenized at 4°C in buffer A (50 mM Tris·HCl, pH 7.5/1 mM EGTA/0.5 M NaCl/1% Triton X-100/1 mM DTT with protease inhibitors) and extracted as described (33). The resulting homogenates were subjected to Bradford protein assays to ensure equal protein loading and resolved on 4–12% SDS/NuPAGE Bis-Tris gels and transferred onto PVDF membranes (Invitrogen). The membranes were blocked in TBST (pH 7.4, 10 mM Tris·HCl/150 mM NaCl/0.1% Tween 20) containing 5% nonfat milk and then probed with anti-Flag antibody (Sigma). Proteins were detected by using enhanced chemiluminescence reagents (NEN).
Optical Neutralization Technique.
Adult fly heads were mounted on microscope slides using clear nail varnish and observed under a light microscope (33). To obtain a semiquantitative and unbiased index of retinal degeneration, the investigator counting the number of visible rhabdomeres did not know the genotype of the samples. The mean number of rhabdomeres per ommatidium was calculated. Rhabdomeres were counted by using cohorts of six flies of each genotype weekly during the lifespan of flies.
Electron Microscopy.
Fly heads were hemisected under a dim red photographic safety light, fixed (2% paraformaldehyde/2% glutaraldehyde in 0.1 M sodium cacodylate/3 mM calcium chloride, pH 7.2) at 4°C overnight, and postfixed in reduced osmium tetroxide for 1 h. Samples were stained en bloc with 2% uranyl acetate (filtered) and dehydrated through a graded series of ethanol. Eyes were oriented in gelatin capsules (size 00) and cured at 50°C for 24 h. Blocks were sectioned on a Riechert Ultracut E with a low-compression Diatome Diamond knife. Eighty-nanometer sections were picked up on copper slot grids and stained with uranyl acetate followed by lead citrate. Grids were viewed on a Hitachi 7600 TEM operating at 80 kV, and digital images were captured with an AMT 1 K × 1 K CCD camera as described (33).
Survival Curve.
Cohorts of 50 flies from each genotype were monitored for survival. Flies were maintained on standard media, and fresh food media were changed every 3–4 days. Mortality was scored daily and analyzed by using Kaplan–Meier survival curves. This experiment was repeated once.
Climbing Assay.
We determined locomotor ability using a climbing assay (negative geotaxis assay) as described previously (23). Cohorts of 60 flies from each genotype were subjected to the assay weekly from 1 week to the time of death. The tested flies were age-matched, randomly selected, anesthetized, and placed in a vertical plastic column (length, 25 cm; diameter, 1.5 cm). After a 30-min recovery from CO2 exposure, flies were gently tapped to the bottom of the column. We counted and calculated the percentage of flies that could climb to or above the median line of the cylinder in 10 seconds. Each week, the assay was repeated three times.
Actometer Test.
Cohorts of 20 flies from each genotype were subjected to the actometer assay at 5 weeks of age. A single fly was placed in a small tube with food at one end and was monitored for 3 days under standard conditions of 12-h light and 12-h darkness intervals. Activity was recorded on the computer every time the fly crossed an infrared beam (locomotion actograms), and the data were grouped into 30-min bins as described (34).
Real-Time RT-PCR.
To determine the expression of CG5483 and human LRRK2 at the mRNA level, primers were designed targeting CG5483 (5′-CGGCCTATTTAAACGCCACAGCAA-3′ and 5′-AACTGAAGTGTTGCGCGAAGAACC-3′) and human LRRK2 (5′-ATTGCGAACCTGGATGTCTCTCGT-3′ and 5′-TCAGGCACGAAGCTCAGCTGATTA-3′), respectively. The semiquantitative real-time RT-PCR was performed by using Stratagene Mx3000P PCR motion and Brilliant II QRT-PCR Master Mix kit according to the manufacturer's protocol.
Immunoprecipitation (IP) and in Vitro Autophosphorylation (Kinase) Assays.
IP experiments from fly head homogenates were performed with anti-FLAG-agarose (Sigma). Precipitates were washed twice with lysis buffer and twice with kinase assay buffer (Cell Signaling Technology). A kinase activity assay was described previously using autophosphorylation because the authentic substrate(s) is not yet known (12). Briefly, kinase reactions were carried out for 90 min at 30°C in 40 μl of kinase assay buffer with the addition of 15 μl of solution containing 50 mM MgCl2, 500 μM ATP, and 10 μCi of [γ-32P]ATP (3,000 Ci/mmol). Reactions were stopped by the addition of Laemmli sample buffer and boiling for 5 min. Samples were separated on 4–12% SDS/PAGE and blotted onto PVDF membranes. Quantification was performed with a phosphoimager (Bio-Rad Molecular Imager FX).
Immunostaining and Cell Counting.
Fluorescent immunostaining was performed on whole-mount dissected adult brain (23, 35) at 1, 21, 35, and 49 weeks of age. Cohorts of six to eight flies per genotype were used at each time point for immunostaining. The dissected brains were mounted in Vectashield (Vector Laboratories).
Rabbit polyclonal anti-TH (Chemicon), mouse monoclonal anti-TH (Immunostar), anti-5-HT (Sigma), anti-elav (Developmental Studies Hybridoma Bank), and anti-Flag-antibodies were used as the primary antibodies. Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-mouse IgG (Invitrogen) were used as secondary antibodies. The numbers of DA and 5-HT neurons were scored in whole-mount brains under fluorescent (Zeiss LSM 250) and/or confocal microscopy (Zeiss LSM 510). For quantification of loss of anti-elav staining in brain, entire brain sections were digitized with an image analysis system. NIH Image J software was used to measure the optical density of anti-elav staining within the entire brain section (six brain sections per experimental group).
Data Analysis.
Quantitative data were expressed as arithmetic means ± SEM based on at least three separate experiments. Statistically significant differences between two groups were analyzed by ANOVA. A P value <0.05 was considered significant.
ACKNOWLEDGMENTS.
This research was funded by the National Institute of Neurological Disorders and Stroke, the National Institutes of Health, the National Parkinson's Foundation, and the American Parkinson's Disease Association (W.W.S.) and by National Eye Institute Grant EY08117 (to C.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Ross OA, Toft M, Whittle AJ, Johnson JL, Papapetropoulos S, Mash DC, Litvan I, Gordon MF, Wszolek ZK, Farrer MJ, et al. Ann Neurol. 2006;59:388–393. doi: 10.1002/ana.20731. [DOI] [PubMed] [Google Scholar]
- 4.Rajput A, Dickson DW, Robinson CA, Ross OA, Dachsel JC, Lincoln SJ, Cobb SA, Rajput ML, Farrer MJ. Neurology. 2006;67:1506–1508. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- 5.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Cookson MR, Dauer W, Dawson T, Fon EA, Guo M, Shen J. J Neurosci. 2007;27:11865–11868. doi: 10.1523/JNEUROSCI.3695-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, et al. Lancet. 2005;365:410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 8.Di Fonzo A, Rohe CF, Ferreira J, Chien HF, Vacca L, Stocchi F, Guedes L, Fabrizio E, Manfredi M, Vanacore N, et al. Lancet. 2005;365:412–415. doi: 10.1016/S0140-6736(05)17829-5. [DOI] [PubMed] [Google Scholar]
- 9.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, et al. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 10.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 13.Luzon-Toro B, de la Torre ER, Delgado A, Perez-Tur J, Hilfiker S. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- 14.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. Hum Mol Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 16.Cauchi RJ, van den Heuvel M. Neurodegener Dis. 2006;3:338–356. doi: 10.1159/000097303. [DOI] [PubMed] [Google Scholar]
- 17.Marsh JL, Thompson LM. Neuron. 2006;52:169–178. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Feany MB, Bender WW. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 19.Lee SB, Kim W, Lee S, Chung J. Biochem Biophys Res Commun. 2007;358:534–539. doi: 10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Haass C, Kahle PJ. Nature. 2000;404:341 doi: 10.1038/35006188. [DOI] [PubMed] [Google Scholar]
- 22.Budnik V, White K. J Comp Neurol. 1988;268:400–413. doi: 10.1002/cne.902680309. [DOI] [PubMed] [Google Scholar]
- 23.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 24.Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Ann Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- 25.Dauer W, Przedborski S. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 26.Forno LS. J Neuropathol Exp Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Mouradian MM. Neurology. 2002;58:179–185. doi: 10.1212/wnl.58.2.179. [DOI] [PubMed] [Google Scholar]
- 28.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 29.Ishihara L, Warren L, Gibson R, Amouri R, Lesage S, Durr A, Tazir M, Wszolek ZK, Uitti RJ, Nichols WC, et al. Arch Neurol. 2006;63:1250–1254. doi: 10.1001/archneur.63.9.1250. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. J Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auluck PK, Meulener MC, Bonini NM. J Biol Chem. 2005;280:2873–2878. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Chaney S, Roberts IJ, Forte M, Hirsh J. Curr Biol. 2000;10:211–214. doi: 10.1016/s0960-9822(00)00340-7. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, Montell C. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothenfluh A, Abodeely M, Price JL, Young MW. Genetics. 2000;156:665–675. doi: 10.1093/genetics/156.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhuri A, Bowling K, Funderburk C, Lawal H, Inamdar A, Wang Z, O'Donnell JM. J Neurosci. 2007;27:2457–2467. doi: 10.1523/JNEUROSCI.4239-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monastirioti M. Microsc Res Tech. 1999;45:106–121. doi: 10.1002/(SICI)1097-0029(19990415)45:2<106::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]