Abstract
Recombinant antigens of Ureaplasma parvum serotypes 3 and 6 were produced in order to develop a serological assay for Ureaplasma antibody detection. The genes of the multiple banded antigen (MBA) were amplified by PCR and cloned in a pTrcHis TOPO plasmid. Purified recombinant proteins were evaluated in Western blotting and enzyme-linked immunosorbent assay (ELISA) with monoclonal antibodies and human sera. Our approach was successful in the production of the recombinant MBAs (rMBAs) for serotypes 3 and 6. The antigens tested positive with serotype-specific monoclonal antibodies in Western blotting and in ELISA. Prominent reactions were detected with the rMBAs and their homologous monoclonal antibodies. Strong cross-reactions were visible in ELISA between rMBA 3 and the monoclonal antibodies from the other U. parvum serotypes. A weak cross-reaction was seen with rMBA 3 and the monoclonal antibody from serotype 4. rMBA 6 showed cross-reaction only with the monoclonal antibody from U. parvum serotype 1. Fifty-one percent of the sera obtained from culture-positive women reacted with one or both rMBAs. Only three (15%) of the sera from culture-negative women reacted with the rMBA. The positive reactions were observed only with rMBA 6. These preliminary tests showed the potential usefulness of the rMBAs produced for detecting an antibody response against Ureaplasma antigens.
Ureaplasma spp. are present in 40 to 80% of sexually mature men and women as a commensal in the lower genital tract. In many cases, the microorganism is not pathogenic. However, it has been shown that in some cases Ureaplasma infection can cause adverse pregnancy outcomes (1, 2, 12, 15, 16, 26, 38, 42).
Ureaplasma consists of two species, U. parvum (serotypes 1, 3, 6, and 14) and U. urealyticum (serotypes 2, 4, 5, and 7 to 13) (20, 35).
No conclusive answer has been found to the question of whether pathogenicity is serotype specific (6, 7, 25, 27, 33) or whether other factors are responsible for the development of disease. It is likely that adverse pregnancy outcome is the consequence of an ascending infection originating from the lower genital tract. The reason why in some patients Ureaplasma spp. cause an ascending infection is not yet known, but it is likely that the etiology is multifactorial and the patient's immunity, the type of strain, and antigen variation may play roles in the disease progression.
Study of the antibody responses in different patient populations might be helpful for further research on the pathogenicity of these microorganisms. In this study, we evaluated whether recombinant antigens of Ureaplasma spp. could be suitable for use in a serological assay. For this purpose, the multiple banded antigen (MBA) of Ureaplasma spp. was chosen, since it is present in all serotypes of Ureaplasma (37) and it has an important role in the immune response (40). Moreover, the MBA contains serotype-specific, as well as non-serotype-specific, epitopes, which could be an advantage in the development of a serotype-specific assay (39, 40). U. parvum serotypes 3 and 6 were selected for the production of recombinant MBAs (rMBAs) because they are the most frequently isolated serotypes (2, 11, 18, 21, 25, 41). Since the repeat sequence from the MBA is the most important epitope for antibodies (40), primers flanking the repeat sequence of the MBA were chosen for PCR amplification.
MATERIALS AND METHODS
Production of the MBA gene.
U. parvum serotype 3 and 6 reference strains (strain designations, 27 and Pi, respectively) were supplied by E. A. Freund (Institute of Medical Microbiology, University of Aarhus, Aarhus, Denmark). The strains were stored at −80°C until they were used. After the strains were cultured on differential agar medium A7 (36), one colony was isolated from the agar medium and grown in 10 ml bromothymol blue broth (32). After centrifugation (25,000 × g; 30 min; 4°C), the cell pellet was resuspended in 100 μl sterile phosphate-buffered saline (PBS), pH 7.3.
After DNA extraction, the selected region of the MBA gene from serotype 3 was amplified by PCR using a primer pair flanking the repeat region of the MBA gene (UMSP88/UMA1586) (Table 1). The region of the MBA gene from serotype 6 was amplified by nested PCR using primer pairs flanking the repeat region of the MBA gene (outer primer pair, UMS-125/UMA1586; inner primer pair, UMSP88/UMAUA) (Table 1).
TABLE 1.
Oligonucleotide primers used in this studya
| Primer | Purpose | Localization | Tm (°C) | Nucleotide sequence (5′→3′) | Reference or source |
|---|---|---|---|---|---|
| UMS-125 | MBA amplification | MBA 5′ | 66 | GTATTTGCAATCTTTATATGTTTTCG | 43 |
| UMA1586 | MBA amplification | MBA 3′ | 72 | GATAATCATTCATCTTCTCTTAATTGTC | 43 |
| UMSP88 | MBA amplification, control positive clones, sequencing | MBA 5′ | 55 | TGTTCTAATTCAACTGTTAAATCT | Present study |
| UMAUA | MBA amplification, control positive clones, sequencing | MBA 3′ | 62 | GGGKWGTTKHACCAYTKCCTGGTT | 19 |
| pTrcHis Forward | Control positive clones, sequencing | Cloning vector | 53 | GAGGTATATATTAATGTATCG | Invitrogen |
| pTrcHis Reverse | Control positive clones, sequencing | Cloning vector | 48 | GATTTAATCTGTATCAGG | Invitrogen |
Oligonucleotide primers were synthesized by Eurogentec (Luik, Belgium).
PCRs were all performed in 50-μl reaction mixtures containing 10 mM Tris HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 1 μM primers, 0.025 U/μl Taq polymerase, 5 μl DNA for single and outer PCRs, and 2 μl of the outer reaction mixture for the inner PCR. The PCR cycles were performed in an iCycler thermal cycler (Bio-Rad, Nazareth-Eke, Belgium).
A 10-min denaturation step at 94°C was followed by 35 cycles of 30 seconds at 94°C, 30 seconds at a primer-specific annealing temperature (53°C for primer pairs UMSP88/UMA1586 and UMSP88/UMAUA and 50°C for primer pair UMS-125/UMA1586), and 2 min at 72°C, with a final elongation step at 72°C for 10 min.
The PCR fragments obtained were purified and concentrated using a QIAquick PCR purification kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's instructions. The PCR products obtained were sequenced to check for the presence of the repeats in the DNA fragments. The sequencing reactions were performed by the Flanders Institute for Biotechnology genetic service facility, Antwerp, Belgium. Sequencing data were processed with Kodon total genome and sequence analysis software, version 2.0 (Applied Maths, Sint-Martens-Latem, Belgium). The primers used for sequencing were UMSP88 and UMAUA/UMA1586 (Table 1).
Cloning and expression of the MBA gene.
Cloning and expression of the MBA gene were performed using a pTrcHis TOPO TA expression kit (Invitrogen, Life Technologies, Paiseley, United Kingdom) according to the manufacturer's instructions. The pTrc His-TOPO vector with the partial MBA gene was transformed in One Shot TOP10 competent Escherichia coli cells. Ten positive transformants were picked for further testing, allowing us to select transformants with the inserts in the correct orientation of the open reading frame. Therefore, PCR was performed using combinations of pTrcHis Forward primer and pTrcHis Reverse primer, recognizing the sequence surrounding the insertion site of the plasmid, and UMSP88 and UMAUA/UMA1586, recognizing the MBA gene. Primer data are shown in Table 1. PCR mixtures and PCR cycles were performed as described above using an annealing temperature of 53°C. Positive transformants were grown in 2 ml LB medium (3) and stored at −80°C in glycerol.
After isopropyl β-d-thiogalactoside (IPTG)-induced protein expression in LB-medium, the E. coli cells were lysed by adding guanidinium-HCl buffer, pH 7.8, followed by a 10-minute sonification. A six-histidine tag was added to the proteins by the vector for further purification and testing in Western blotting. After centrifugation (3,800 × g; 15 min; room temperature), the supernatant was stored at −20°C until purification.
Sequencing of the positive clones was performed by the Flanders Institute for Biotechnology genetic service facility, Antwerp, Belgium. The sequencing results of the positive clones were aligned with the GenBank sequences of the MBAs from U. parvum serotypes 3 (AF222894) (14) and 6 (AF056984) (22). Sequencing analysis and sequence alignments were processed with Kodon total genome and sequence analysis software, version 2.0 (Applied Maths, Sint-Martens-Latem, Belgium). The primers used for sequencing were pTrcHis Forward and pTrcHis Reverse; primer data are shown in Table 1.
Purification of the rMBAs.
Purification of the rMBAs was performed using ion metal affinity chromatography. Purification was performed with Ni Sepharose 6 Fast Flow (GE Healthcare, Brussels, Belgium). Briefly, a sample was added to the column and incubated for at least 30 min; after centrifugation (1,600 × g; 5 min; room temperature), the supernatant was removed. After three wash steps with phosphate buffer containing 20 mM imidazole, pH 7.4, for 5 minutes each time, the proteins were eluted using phosphate buffer containing 500 mM imidazole, pH 7.2. The purified rMBAs were evaluated in Western blotting with an anti-histidine antibody conjugated with horseradish peroxidase (Sigma, Belgium). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed as previously described (5).
Evaluation of the rMBA with MAbs.
The purified rMBAs were evaluated in Western blotting and enzyme-linked immunosorbent assay (ELISA) with a complete set of serotype-specific monoclonal antibodies (MAbs) available in the laboratory (9, 10, 24). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were performed as previously described (5).
ELISA was performed by coating a Nunc-Immuno Polysorp Plate with 100 μl rMBA using Na2CO3/NaHCO3 as a coating buffer. Optimal rMBA concentrations were determined by checkerboard titration (2.5 μg/ml and 1.25 μg/ml for serotypes 3 and 6, respectively). After 2 h of incubation at 37°C, the wells were washed twice with 0.1% PBS-Tween 20 (PBST) and blocked with bovine serum albumin-PBS (3% bovine serum albumin) at 37°C for 1 h. After another two wash steps, 100 μl of diluted MAb was added to the wells and incubated for 1 hour at 37°C. Optimal MAb dilutions were determined by checkerboard titration (1/800 for MAb-3 and 1/3,200 for MAb-6). Three wash steps followed the incubation, and 100 μl horseradish peroxidase-labeled anti-mouse antibody (1/1,000 in 0.05% PBST) was incubated in the wells for 1 h at 37°C. The wells were washed four times before substrate was added (orthophenylenediamine-2HCl), and the mixture was incubated for 30 min at room temperature. The reaction was stopped with 50 μl 2 M H2SO4, and the optical density (OD) was read at 492 nm.
Evaluation of the rMBAs with human sera.
In order to evaluate the potential usefulness of the rMBAs in the detection of Ureaplasma antibodies, a preliminary ELISA was elaborated and tested with 20 sera obtained from women in whom the cervical culture was negative for Ureaplasma spp. and with 51 sera from women with a positive cervical culture. The genital tract specimens were obtained from healthy pregnant women at the first prenatal consultation in the Universitair Ziekenhuis Brussels, and none of the women had signs or symptoms of genital tract infection. Cervical swabs were transported in Amies transport medium supplemented with charcoal (International Medical, Brussels, Belgium). The cultures were performed as previously described (26) and were inoculated on differential agar medium A7 and in bromothymol blue broth.
After all runs were performed, the initial cutoff value for positivity was calculated as the mean OD plus 2 standard deviations (SD) obtained in the sera from culture-negative women. If this initial value gave rise to a positive result in one of the sera from a culture-negative woman, the initial cutoff value was recalculated after excluding the value of this serum for cutoff calculation, and a new “adjusted” cutoff value was calculated. The ELISA was performed as described above using rMBA dilutions of 1/1,600 for rMBA 3 and 1/800 for rMBA 6. Human sera and anti-human immunoglobulin G-peroxidase conjugate were diluted 1/1,000 in 0.05% PBST.
Nucleotide sequence accession number.
The obtained consensus sequence of the clone 6G1 was submitted to the GenBank database (http://www.ncbi.nlm.nih.gov) under accession number EU095525. With these sequences, we could partially supply the lacking sequence of the 3′ end of the MBA gene serotype 6.
RESULTS
Cloning and expression of the MBA gene.
DNA extraction of U. parvum serotypes 3 and 6 was performed and used for PCR as described in Materials and Methods. After nested PCR, a DNA fragment from the MBA of ca. 400 bp was obtained for serotype 6. For serotype 3, a PCR product of 1,500 bp was obtained. Sequencing of the PCR products showed the presence of 15 repeat fragments from U. parvum serotype 6 and 36 for U. parvum serotype 3. The PCR products from serotypes 3 and 6 were cloned successfully. Two positive transformants were obtained for U. parvum serotype 3 (3C4 and 3C9) and three for serotype 6 (6G1, 6G2, and 6G3).
Sequence alignment between sequencing results from the clones and sequences published in GenBank for U. parvum serotypes 3 and 6 showed 100% identity. In Western blotting, rMBAs 3 and 6 showed a positive reaction with the anti-histidine antibody at the expected molecular mass according to the PCR product length (60 and 35 kDa, respectively).
Evaluation of the rMBAs with MAbs.
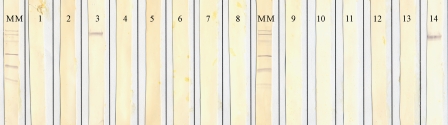
In Western blotting, rMBAs 3 and 6 showed reactivity with their homologous MAbs. In addition, rMBA 3 showed reactivity with the MAb directed to serotype 14 (Fig. 1 and 2).
FIG. 1.
Western blot showing the reaction patterns of rMBA 3 with all the serotype-specific MAbs. MM, molecular mass ladder (97.4, 66.2, 45, 31, 21.5, and 14.4 kDa). Lanes 1 to 14 contain rMBA 3 reactions with all 14 serotype-specific MAbs of Ureaplasma spp.
FIG. 2.
Western blot showing the reaction patterns of rMBA 6 with all the serotype-specific MAbs. MM, molecular mass ladder (97.4, 66.2, 45, 31, 21.5, and 14.4 kDa). Lanes 1 to 14 contain rMBA 6 reactions with all 14 serotype-specific MAbs of Ureaplasma spp.
In ELISA, rMBA 3 reacted strongly with its serotype-specific MAb. In addition to this reaction, relatively strong reactions were also observed with MAbs directed against the other U. parvum serotypes, 1, 6, and 14. A weaker reaction was seen with the MAb directed against serotype 4 (data not shown). rMBA 6 reacted strongly with its homologous MAb and with the MAb against U. parvum serotype 1.
Evaluation of the rMBAs with human sera.
The mean OD after background subtraction obtained in serum samples from culture-negative women was 0.138 (range, −0.007 to 0.355; SD, 0.111) for rMBA 3 and 0.297 (range, 0.034 to 0.943; SD, 0.221) for rMBA 6. Cutoff values were calculated at 0.360 for rMBA 3 and 0.739 for rMBA 6. None of the 20 serum samples from culture-negative women showed a positive reaction in ELISA with rMBA 3. However, two of these serum samples showed OD values above the initial cutoff value in ELISA with rMBA 6. After elimination of these values in the cutoff calculation, the adjusted cutoff value for rMBA 6 was set at 0.480. This adjusted cutoff gave rise to three positive serum samples (15%) among the culture-negative women with rMBA 6. From the 51 serum samples obtained from culture-positive patients, 26 (51%) were positive with at least one of the two rMBAs (3 or 6): 19 in ELISA using rMBA 3 and 19 using rMBA 6 as the antigen. Twelve sera reacted with both antigens (Table 2).
TABLE 2.
Summary of ELISA results for serum samples from culture-positive and culture-negative women
| ELISA result | No. (%) of patients positive
|
|
|---|---|---|
| Culture-positive patients (51 samples) | Culture-negative patients (20 samples) | |
| rMBA3 + | 19 (37.3) | 0 (0) |
| rMBA6 + | 19 (37.3) | 3 (15) |
| rMBA3 or rMBA6 + | 26 (51.0) | 3 (15) |
DISCUSSION
The high colonization rate of Ureaplasma has elicited a long discussion about the pathogenicity of the organism in reproduction pathology. However, well-defined case reports and studies have shown that Ureaplasma can interfere with fetal development. A variety of serological techniques have been used to detect the antibody response against Ureaplasma spp. (8, 13, 17, 28-31, 31, 34). However, these tests have a number of disadvantages. Immunoblotting cannot quantify results, the metabolic inhibition test is complex and requires large quantities of serum samples, and the reading of immunofluorescence assays may be subject to interpretation problems.
Previously described ELISAs with nonpurified antigens showed promising results for Ureaplasma antibody detection with human sera. However, the use of nonpurified antigens could hamper the detection of serotype-specific antibody responses (4, 23). The use of highly purified serotype-specific antigens offers the advantage of working according to standardized methods. Pure recombinant antigens are stable and can be produced in large quantities. Whole-cell antigenic preparations are often contaminated with medium components and show wide variation in antigenic content.
For amplification of the MBA gene for serotypes 3 and 6, forward primer UMSP88 was designed in the laboratory. Due to the unknown 3′ sequence of the MBA gene from U. parvum serotype 6, primer design in this region was not possible. The reverse primer UMA1586, also used for serotype 3 MBA amplification, was used for serotype 6 in the outer PCR. The reverse primer UMAUA, previously described for U. parvum serotype 3, was used for the inner PCR for serotype 6 MBA amplification. The primers used for U. parvum serotypes 3 and 6 yielded satisfactory results.
We were successful in the production of rMBAs for serotypes 3 and 6. Evaluation of these rMBAs in ELISA and in Western blotting showed reactivity with their homologous antibodies and with some heterologous MAbs. All but one of the cross-reactions were observed with MAbs of the U. parvum species. For some MAbs, different cross-reactivity patterns were observed according to the technique used. Cross-reactivity was more frequent when the rMBAs were tested in ELISA than when they were tested in Western blotting. Such differences in cross-reactivity between different techniques have been described (9, 24). This is probably due to differences in the native forms of the proteins used in ELISA, whereas in Western blotting, the denatured structures of the proteins are used, resulting in a lower accessibility for the antibodies.
These rMBAs were further tested with human sera obtained from culture-negative and culture-positive women. Presuming that culture-negative women were not infected and would show no seroreactivity, the initial cutoff value was calculated at the mean OD (plus 2 SD) values obtained in culture-negative women. However, since the antibody response might still be detected after elimination of the microorganism and since culture might have failed to detect low numbers of microorganisms in some women, we adjusted the cutoff after excluding the OD values of initially reactive samples obtained in culture-negative women. This adjustment of the cutoff value was necessary only for rMBA 6.
After defining the cutoff value for positivity, we found that 51% of the sera obtained from culture-positive women reacted with one or both of the rMBAs. Considering that the serological test was performed with antigens specific for serotypes 3 and 6 and that women colonized with other serotypes might not be reactive, the positivity rate obtained in culture-positive women is promising. Positive reactions in sera from culture-negative women were observed only when using rMBA 6: 15% of the sera reacted positively with rMBA 6. These positive reactions may be the consequence of a persistent antibody response in a previously colonized patient, a cross-reaction with some epitopes present in the rMBA, or a false-negative culture result.
Further research, however, is necessary before the serological test will be clinically useful. Since it is not yet known whether serotype-specific antibody responses or global antibody responses will be clinically relevant, more rMBAs of other serotypes will be produced and their usefulness as single antigenic preparations or in combination with other antigens will be evaluated in a large clinical setting. In addition, the kinetics of the antibody response should be evaluated, and their possible use in distinguishing between colonization and infection must be investigated.
In conclusion, recombinant antigens of the MBAs of Ureaplasma spp. were successfully produced. Preliminary tests have indicated their potential usefulness as antigenic preparations in ELISA.
Acknowledgments
This work was supported by a grant of the Steunfonds Marguerite-Marie Delacroix.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Abele-Horn, M., J. Peters, O. Genzel-Boroviczeny, C. Wolff, A. Zimmermann, and W. Gottschling. 1997. Vaginal Ureaplasma urealyticum colonization: influence on pregnancy outcome and neonatal morbidity. Infection 25:286-291. [DOI] [PubMed] [Google Scholar]
- 2.Abele-Horn, M., C. Wolff, P. Dressel, F. Pfaff, and A. Zimmermann. 1997. Association of Ureaplasma urealyticum biovars with clinical outcome for neonates, obstetric patients, and gynecological patients with pelvic inflammatory disease. J. Clin. Microbiol. 35:1199-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. B., G. H. Cassell, D. Taylor-Robinson, and M. C. Shepard. 1983. Measurement of antibody to Ureaplasma urealyticum by an enzyme-linked immunosorbent assay and detection of antibody responses in patients with nongonococcal urethritis. J. Clin. Microbiol. 17:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, X., A. Naessens, and S. Lauwers. 1993. Identification and characterization of serotype 4-specific antigens of Ureaplasma urealyticum by use of monoclonal antibodies. Infect. Immun. 61:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cracea, E., D. Botez, S. Constantinescu, and M. Georgescu-Braila. 1982. Ureaplasma urealyticum serotypes isolated from cases of female sterility. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 252:535-539. [PubMed] [Google Scholar]
- 7.Cracea, E., S. Constantinescu, and M. Lazar. 1985. Serotypes of Ureaplasma urealyticum isolated from patients with nongonococcal urethritis and gonorrhea and from asymptomatic urethral carriers. Sex. Transm. Dis. 12:219-223. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, C. K., C. A. Bonville, J. H. Hagen, J. L. Belkowitz, R. M. Kawatu, A. M. Higgins, and L. B. Weiner. 1996. Immunoblot analysis of anti-Ureaplasma urealyticum antibody in pregnant women and newborn infants. Clin. Diagn. Lab. Immunol. 3:487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echahidi, F., G. Muyldermans, S. Lauwers, and A. Naessens. 2000. Development of monoclonal antibodies against Ureaplasma urealyticum serotypes and their use for serotyping clinical isolates. Clin. Diagn. Lab. Immunol. 7:563-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echahidi, F., G. Muyldermans, S. Lauwers, and A. Naessens. 2001. Development of an enzyme-linked immunosorbent assay for serotyping Ureaplasma urealyticum strains using monoclonal antibodies. Clin. Diagn. Lab. Immunol. 8:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez, M. C., M. A. Latino, M. Y. Zamora, M. Pellecchia, V. Neve, R. Llanes, R. Macfarlane, and L. Balbo. 2003. Development of a multiple PCR method for the identification of Ureaplasma parvum and Ureaplasma urealyticum. Rev. Argent. Microbiol. 35:138-142. [PubMed] [Google Scholar]
- 12.Foulon, W., D. Van Liedekerke, C. Demanet, L. Decatte, M. Dewaele, and A. Naessens. 1995. Markers of infection and their relationship to preterm delivery. Am. J. Perinatol. 12:208-211. [DOI] [PubMed] [Google Scholar]
- 13.Gallo, D., K. W. Dupuis, N. J. Schmidt, and G. E. Kenny. 1983. Broadly reactive immunofluorescence test for measurement of immunoglobulin M and G antibodies to Ureaplasma urealyticum in infant and adult sera. J. Clin. Microbiol. 17:614-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 15.Grattard, F., B. Soleihac, B. De Barbeyrac, C. Bebear, P. Seffert, and B. Pozzetto. 1995. Epidemiologic and molecular investigations of genital mycoplasmas from women and neonates at delivery. Pediatr. Infect. Dis. J. 14:853-858. [DOI] [PubMed] [Google Scholar]
- 16.Gray, D. J., H. B. Robinson, J. Malone, and R. B. Thomson, Jr. 1992. Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat. Diagn. 12:111-117. [DOI] [PubMed] [Google Scholar]
- 17.Kenny, G. E., and F. D. Cartwright. 1984. Immunoblotting for determination of the antigenic specificities of antibodies to the Mycoplasmatales. Isr. J. Med. Sci. 20:908-911. [PubMed] [Google Scholar]
- 18.Kim, M., G. Kim, R. Romero, S. S. Shim, E. C. Kim, and B. H. Yoon. 2003. Biovar diversity of Ureaplasma urealyticum in amniotic fluid: distribution, intrauterine inflammatory response and pregnancy outcomes. J. Perinat. Med. 31:146-152. [DOI] [PubMed] [Google Scholar]
- 19.Kong, F., et al. 2000. Molecular genotyping of human Ureaplsama species based on multiple-banded antigen (MBA) gene sequences. Int. J. Syst. Evol. Microbiol. 50:1921-1929. [DOI] [PubMed] [Google Scholar]
- 20.Kong, F., G. James, Z. Ma, S. Gordon, W. Bin, and G. L. Gilbert. 1999. Phylogenetic analysis of Ureaplasma urealyticum—support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 49:1879-1889. [DOI] [PubMed] [Google Scholar]
- 21.Kong, F., Z. Ma, G. James, S. Gordon, and G. L. Gilbert. 2000. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J. Clin. Microbiol. 38:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong, F., X. Zhu, W. Wang, X. Zhou, S. Gordon, and G. L. Gilbert. 1999. Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1. J. Clin. Microbiol. 37:538-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liepmann, M. F., P. Wattre, A. Dewilde, G. Papierok, and M. Delecour. 1988. Detection of antibodies to Ureaplasma urealyticum in pregnant women by enzyme-linked immunosorbent assay using membrane antigen and investigation of the significance of the antibodies. J. Clin. Microbiol. 26:2157-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naessens, A., X. Cheng, S. Lauwers, and J. A. Robertson. 1998. Development of a monoclonal antibody to a Ureaplasma urealyticum serotype 9 antigen. J. Clin. Microbiol. 36:1125-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naessens, A., W. Foulon, J. Breynaert, and S. Lauwers. 1988. Serotypes of Ureaplasma urealyticum isolated from normal pregnant women and patients with pregnancy complications. J. Clin. Microbiol. 26:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naessens, A., W. Foulon, H. Cammu, A. Goossens, and S. Lauwers. 1987. Epidemiology and pathogenesis of Ureaplasma urealyticum in spontaneous abortion and early preterm labor. Acta Obstet. Gynecol. Scand. 66:513-516. [DOI] [PubMed] [Google Scholar]
- 27.Piot, P. 1976. Distribution of eight serotypes of Ureaplasma urealyticum in cases of non-gonococcal urethritis and of gonorrhoea, and in healthy persons. Br. J. Vener. Dis. 52:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn, P. A. 1986. Evidence of an immune response to Ureaplasma urealyticum in perinatal morbidity and mortality. Pediatr. Infect. Dis. 5:S282-S287. [DOI] [PubMed] [Google Scholar]
- 29.Quinn, P. A., H. C. Li, C. Th'ng, M. Dunn, and J. Butany. 1993. Serological response to Ureaplasma urealyticum in the neonate. Clin. Infect. Dis. 17(Suppl. 1):S136-S143. [DOI] [PubMed] [Google Scholar]
- 30.Quinn, P. A., S. Rubin, D. M. Nocilla, S. E. Read, and M. Chipman. 1983. Serological evidence of Ureaplasma urealyticum infection in neonatal respiratory disease. Yale J. Biol. Med. 56:565-572. [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn, P. A., A. B. Shewchuk, J. Shuber, K. I. Lie, E. Ryan, M. Sheu, and M. L. Chipman. 1983. Serologic evidence of Ureaplasma urealyticum infection in women with spontaneous pregnancy loss. Am. J. Obstet. Gynecol. 145:245-250. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, J. A. 1978. Bromothymol blue broth: improved medium for detection of Ureaplasma urealyticum (T-strain mycoplasma). J. Clin. Microbiol. 7:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson, J. A., L. H. Honore, and G. W. Stemke. 1986. Serotypes of Ureaplasma urealyticum in spontaneous abortion. Pediatr. Infect. Dis. 5:S270-S272. [DOI] [PubMed] [Google Scholar]
- 34.Robertson, J. A., and G. W. Stemke. 1979. Modified metabolic inhibition test for serotyping strains of Ureaplasma urealyticum (T-strain mycoplasma). J. Clin. Microbiol. 9:673-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson, J. A., G. W. Stemke, J. W. Davis, Jr., R. Harasawa, D. Thirkell, F. Kong, M. C. Shepard, and D. K. Ford. 2002. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001. Int. J. Syst. Evol. Microbiol. 52:587-597. [DOI] [PubMed] [Google Scholar]
- 36.Shepard, M. C., and C. D. Lunceford. 1976. Differential agar medium (A7) for identification of Ureaplasma urealyticum (human T mycoplasmas) in primary cultures of clinical material. J. Clin. Microbiol. 3:613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng, L., X. Zheng, J. Glass, H. Watson, J. Tsai, and G. Cassell. 1994. Ureaplasma urealyticum biovar specificity and diversity are encoded in multiple banded antigen gene. J. Clin. Microbiol. 32:1464-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel, I., P. Thorsen, V. K. Hogan, L. A. Schieve, B. Jacobsson, and C. D. Ferre. 2006. The joint effect of vaginal Ureaplasma urealyticum and bacterial vaginosis on adverse pregnancy outcomes. Acta Obstet. Gynecol. Scand. 85:778-785. [DOI] [PubMed] [Google Scholar]
- 39.Watson, H. L., D. K. Blalock, and G. H. Cassell. 1990. Variable antigens of Ureaplasma urealyticum containing both serovar-specific and serovar-cross-reactive epitopes. Infect. Immun. 58:3679-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson, H. L., X. Zheng, and G. H. Cassell. 1993. Structural variations and phenotypic switching of mycoplasmal antigens. Clin. Infect. Dis. 17(Suppl. 1):S183-S186. [DOI] [PubMed] [Google Scholar]
- 41.Yi, J., B. H. Yoon, and E. C. Kim. 2005. Detection and biovar discrimination of Ureaplasma urealyticum by real-time PCR. Mol. Cell Probes. 19:255-260. [DOI] [PubMed] [Google Scholar]
- 42.Yoon, B. H., J. W. Chang, and R. Romero. 1998. Isolation of Ureaplasma urealyticum from the amniotic cavity and adverse outcome in preterm labor. Obstet. Gynecol. 92:77-82. [DOI] [PubMed] [Google Scholar]
- 43.Zheng, X., L. J. Teng, H. L. Watson, J. I. Glass, A. Blanchard, and G. H. Cassell. 1995. Small repeating units within the Ureaplasma urealyticum MB antigen gene encode serovar specificity and are associated with antigen size variation. Infect. Immun. 63:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]