Abstract
The stable incidence of new leprosy cases suggests that transmission of infection is continuing despite the worldwide implementation of multidrug therapy programs. Highly specific tools are required to accurately diagnose asymptomatic and early stage Mycobacterium leprae infections which are the likely sources of transmission and cannot be identified by using the detection of antibodies against phenolic glycolipid I. One of the hurdles hampering T-cell-based diagnostic tests is that M. leprae antigens cross-react at the T-cell level with antigens present in other mycobacteria, like M. tuberculosis or M. bovis bacillus Calmette-Guerin (BCG). Using comparative genomics, we previously identified five candidate proteins highly restricted to M. leprae which showed promising features with respect to application in leprosy diagnostics. However, despite the lack of overall sequence homology, the use of recombinant proteins includes the risk of detecting T-cell responses that are cross-reactive with other antigens. To improve the diagnostic potential of these M. leprae sequences, we used 50 synthetic peptides spanning the sequences of all five proteins for the induction of T-cell responses (gamma interferon) in leprosy patients, healthy household contacts (HHC) of leprosy patients, and healthy controls in Brazil, as well as in tuberculosis patients, BCG vaccinees, and healthy subjects from an area of nonendemicity. Using the combined T-cell responses toward four of these peptides, all paucibacillary patients and 13 out of 14 HHC were detected without compromising specificity. The peptides contain HLA binding motifs for various HLA class I and II alleles, thereby meeting an important requirement for the applicability of diagnostic tools in genetically diverse populations. Thus, this study provides the first evidence for the possibility of immunodiagnostics for leprosy based on mixtures of peptides recognized in the context of different HLA alleles.
Mycobacterium leprae is the etiologic agent of leprosy, an infectious, neurodegenerative human disease. Leprosy and its associated disabilities are unlikely to disappear soon, as recognized in the World Health Organization's (WHO's) Global Strategy 2006-2010 (8). Despite a spectacular decrease in global prevalence of leprosy since 1982 to 219,826 cases in 2005, the incidence appears not to have changed significantly, and in fact, leprosy is more common than expected (17), with 296,499 new cases in 2005 (158,728 multibacillary [MB], 90,122 females, 28,005 children, 13,361 cases of grade 2 disability) (http://www.who.int/wer) (2). The continued incidence of leprosy in countries where it is endemic is thought to result from the perpetuating reservoir of M. leprae-infected contacts or persons with subclinical leprosy. However, diagnostic tools for the early detection of asymptomatic M. leprae infection are not available yet. Assays have been developed that detect M. leprae-specific immunoglobulin M antibodies against phenolic glycolipid I (PGL-I) and which are able to identify MB leprosy patients (with strong humoral immunity to M. leprae), but these fail to detect most paucibacillary (PB) leprosy patients and leprosy patients' contacts, as these typically develop strong cellular, but not humoral, immunity. Furthermore, MB leprosy is hard to detect at an initial stage when the patient is already an active source of infection. Thus, to complement classical, clinical methods for the diagnosis of infection, improved diagnostic tools are required that detect M. leprae infection before clinical manifestations arise and distinguish M. leprae infection from infection with other mycobacteria, such as M. tuberculosis and environmental mycobacteria.
Recently, two commercially available gamma interferon (IFN-γ) release assays have been developed that specifically detect M. tuberculosis infection by exploiting antigens that are selectively expressed by M. tuberculosis and deleted from all (nonvirulent) Mycobacterium bovis bacillus Calmette-Guerin (BCG) strains and most other nontuberculous mycobacteria (7). This has inspired research into the feasibility of developing similar cell-mediated immunity (CMI)-based assays for the identification of asymptomatic leprosy. We (10) and others (3, 4, 21) have used comparative genomics to identify putative open reading frames that are only found in the M. leprae genome and lack homologues in any of the available (myco-)bacterial databases. Using an HLA-based bioinformatics approach, we identified 12 candidate genes that are unique to M. leprae and were predicted to contain T-cell epitopes restricted via several major HLA-DR alleles. Expressed as recombinant proteins, five of these antigens (ML0576, ML1989, ML1990, ML2283, and ML2567) were indeed able to induce significant T-cell responses in PB leprosy patients and M. leprae-exposed healthy controls, but not in most MB leprosy patients, tuberculosis (TB) patients, or controls from areas of leprosy endemicity (10). These proteins have potential added value in diagnosing early infection since 71% of M. leprae-exposed healthy controls that did not have antibodies to the M. leprae-specific PGL-I responded to one to five of the M. leprae antigens.
In a parallel study on the quest for suitable diagnostic tools for leprosy, it was observed that although M. leprae proteins induced higher levels of IFN-γ, the responses to peptides were more specific (21). This is in agreement with the finding that the use of recombinant proteins coincides with an increased risk of detecting cross-reactive T-cell responses, despite the lack of overall sequence homology, due to smaller segment homologies. In addition, purification and quality control assays for recombinant proteins are more labor-intensive than is the case for synthetic peptides. Despite these advantages, however, T-cell responses to peptides are HLA restricted, which may limit the applicability of single peptides with respect to diagnostic T-cell-based assays in genetically diverse populations. This was also clear from the results of a previous study analyzing T-cell responses toward M. leprae-derived peptides, as considerable variations in peptide reactivity were observed in samples from different regions included in the study (5). Therefore, the rationale of the current study was to identify and combine several M. leprae-specific peptide epitopes derived from the five M. leprae candidate proteins that we have previously identified. Since peptide responses are HLA restricted, the use of a combination of several different M. leprae peptides, as applied in the commercial diagnostics tests for TB, could increase sensitivity while maintaining specificity and avoiding cross-reactivity. Thus, in our screening strategy, we selected peptides that are immunogenic in leprosy patients, but not in M. leprae-unexposed individuals, such as Dutch TB patients, Dutch BCG vaccinees, purified protein derivative of M. tuberculosis (PPD)-negative Dutch individuals, and healthy Brazilian controls. The resulting peptides may then be used to distinguish exposure to M. leprae from BCG vaccination and exposure to other mycobacteria, especially in individuals at risk for developing leprosy, like occupational or healthy household contacts of leprosy patients (HHC).
In Brazil, the number of new cases detected during 2006 was 44,436, resulting in a registered prevalence of 60,567 cases at the beginning of 2007 (2). Also, the fact that children are still developing leprosy suggests that multidrug therapy (MDT) has not substantially reduced transmission (16). The genetic diversity in its population (approximately one-third each Caucasian, indigenous, and African descended) and the areas of extraordinarily high leprosy endemicity, compounded by poverty, make Brazil ideal for the development of globally applicable, T-cell-based diagnostic tools.
MATERIALS AND METHODS
Production and testing of M. leprae-specific recombinant antigens.
Five M. leprae candidate genes were amplified by PCR from genomic DNA of M. leprae and cloned using the Gateway technology platform (Invitrogen, Carlsbad, CA) with the expression vector pDEST17 containing an N-terminal histidine tag (Invitrogen) (9). Sequencing was performed on selected clones to confirm the identity of all cloned DNA fragments. The recombinant proteins were overexpressed in Escherichia coli BL21(DE3) and purified as described previously (9) to remove any traces of endotoxin. Each purified M. leprae protein was analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining and Western blotting with an anti-His antibody (Invitrogen) to confirm size and purity. The endotoxin contents were below 50 IU/mg recombinant protein as tested using a Limulus amebocyte lysate (LAL) assay (Cambrex, East Rutherford, NJ). All proteins were tested to exclude antigen-nonspecific T-cell stimulation and cellular toxicity in IFN-γ release assays using peripheral blood mononuclear cells (PBMC) of M. leprae-unexposed, BCG-negative, Mantoux skin test-negative healthy donors.
Synthetic peptides.
Synthetic peptides (20-mers overlapping by 10 amino acids) were produced at the Leiden University Medical Center facility by simultaneous multiple peptide synthesis as described previously (14). Their homogeneity and purity were confirmed by analytical high-performance liquid chromatography and by mass spectrometry. The purity of all peptides was ≥75%.
Study subjects.
Brazilian leprosy patients were recruited from the leprosy outpatient unit, leprosy laboratory (Oswaldo Cruz Institute, Rio de Janeiro). Among these were six PB leprosy patients, four MB leprosy patients, and four MB patients with reversal reaction (Rx). Leprosy patients were diagnosed and classified based on clinical, bacteriological, and if possible, histopathological findings. The leprosy patient treatments were according to the WHO guidelines: MB patients were treated with rifampin, dapsone, and clofazimine, and PB patients were treated with rifampin and dapsone. All MB patients were skin slit smear positive, whereas PB patients were all skin slit smear negative. Patients with leprosy reactions were treated with prednisone (1 mg/kg body weight). As controls, 14 HHC of MB leprosy patients and 12 healthy individuals from the same area in Brazil were recruited. The latter group were blood donor volunteers attending the Blood Bank of the Santa Casa da Misericordia, Rio de Janeiro. They consisted of five M. leprae T-cell nonresponders and seven M. leprae T-cell responders, as determined by their T-cell response (IFN-γ) to whole-M. leprae bacterial extracts (data not shown). All controls were examined for clinical signs of leprosy and the presence of a BCG vaccination scar. As negative control groups, 10 Dutch TB patients, 11 Dutch BCG-vaccinated individuals, and 10 Dutch healthy individuals were recruited at the Leiden University Medical Center. Informed consent was obtained from all individuals before venipuncture. Ethical approval of the study protocol was obtained through the appropriate local ethics committees.
Cell culture and stimulation.
Venous blood from study participants was obtained in heparinized tubes, and PBMC were isolated by Ficoll density centrifugation. PBMC (2 × 106 cells/ml) were plated in triplicate cultures in 96-well round-bottom plates (Costar Corporation, Cambridge, MA) in 200 μl/well of adoptive immunotherapy medium (AIM-V; Gibco) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Invitrogen). Synthetic peptides, recombinant protein, or irradiated armadillo-derived M. leprae sonicate (Colorado State University, Fort Collins, CO) was added at a final concentration of 10 μg/ml. After six days of culture at 37°C at 5% CO2 and 90% relative humidity, 75 μl supernatant was removed from each well, pooled, and frozen in aliquots at −20°C until further analysis.
IFN-γ enzyme-linked immunosorbent assay.
IFN-γ levels were determined by enzyme-linked immunosorbent assay (UCytech, The Netherlands). The cutoff value to define positive responses was set beforehand at 100 pg/ml. The assay's sensitivity level was 20 pg/ml. The values for unstimulated cell cultures were typically <20 pg/ml. As a positive control, phytohemagglutinin (1%) and PPD (Statens Serum Institute, Copenhagen, Denmark) were used (10 μg/ml).
Statistical analysis.
The differences in IFN-γ levels between test groups were analyzed with the two-tailed Mann-Whitney U test for nonparametric distribution using Graph Pad Prism (version 4). The P values were corrected for multiple comparisons. The statistical significance level used was a P value of <0.05.
RESULTS
T-cell recognition of M. leprae peptides in Brazilian leprosy patients and HHC.
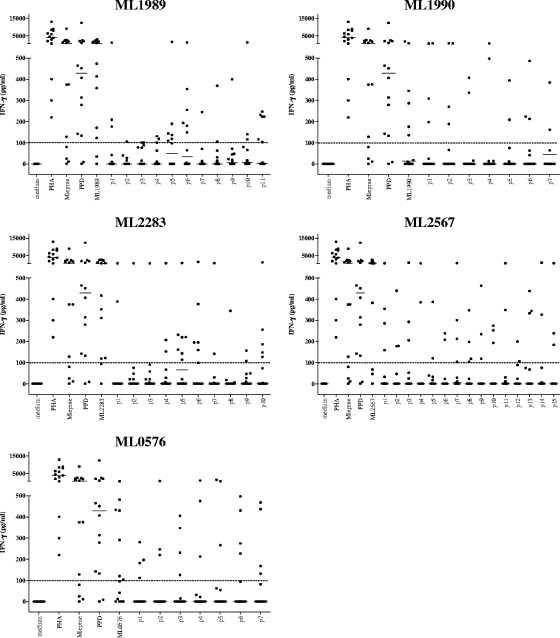
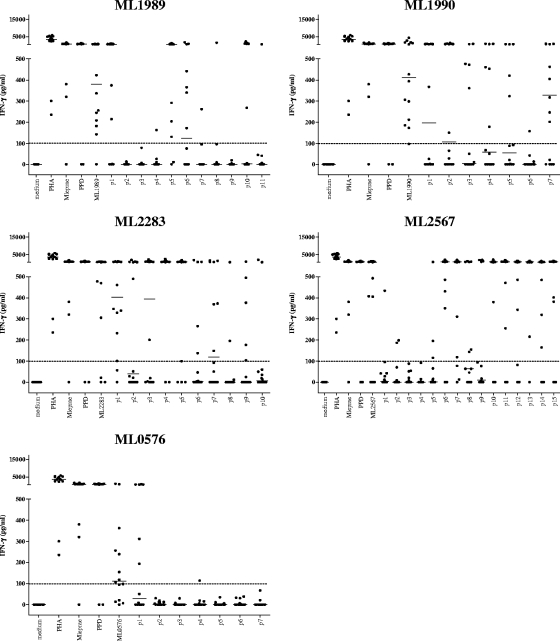
The overall aim of our study was to identify peptide sequences derived from M. leprae that can help to improve specificity in identifying M. leprae exposure using cellular immunity-based assays. In order to achieve this, we decided to use synthetic peptides rather than whole recombinant proteins, to minimize the chance of potential immune cross-reactivity to smaller stretches within the proteins despite the latter being classified as unique to M. leprae. Fifty overlapping peptides were synthesized (7 from ML0576-encoded protein, 11 from ML1989-encoded protein, 7 from ML1990-encoded protein, 10 from ML2283-encoded protein, and 15 from ML2567-encoded protein), spanning the complete sequences of the five M. leprae proteins that we identified previously as being promising diagnostic antigens (10). Peptide recognition by T cells was evaluated by measuring the IFN-γ production of PBMC derived from 14 Brazilian leprosy patients (six PB, four MB, and four Rx) and 14 HHC of MB leprosy patients in response to single M. leprae peptides or their corresponding recombinant proteins (Fig. 1 and 2, respectively). Overall, T-cell responses were induced most frequently by the recombinant proteins. As expected, most of the MB patients' PBMC hardly responded to M. leprae sonicate or to the peptides, whereas both PB and Rx leprosy patients' PBMC recognized M. leprae sonicate and M. leprae proteins as well as the peptides. In the HHC group, peptides derived from ML1989, ML1990, ML2283, and ML2567 were very immunogenic, inducing high levels of IFN-γ, while responses to ML0576 were only observed to ML0576 p1.
FIG. 1.
IFN-γ production of PBMC derived from Brazilian leprosy patients (PB, n = 6; Rx, n = 4; MB, n = 4) induced by five recombinant M. leprae unique candidate proteins or 20-mer peptides overlapping the entire sequence of each corresponding protein. Horizontal bars indicate median responses. Dotted line shows 100-pg/ml cutoff value defining a positive response. PHA, phytohemagglutinin.
FIG. 2.
IFN-γ production of PBMC derived from Brazilian HHC of MB leprosy patients (n = 14) induced by five recombinant M. leprae unique candidate proteins or 20-mer peptides overlapping the entire sequence of each corresponding protein. Horizontal bars indicate median responses. Dotted line shows 100-pg/ml cutoff value defining a positive response. PHA, phytohemagglutinin.
T-cell recognition of M. leprae peptides in Dutch controls.
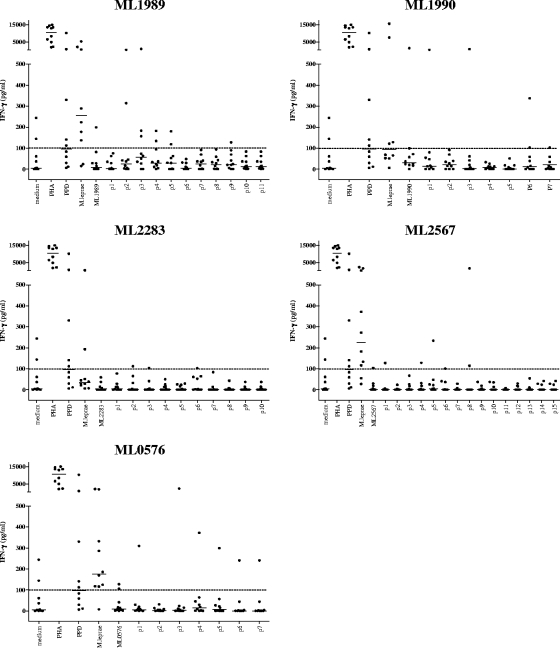
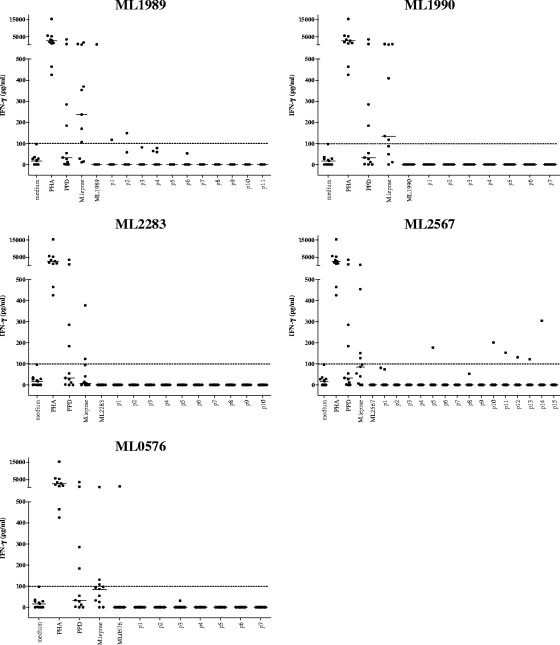
In order to select those peptides recognized in association with M. leprae exposure, but not with BCG vaccination or exposure to M. tuberculosis or other mycobacteria, we tested all 50 M. leprae peptides and the five recombinant proteins in Dutch TB patients and Dutch BCG-vaccinated individuals. Although M. leprae proteins and M. leprae sonicate were sometimes recognized, the level of T-cell activation observed in response to the M. leprae peptides in these two Dutch control groups was low (Fig. 3 and 4) and the number of responsive individuals was limited compared to the leprosy patient and HHC groups. For two different Dutch TB patients, three peptides from ML2283 were slightly positive (100 to 143 pg/ml) after correction for medium values; two other TB patients' PBMC recognized three and two peptides of ML0567; and two other TB patients' PBMC recognized three and one peptide from ML2567, respectively. For ML1989, p2 to p5 induced responses in two or three individuals, and in two TB patients, ML1990 p1, p3, and p6 were recognized by one individual's PBMC (Fig. 3).
FIG. 3.
IFN-γ production of PBMC derived from Dutch TB patients (n = 10) induced by five recombinant M. leprae unique candidate proteins or 20-mer peptides overlapping the entire sequence of each corresponding protein. Horizontal bars indicate median responses. Dotted line shows 100-pg/ml cutoff value defining a positive response. PHA, phytohemagglutinin.
FIG. 4.
IFN-γ production of PBMC derived from Dutch BCG vaccinees (n = 11) induced by five recombinant M. leprae unique candidate proteins or 20-mer peptides overlapping the entire sequence of each corresponding protein. Dotted line shows 100-pg/ml cutoff value defining a positive response. Horizontal bars indicate median responses. PHA, phytohemagglutinin.
After correction for medium values, none of the peptides of ML1990, ML2283, and ML0576 were recognized by PBMC of the Dutch BCG-vaccinated individuals; one individual's PBMC recognized p1 of ML1989, and another BCG vaccinee's PBMC recognized five peptides derived from ML2567 (Fig. 4).
In combination with the data for the Brazilian leprosy patients and HHC (Fig. 1 and 2), this supports the possibility that several of these 50 peptides may indeed induce M. leprae-specific T-cell responses not present in individuals exposed to other, highly related mycobacterial species.
Cumulative T-cell responses against selected M. leprae peptides.
In a fashion similar to that described above for the two Brazilian and two Dutch groups (Fig. 1 to 4), we tested two additional control groups, namely, Dutch healthy individuals who were not vaccinated with BCG and had no known prior contact with leprosy or TB (see Fig. S1 in the supplemental material) and healthy Brazilian individuals (see Fig. S2 in the supplemental material). None of the peptides of ML2283 were recognized in the Dutch negative-control groups. Only five individuals responded marginally to a very limited set of peptides derived from the other four M. leprae proteins: for ML0567 and ML1990, two persons' PBMC each recognized one different peptide; for ML1989, one person's PBMC recognized three peptides; and for ML2567, one peptide was recognized by three individuals' PBMC, one of which reacted to two other peptides as well (data not shown). The healthy Brazilian control group consisted of five M. leprae nonresponders and seven M. leprae responders, as determined by their in vitro T-cell response (IFN-γ) to whole-M. leprae bacterial extracts. The M. leprae nonresponders showed hardly any recognition of the peptides, whereas several peptides activated PBMC of the M. leprae-responding controls from the area of endemicity (see Fig. S2 in the supplemental material).
The use of peptides as diagnostic tools is complicated by the HLA restriction of peptide recognition by T cells, which limits the applicability of single peptides. This prompted us to investigate whether, based on the analysis of single-peptide responses in the six test groups (Fig. 1 to 4 and data not shown), we could select peptides that induced IFN-γ responses in leprosy patients and HHC, but not in Dutch TB patients, Dutch BCG-vaccinated individuals, or Dutch healthy, PPD-negative individuals. In order to maximize the coverage of the number of HLA alleles in the context of which such selected peptides are restricted, peptides were selected that each induced IFN-γ responses in PBMC of different individuals. This selection method makes it likely that different HLA restriction elements are involved, allowing for the majority of M. leprae-infected individuals to be detected when the analysis of the combined peptide responses is eventually used in one diagnostic test. Since such increased sensitivity should not affect the specificity, peptides that induced T-cell responses in the Dutch test groups were excluded from further analysis. Finally, this procedure resulted in the selection of four peptides, derived from ML1989, ML1990, ML2283, and ML2567 (Fig. 5).
FIG. 5.
IFN-γ production against four selected peptides derived from ML2283, ML1989, ML1990, or ML2567 in PBMC derived from PB leprosy patients (n = 6); MB leprosy patients (n = 4); patients with leprosy Rx (n = 4); HHC (n = 14); healthy controls from the area of endemicity without (EC Mlep−) (n = 5) or with (EC Mlep+) (n = 7) in vitro T-cell responses to M. leprae extracts; Dutch healthy, PPD-negative control individuals (NEC) (n = 10); Dutch TB patients (n = 10); and Dutch BCG-vaccinated individuals (n = 11). For each test group, the number of IFN-γ responders versus the total number of individuals in the group is indicated below the x axis. Horizontal bars indicate median responses. Dotted line shows 100-pg/ml cutoff value defining a positive response.
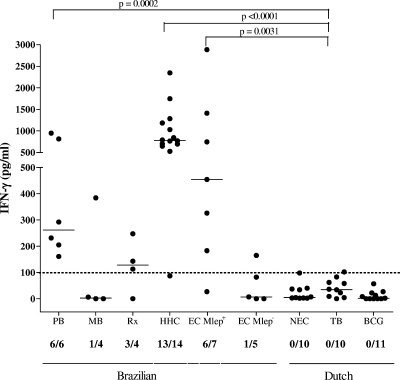
The cumulative T-cell responses, as calculated by the addition of the IFN-γ responses induced against these four peptides separately, improved the sensitivity in the PB group to 100%, compared to a maximum of 50% when assessing the response to only single peptides (Fig. 6 and Table 1). Similarly, in the HHC group, the detection rate increased from 79% (11/14) in the case of individual peptide results to 93% (13/14 individuals) for the combined peptide results. This also resulted in significant differences between the PB group and the TB group (P = 0.002) and the HHC and the TB groups (P < 0.0001).
FIG. 6.
IFN-γ production patterns of four selected M. leprae peptides from ML2283, ML1989, ML1990, or ML2567 induced in PBMC derived from all test groups. For each test group, the number of IFN-γ responders versus the total number of individuals in the group is indicated below the x axis. Horizontal bars indicate median responses. Dotted line shows 100-pg/ml cutoff value defining a positive response. P values for comparison between PB/TB, HHC/TB, and EC Mlep+/TB groups are shown. The MB group did not differ significantly from the control groups. Patient group abbreviations are defined in the text or in the Fig. 5 legend.
TABLE 1.
Cumulative outcome of IFN-γ responsesa
| Sample sourceb | IFN-γ response (pg/ml) tod:
|
Peak IFN-γ response (pg/ml) to any one peptide | |||
|---|---|---|---|---|---|
| ML1989 p11 | ML1990 p7 | ML2283 p5 | ML2567 p3 | ||
| PB1 | 0 | 0 | 813 | 52 | 813 |
| PB2 | 0 | 0 | 0 | 946 | 946 |
| PB3 | 231 | 60 | 230 | 0 | 231 |
| PB4 | 0 | 0 | 0 | 205 | 205 |
| PB5 | 222 | 63 | 219 | 292 | 292 |
| PB6 | 102 | 161 | 20 | 0 | 161 |
| Total PB | 3/6 | 1/6 | 3/6 | 3/6 | 6/6c |
| Rx1 | 247 | 0 | 161 | 24 | 247 |
| Rx2 | 0 | 30 | 113 | 0 | 113 |
| Rx3 | 115 | 40 | 143 | 0 | 143 |
| Rx4 | 0 | 0 | 0 | 0 | 0 |
| Total Rx | 2/4 | 0/4 | 3/4 | 0/4 | 3/4c |
| MB1 | 0 | 0 | 0 | 0 | 0 |
| MB2 | 222 | 384 | 220 | 0 | 384 |
| MB3 | 0 | 0 | 0 | 0 | 0 |
| MB4 | 6 | 0 | 1 | 0 | 6 |
| Total MB | 1/4 | 1/4 | 1/4 | 0/4 | 1/4c |
| HHC1 | 529 | 246 | 1,747 | 8 | 1,747 |
| HHC2 | 0 | 201 | 1,284 | 0 | 1,284 |
| HHC3 | 44 | 250 | 2,345 | 20 | 2,345 |
| HHC4 | 0 | 404 | 842 | 0 | 842 |
| HHC5 | 0 | 636 | 694 | 54 | 694 |
| HHC6 | 5 | 462 | 701 | 0 | 701 |
| HHC7 | 0 | 21 | 526 | 51 | 526 |
| HHC8 | 6 | 765 | 0 | 0 | 765 |
| HHC9 | 0 | 317 | 789 | 0 | 789 |
| HHC10 | 0 | 0 | 770 | 0 | 770 |
| HHC11 | 0 | 912 | 1,031 | 0 | 1,031 |
| HHC12 | 40 | 642 | 646 | 0 | 646 |
| HHC13 | 0 | 30 | 0 | 87 | 87 |
| HHC14 | 0 | 500 | 1,183 | 0 | 1,183 |
| Total HHC | 1/14 | 11/14 | 12/14 | 0/14 | 13/14c |
| EC M. leprae+1 | 8 | 15 | 12 | 326 | 326 |
| EC M. leprae+2 | 0 | 0 | 27 | 22 | 27 |
| EC M. leprae+3 | 25 | 287 | 746 | 3 | 746 |
| EC M. leprae+4 | 68 | 34 | 2,885 | 935 | 2,885 |
| EC M. leprae+5 | 280 | 205 | 1,410 | 1,360 | 1,410 |
| EC M. leprae+6 | 77 | 183 | 152 | 137 | 183 |
| EC M. leprae+7 | 29 | 269 | 454 | 0 | 454 |
| Total EC M. leprae+ | 1/7 | 4/7 | 5/7 | 4/7 | 6/7c |
| EC M. leprae−1 | 0 | 0 | 90 | 82 | 90 |
| EC M. leprae−2 | 133 | 183 | 33 | 165 | 165 |
| EC M. leprae−3 | 0 | 0 | 0 | 0 | 0 |
| EC M. leprae−4 | 0 | 0 | 22 | 0 | 0 |
| EC M. leprae−5 | 7 | 0 | 4 | 1 | 7 |
| Total EC M. leprae− | 1/5 | 1/5 | 0/5 | 1/5 | 2/5c |
| NEC1 | 2 | 39 | 0 | 1 | 39 |
| NEC2 | 0 | 1 | 2 | 0 | 2 |
| NEC3 | 0 | 0 | 0 | 0 | 0 |
| NEC4 | 0 | 0 | 0 | 0 | 0 |
| NEC5 | 0 | 12 | 0 | 0 | 12 |
| NEC6 | 0 | 0 | 1 | 0 | 1 |
| NEC7 | 0 | 0 | 0 | 0 | 0 |
| NEC8 | 0 | 0 | 0 | 98 | 98 |
| NEC9 | 0 | 0 | 4 | 38 | 38 |
| NEC10 | 0 | 0 | 0 | 0 | 0 |
| Total NEC | 0/10 | 0/10 | 0/10 | 0/10 | 0/10c |
| TB1 | 3 | 34 | 2 | 0 | 34 |
| TB2 | 0 | 2 | 1 | 0 | 2 |
| TB3 | 62 | 0 | 0 | 0 | 62 |
| TB4 | 0 | 0 | 0 | 0 | 0 |
| TB5 | 15 | 99 | 0 | 0 | 99 |
| TB6 | 0 | 0 | 0 | 0 | 0 |
| TB7 | 5 | 10 | 7 | 17 | 17 |
| TB8 | 5 | 4 | 0 | 0 | 5 |
| TB9 | 0 | 0 | 0 | 0 | 0 |
| TB10 | 0 | 0 | 0 | 0 | 0 |
| Total TB | 0/10 | 1/10 | 0/10 | 0/10 | 0/10c |
| BCG1 | 0 | 0 | 0 | 7 | 7 |
| BCG2 | 0 | 0 | 0 | 0 | 0 |
| BCG3 | 0 | 0 | 0 | 10 | 10 |
| BCG4 | 0 | 0 | 0 | 0 | 0 |
| BCG5 | 0 | 0 | 0 | 22 | 22 |
| BCG6 | 0 | 0 | 0 | 0 | 0 |
| BCG7 | 0 | 0 | 0 | 0 | 0 |
| BCG8 | 0 | 0 | 0 | 0 | 0 |
| BCG9 | 0 | 0 | 0 | 0 | 0 |
| BCG10 | 0 | 0 | 0 | 0 | 0 |
| BCG11 | 0 | 0 | 0 | 0 | 0 |
| Total BCG | 0/11 | 0/11 | 0/11 | 0/11 | 0/11c |
All IFN-γ values are corrected for medium values. For each individual, the highest IFN-γ value is depicted as the peak IFN-γ response. Italics show values above the 100-pg/ml cutoff value defining a positive response. EC M. leprae+/−, healthy controls from areas of leprosy endemicity without (−) or with (+) in vitro T-cell responses to M. leprae extracts; NEC, Dutch healthy control individuals.
The sample source is identified by test group and donor.
The value is the cumulative number of patients with a positive response to any one peptide/total number tested per group.
Totals are the numbers of responding donors/total number tested per group.
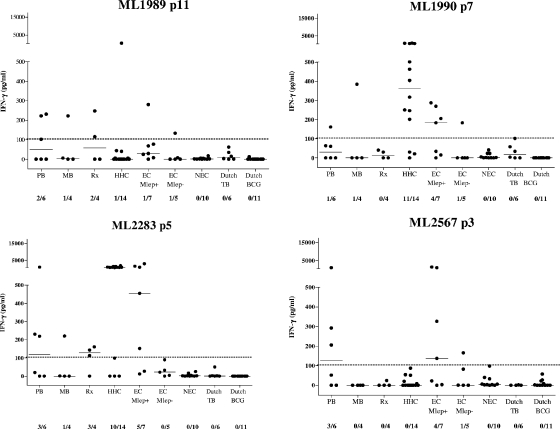
Among the Brazilian healthy controls, from the M. leprae-nonresponsive group, only one individual from the M. leprae-nonresponsive area of endemicity control group responded to the selected peptides from ML1989, ML1990, and ML2567, but the IFN-γ levels were much lower than for the M. leprae-responsive area of endemicity control group (133, 183, and 165 pg/ml, respectively, for this individual). None of the nonresponsive Brazilian healthy controls' PBMC reacted to ML2283 p5, which, interestingly, represents the sequence inducing high levels of IFN-γ (around 2,000 pg/ml) in most HHC. This indicates that these four peptides, particularly ML2283 p5, induce T-cell reactivity that is highly specific for M. leprae exposure (Fig. 5). In contrast, the M. leprae-responsive Brazilian healthy controls responded more often to the peptides, ranging from one out of seven (for ML1989) to five out of seven (for ML2283), and this group differed significantly from the TB group (P = 0.0031). Since M. leprae exposure in the M. leprae-responsive Brazilian healthy controls cannot be ruled out in this area of endemicity, T-cell responses toward these peptides in this test group may very well reflect previous exposure to M. leprae.
When analyzed in a combined fashion, the four peptides induced high levels of IFN-γ responses (183, 326, 454, 746, 1,410, and 2,885 pg/ml) in PBMC from six out of seven of the M. leprae-responsive control individuals from the area of endemicity, while PBMC of one out of five of the nonresponsive healthy controls from the area of endemicity were slightly positive (165 pg/ml) (Fig. 6).
HLA binding motifs and sequence similarity in M. leprae peptides.
The four selected peptide sequences were screened for the presence of peptide binding motifs for common HLA alleles using the ProPred server for HLA class II motifs (19, 24) and HLA class I motifs (20). Interestingly, all four peptides contained binding motifs for multiple class I and class II alleles (Table 2), suggesting that these peptides could indeed have the potential to induce T-cell responses in the context of multiple HLA alleles.
TABLE 2.
Presence of HLA-peptide binding motifsa
| Amino acid sequence | Accession no. | Peptide binding motifs present from:
|
|
|---|---|---|---|
| HLA class II | HLA class I | ||
| STNYVNLSTIGIKLKI | ML1989 | DRB1*0101, 1501, 0301, 0401, 1301, 0701, 0801 | B*5401 |
| VGFVNAWSLLFYPPQGWESP | ML1990 | DRB1*0101, 1501, 0401, 1301, 0701, 0801 | A*24, B*5401, C*0401, C*0702 |
| PLARELYRKIDCYSEEDLVE | ML2283 | DRB1*0101, 0701, 0801 | B40, B*5401, B8, B61, B62 |
| TRKYPGCTSQRNRKHMQVRP | ML2567 | DRB1*0301 | B*2702, B*5401 |
To confirm the M. leprae specificity of the four selected peptides, we evaluated the amino acid sequences using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi). After scanning for short, nearly exact matches, the input peptide sequences produced significant alignments only with sequences from the group 8 (Leproma database, http://genolist.pasteur.fr/Leproma/) M. leprae proteins themselves, as expected. Thus, no sequence similarities were found with any other sequences deposited in the current databases, including other (myco)bacteria.
DISCUSSION
The elimination of leprosy requires, in addition to MDT, the development of novel diagnostic tools. In particular, tests for the early detection of preclinical M. leprae infection, likely the major source of unidentified transmission, have been an important topic in leprosy research for the last two decades. The successful use of T-cell-based IFN-γ release assays using M. tuberculosis-specific peptides of ESAT-6 (Rv3875), CFP-10 (Rv3874), and TB7.7 (Rv2654) for TB diagnostics in humans (7) seems to hold promising possibilities for the use of peptides in CMI-based diagnostics tests for leprosy. In combination with the classical detection of anti-PGL-I immunoglobulin M antibodies, similar T-cell-based tests for leprosy may allow the detection of most forms of leprosy (PB and MB) but may also detect preclinical forms of this disease, thereby enabling the initiation of MDT at an early stage.
We previously reported that neither M. leprae ESAT-6 (ML0049)- nor M. leprae CFP-10 (ML0050)-derived peptides showed sufficient specificity in T-cell assays, as they were recognized strongly by samples from TB patients as well (11, 12). Similar data were observed when ESAT-6 peptides derived from Mycobacterium kansasii were compared with their homologues in M. bovis: all peptides tested contained distinct sequences, yet all, with the exception of one ESAT-6-derived peptide, were immunologically cross-reactive (25). However, using comparative genomics, we subsequently selected five M. leprae-unique candidate proteins that triggered specific IFN-γ responses from PBMC of M. leprae-exposed individuals only (10). In addition, these proteins may become useful in diagnosing early infection, as CMI against these proteins was detected in 71% of M. leprae-exposed healthy controls that did not have antibodies to PGL-I. However, we also observed T-cell reactivity to these M. leprae proteins in some Brazilian TB patients. This might have been due to previous M. leprae exposure, but analogous to a previous observation (1), cross-reactivity with stretches of other, unknown antigens cannot be formally excluded: M. leprae-derived peptides can be cross-reactive with epitopes having sequence homology in as-yet-unsequenced (myco)bacteria or with mimicry epitopes in other antigens. Moreover, it is possible that a single T-cell receptor is able to recognize two different HLA-peptide complexes, each composed of a distinct HLA allele and a different peptide (13, 15). Cross-reactive M. leprae peptides can be completely distinct from the M. leprae-specific T-cell epitopes yet located on the same protein. Irrespective of the precise explanation accounting for these observations, the identification of relevant peptide sequences that are uniquely recognized by M. leprae-exposed individuals will increase the diagnostic performance of T-cell-based tests.
In this study, we describe a rational approach to selecting peptides that reduce the risk of T-cell cross-reactivity (11, 12) and increase the detection of M. leprae infection by combining comparative genomics with bioinformatic HLA-peptide binding algorithms. In order to identify discriminating peptides that detect M. leprae infection with high sensitivity and specificity, we produced 50 synthetic peptides covering the sequences of the proteins encoded by ML0576, ML1989, ML1990, ML2283, and ML2567. These five M. leprae-unique candidate proteins had previously been selected based on the presence of amino acid motifs predicting high-affinity binding to multiple HLA class II molecules, thereby increasing the likelihood of their recognition by T cells from multiple donors (10). In the current study, all 50 peptides were tested for the induction of IFN-γ responses in PBMC of Brazilian leprosy patients, healthy contacts of leprosy patients, and healthy controls, as well as in PBMC of Dutch TB patients, BCG vaccinees, and healthy subjects without previous exposure to leprosy (Fig. 1 to 4). This analysis showed that various single peptides were recognized by PBMC of multiple M. leprae-exposed individuals, with particularly high responses in PBMC of HHC.
Next, 4 of the 50 peptides that induced IFN-γ responses in PBMC of leprosy patients and HHC, but not in PBMC of Dutch controls (TB patients or BCG-vaccinated or PPD-negative individuals), were selected (Table 2). These peptides activated the PBMC of distinct M. leprae-exposed individuals, each covering from 1 out of 6 to 3 out of 6 PB patients, 0 out of 4 to 3 out of 4 patients with leprosy Rx, and 0 out of 14 to 11 out of 14 HHC, likely reflecting the variety in the HLA restriction profiles of each peptide (Fig. 5). Interestingly, when the cumulative responses toward these four peptides were analyzed, all 6 PB patients and 13 out of 14 HHC were detected, whereas the specificity was not compromised since none of the negative-control individuals' PBMC were responsive (Fig. 5 and 6).
A previously extensive variation in peptide reactivity was observed at different test sites using M. leprae-derived peptides (5). Thus, the promising M. leprae peptides described in this study need to be tested in a larger number of individuals and in T-cell-based assays in other ethnic populations in areas of leprosy endemicity. Nevertheless, the Brazilian population in the region of Rio de Janeiro investigated in this study already encompasses a considerable variety in ethnic origins (one-third each Caucasian, indigenous, and African descended). Combined with the presence of multiple HLA-peptide binding motifs in the four selected M. leprae-unique peptides, this has promise for a wider use of these unique peptides and mixtures thereof to design tests with increased sensitivity (compared to using single peptides) and specificity (by lowering the risk of T-cell cross-reactivity compared to using whole proteins). In addition, it is important to evaluate the T-cell responses detected in HHC directed against M. leprae-unique proteins or peptides in a prospective manner in order to relate such responses to M. leprae infection and the development of leprosy.
Thus, this approach may represent a first step toward the identification of additional M. leprae-specific peptides from the M. leprae genome sequence, based on predicted HLA-peptide binding profiles as well as functional screening.
Recently, the use of a combinatorial approach in serological assays also showed increased sensitivity compared to the standard PGL-I assay, while maintaining specificity (18). Eventually the use of a combination of antigens (peptides and/or proteins) for both CMI- and serology-based diagnostics (6, 22, 23) may allow the specific and sensitive detection of most forms (PB and MB) of leprosy and, especially, preclinical forms of this disease. Such early detection of infectious cases, as among HHC who are highly exposed to M. leprae and at risk of contracting leprosy, will allow for early treatment and thereby help to break the transmission cycle.
Supplementary Material
Acknowledgments
This study was supported by The Netherlands Leprosy Relief Foundation (NLR) ILEP grant 702.02.65 and the Q. M. Gastmann-Wichers Foundation.
We thank N. D. L. Savage for critically reading the manuscript, J. A. C. Nery and A. M. Salles as the attending physicians at the leprosy outpatient unit, FIOCRUZ, Rio de Janeiro, Brazil, and the members of the IDEAL (Initiative for Diagnostic and Epidemiological Assays for Leprosy) consortium for stimulating discussions.
Footnotes
Published ahead of print on 16 January 2008.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Aagaard, C., I. Brock, A. Olsen, T. H. Ottenhoff, K. Weldingh, and P. Andersen. 2004. Mapping immune reactivity toward Rv2653 and Rv2654: two novel low-molecular-mass antigens found specifically in the Mycobacterium tuberculosis complex. J. Infect. Dis. 189:812-819. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2006. Global leprosy situation. 2006. Wkly. Epidemiol. Rec. 81:309-316. [PubMed] [Google Scholar]
- 3.Araoz, R., N. Honore, S. Banu, C. Demangel, Y. Cissoko, C. Arama, M. K. Uddin, S. K. Hadi, M. Monot, S. N. Cho, B. Ji, P. J. Brennan, S. Sow, and S. T. Cole. 2006. Towards an immunodiagnostic test for leprosy. Microbes Infect. 8:2270-2276. [DOI] [PubMed] [Google Scholar]
- 4.Araoz, R., N. Honore, S. Cho, J. P. Kim, S. N. Cho, M. Monot, C. Demangel, P. J. Brennan, and S. T. Cole. 2006. Antigen discovery: a postgenomic approach to leprosy diagnosis. Infect. Immun. 74:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dockrell, H. M., S. Brahmbhatt, B. D. Robertson, S. Britton, U. Fruth, N. Gebre, M. Hunegnaw, R. Hussain, R. Manandhar, L. Murillo, M. C. Pessolani, P. Roche, J. L. Salgado, E. Sampaio, F. Shahid, J. E. Thole, and D. B. Young. 2000. A postgenomic approach to identification of Mycobacterium leprae-specific peptides as T-cell reagents. Infect. Immun. 68:5846-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duthie, M. S., W. Goto, G. C. Ireton, S. T. Reece, L. P. Cardoso, C. M. Martelli, M. M. Stefani, M. Nakatani, R. C. de Jesus, E. M. Netto, M. V. Balagon, E. Tan, R. H. Gelber, Y. Maeda, M. Makino, D. Hoft, and S. G. Reed. 2007. Use of protein antigens for early serological diagnosis of leprosy. Clin. Vaccine Immunol. 14:1400-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrara, G., M. Losi, R. D'Amico, P. Roversi, R. Piro, M. Meacci, B. Meccugni, I. M. Dori, A. Andreani, B. M. Bergamini, C. Mussini, F. Rumpianesi, L. M. Fabbri, and L. Richeldi. 2006. Use in routine clinical practice of two commercial blood tests for diagnosis of infection with Mycobacterium tuberculosis: a prospective study. Lancet 367:1328-1334. [DOI] [PubMed] [Google Scholar]
- 8.Fine, P. E. 2007. Leprosy: what is being “eliminated”? Bull. W. H. O. 85:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franken, K. L., H. S. Hiemstra, K. E. van Meijgaarden, Y. Subronto, J. den Hartigh, T. H. Ottenhoff, and J. W. Drijfhout. 2000. Purification of his-tagged proteins by immobilized chelate affinity chromatography: the benefits from the use of organic solvent. Protein Expr. Purif. 18:95-99. [DOI] [PubMed] [Google Scholar]
- 10.Geluk, A., M. R. Klein, K. L. Franken, K. E. van Meijgaarden, B. Wieles, K. C. Pereira, S. Buhrer-Sekula, P. R. Klatser, P. J. Brennan, J. S. Spencer, D. L. Williams, M. C. Pessolani, E. P. Sampaio, and T. H. Ottenhoff. 2005. Postgenomic approach to identify novel Mycobacterium leprae antigens with potential to improve immunodiagnosis of infection. Infect. Immun. 73:5636-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geluk, A., K. E. van Meijgaarden, K. L. M. C. Franken, Y. W. Subronto, B. Wieles, S. M. Arend, E. P. Sampaio, T. de Boer, W. R. Faber, B. Naafs, and T. H. M. Ottenhoff. 2002. Identification and characterization of the ESAT-6 homologue of Mycobacterium leprae and T-cell cross-reactivity with Mycobacterium tuberculosis. Infect. Immun. 70:2544-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geluk, A., K. E. van Meijgaarden, K. L. Franken, B. Wieles, S. M. Arend, W. R. Faber, B. Naafs, and T. H. Ottenhoff. 2004. Immunological crossreactivity of the Mycobacterium leprae CFP-10 with its homologue in Mycobacterium tuberculosis. Scand. J. Immunol. 59:66-70. [DOI] [PubMed] [Google Scholar]
- 13.Geluk, A., K. E. van Meijgaarden, and T. H. Ottenhoff. 1997. Flexibility in T-cell receptor ligand repertoires depends on MHC and T-cell receptor clonotype. Immunology 90:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiemstra, H. S., G. Duinkerken, W. E. Benckhuijsen, R. Amons, R. R. de Vries, B. O. Roep, and J. W. Drijfhout. 1997. The identification of CD4+ T cell epitopes with dedicated synthetic peptide libraries. Proc. Natl. Acad. Sci. USA 94:10313-10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang, H. L., H. Jacobsen, S. Ikemizu, C. Andersson, K. Harlos, L. Madsen, P. Hjorth, L. Sondergaard, A. Svejgaard, K. Wucherpfennig, D. I. Stuart, J. I. Bell, E. Y. Jones, and L. Fugger. 2002. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat. Immunol. 3:940-943. [DOI] [PubMed] [Google Scholar]
- 16.Lockwood, D. N., and S. Suneetha. 2005. Leprosy: too complex a disease for a simple elimination paradigm. Bull. W. H. O. 83:230-235. [PMC free article] [PubMed] [Google Scholar]
- 17.Meima, A., W. C. Smith, G. J. van Oortmarssen, J. H. Richardus, and J. D. Habbema. 2004. The future incidence of leprosy: a scenario analysis. Bull. W. H. O. 82:373-380. [PMC free article] [PubMed] [Google Scholar]
- 18.Parkash, O., A. Kumar, R. Pandey, B. K. Girdhar, K. L. Franken, and T. H. Ottenhoff. 2007. Serological heterogeneity against various Mycobacterium leprae antigens and its use in serodiagnosis of leprosy patients. J. Med. Microbiol. 56:1259-1261. [DOI] [PubMed] [Google Scholar]
- 19.Singh, H., and G. P. Raghava. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236-1237. [DOI] [PubMed] [Google Scholar]
- 20.Singh, H., and G. P. Raghava. 2003. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics 19:1009-1014. [DOI] [PubMed] [Google Scholar]
- 21.Spencer, J. S., H. M. Dockrell, H. J. Kim, M. A. Marques, D. L. Williams, M. V. Martins, M. L. Martins, M. C. Lima, E. N. Sarno, G. M. Pereira, H. Matos, L. S. Fonseca, E. P. Sampaio, T. H. Ottenhoff, A. Geluk, S. N. Cho, N. G. Stoker, S. T. Cole, P. J. Brennan, and M. C. Pessolani. 2005. Identification of specific proteins and peptides in mycobacterium leprae suitable for the selective diagnosis of leprosy. J. Immunol. 175:7930-7938. [DOI] [PubMed] [Google Scholar]
- 22.Spencer, J. S., H. J. Kim, A. M. Marques, M. Gonzalez-Juarerro, M. C. Lima, V. D. Vissa, R. W. Truman, M. L. Gennaro, S. N. Cho, S. T. Cole, and P. J. Brennan. 2004. Comparative analysis of B- and T-cell epitopes of Mycobacterium leprae and Mycobacterium tuberculosis culture filtrate protein 10. Infect. Immun. 72:3161-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer, J. S., M. A. Marques, M. C. Lima, A. P. Junqueira-Kipnis, B. C. Gregory, R. W. Truman, and P. J. Brennan. 2002. Antigenic specificity of the Mycobacterium leprae homologue of ESAT-6. Infect. Immun. 70:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sturniolo, T., E. Bono, J. Ding, L. Raddrizzani, O. Tuereci, U. Sahin, M. Braxenthaler, F. Gallazzi, M. P. Protti, F. Sinigaglia, and J. Hammer. 1999. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 17:555-561. [DOI] [PubMed] [Google Scholar]
- 25.Vordermeier, H. M., J. Brown, P. J. Cockle, W. P. Franken, S. M. Arend, T. H. Ottenhoff, K. Jahans, and R. G. Hewinson. 2007. Assessment of cross-reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-cell epitope level. Clin. Vaccine Immunol. 14:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.