Abstract
Summary: Although the phenomenon of bacterial cell death and lysis has been studied for over 100 years, the contribution of these important processes to bacterial physiology and development has only recently been recognized. Contemporary study of cell death and lysis in a number of different bacteria has revealed that these processes, once thought of as being passive and unregulated, are actually governed by highly complex regulatory systems. An emerging paradigm in this field suggests that, analogous to programmed cell death in eukaryotes, regulated cell death and lysis in bacteria play an important role in both developmental processes, such as competence and biofilm development, and the elimination of damaged cells, such as those irreversibly injured by environmental or antibiotic stress. Further study in this exciting field of bacterial research may provide new insight into the potential evolutionary link between control of cell death in bacteria and programmed cell death (apoptosis) in eukaryotes.
INTRODUCTION
The phenomenon of bacterial cell death and lysis has been the subject of study for well over 100 years. Because of its readily observable nature, work in the early 1900s focused on the self-destructive end stage of the bacterial life cycle, termed “autolysis,” and began with the identification of bacterial species that exhibited this seemingly counterproductive and enigmatic process (reviewed in reference 238). Although it was initially believed that secreted proteases were responsible for cell lysis, the lack of correlation between the organisms known to secrete protease activity and their propensity to undergo autolysis suggested otherwise (238). In 1919, Lord and Nye (155) suggested that the enzymes responsible for the autolysis of Streptococcus pneumoniae were intrinsically produced and sensitive to extremes in pH. Avery and Cullen (9) confirmed those findings and went on to identify specialized “bacteriolytic enzymes” that catalyzed this process, thus launching the era of the “autolysin.” Subsequent studies revealed that these enzymes (also referred to as murein or peptidoglycan hydrolases) were distinct from other commonly studied enzymes at the time, such as protease, pancreatic RNase, and hyaluronidase (179), and were shown to exhibit specificity for the species from which they were isolated (95). Furthermore, Nomura and Hosoda (180) demonstrated that isolated cell walls could be dissolved by treatment with purified autolysin and that protoplasts could be generated by degrading the cell wall in the presence of high concentrations of sucrose or polyethylene glycol, which provide an osmostabilizing effect. In addition to demonstrating the specificity of these enzymes for the cell wall, these data provided some of the first evidence for the structural and protective roles of peptidoglycan.
In the mid-1900s, growth conditions that induced the autolysis of Mycobacterium tuberculosis were investigated, providing clues to the physiological requirements needed for optimal murein hydrolase activity. Redmond and Bowman (210) demonstrated that autolysis could be inactivated by incubating the bacteria at high temperatures, indicating that the autolytic system was temperature sensitive. A year later, that same group showed that autolysis was induced by growth in the presence of oxygen and excess glucose (28) as well as by limited nitrogen availability (209). Subsequent investigations revealed that the autolysis phenomenon in bacteria is influenced by a variety of different factors including NaCl, pH, growth phase, proteases, cardiolipin, teichoic acids, and sodium polyanethole sulfonate (84, 89, 182, 205, 248, 269, 272, 278). Despite the numerous basic physiological characterizations of autolysis, few attempts were made to carefully assess the biological role of this process.
Today, we have essentially taken bacterial cell death and lysis for granted. It has been traditionally thought of as the consequences of “unbalanced growth” or the end stage of the bacterial life cycle that occurs after all of the perceived more interesting physiological processes have run their course. However, several studies suggested that these processes are much more complex than previously thought, processes that might be fundamental to bacterial physiology and essential to our understanding of how bacteria develop within complex communities (e.g., biofilms), just as knowledge of programmed cell death (PCD) (apoptosis) is essential to an understanding of the development of more complex eukaryotic organisms. Despite the fact that bacterial autolysis has been studied for over a century, only recently has progress in understanding this phenomenon been achieved. Thus, this review will focus on established mechanisms involved in the control of bacterial cell death and lysis and will highlight recent advances that illuminate the true biological functions of these enigmatic processes.
THE CELL WALL AND INTRINSIC CONTROL OF LYSIS
Peptidoglycan
The bacterial cell wall is an incredibly complex “superstructure” that remains ill defined despite having been studied for over half a century. Because of its complexity, it is one of the final frontiers that relatively few have dared to enter. It has been described as a three-dimensional fabric that completely encases the bacterial cell, giving it shape and resistance to internal osmotic forces. Remarkably, the cell wall of gram-negative bacteria is thin (approximately 10 nm in thickness), comprised of only two to five layers of peptidoglycan, depending on the growth stage (86, 146, 204). In gram-positive bacteria, the cell wall is much thicker (20 to 40 nm thick) (see reference 82) and comprised of two high-molecular-weight polymers known as peptidoglycan and teichoic acid. Whereas the peptidoglycan provides the structural framework of the cell wall, the primary function of teichoic acids (discussed below), which make up roughly 50% of the cell wall material, is thought to control the overall surface charge, affecting murein hydrolase activity, resistance to antibacterial peptides, and adherence to surfaces. Although both of these molecules are polymerized on the surface of the cytoplasmic membrane, their precursors are assembled in the cytoplasm. In the early stages of peptidoglycan assembly, so-called “muropeptide” precursors (comprised of the N-acetylmuramic acid, N-acetylglucosamine disaccharide, and a 5-amino-acid stem peptide) (Fig. 1) are synthesized and then presented on the outer face of the cytoplasmic membrane to penicillin-binding proteins (PBPs), where they are polymerized into peptidoglycan. Two enzyme activities have been attributed to PBPs: transpeptidase and transglycosylase. The transglycosylase activity of these proteins catalyzes the formation of (β1→4)-glycosidic linkages between two muropeptide units, resulting in the formation of glycan chains that are 21 units in length on average in Escherichia coli, although this varies from species to species (144). In contrast, the transpeptidase activity cross-links the muropeptide chains together by catalyzing the formation of a peptide bond between the carboxyl group of the penultimate d-alanine of one stem peptide to the ɛ amino group of a dibasic amino acid (via a peptide bridge) at position 2 or 3 of the stem peptide. Although the peptidoglycan of E. coli is relatively weakly cross-linked (with only 50% of the stem peptides involved in cross-linking), Staphylococcus aureus peptidoglycan is extensively cross-linked, with greater than 90% of the stem peptides linked together, forming a network where there is an average of 15 interlinked stem peptides (144). Recent studies have shown that this fabric comprises a patchwork of different molecules with dramatic species-to-species variation as well as molecular diversity within a single species (69, 226). The diversity of molecules incorporated into peptidoglycan likely reflects the versatility of the PBPs that catalyze peptidoglycan, but the biological function of this variability remains unknown.
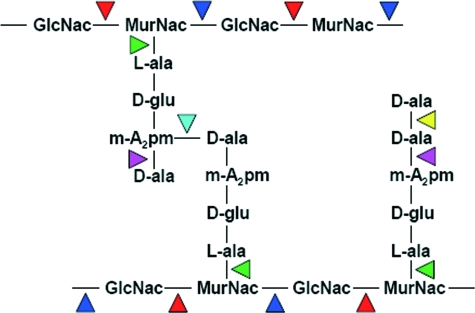
FIG. 1.
Murein hydrolase targets within E. coli peptidoglycan. The peptidoglycan network is comprised of interlinked muropeptide chains containing alternating subunits of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). Stem peptides comprised of l- and d-amino acids, as well as m-diaminopimelic acid (m-A2pm), are attached to N-acetylmuramic acid and are cross-linked to stem peptides from adjacent muropeptide strands as indicated. Different murein hydrolases that cleave the various bonds within peptidoglycan have been identified and include β-N-acetylglucosaminidase (red), lytic transglycosylase (blue), N-acetylmuramyl-l-alanine-amidase (green), d,d-carboxypeptidase (yellow), and l,d-carboxypeptidase (pink). (Adapted from reference 111 with permission of the publisher.)
Murein Hydrolases
Bacterial murein hydrolases are also widely diverse (Fig. 1), reflecting the different bonds found in peptidoglycan. They are a unique family of enzymes that specifically cleave structural components of peptidoglycan and have been shown to participate in a number of important biological processes during cell growth and division, including daughter cell separation, cell wall growth, and peptidoglycan recycling and turnover (8, 111, 195, 229, 230, 267). In addition, these enzymes contribute to the pathogenicity of bacteria and are required for susceptibility to antibiotics (111). Biochemical analyses of murein hydrolases reveal that they have hydrolytic activities that are specific for various structural components of the peptidoglycan, including N-acetylmuramidase, N-acetylglucosaminidase, N-acetylmuramyl-l-alanine amidase, and endotransglycosidase activities that presumably have specific roles in the biosynthesis and processing of the bacterial cell wall (111, 230, 267). Those murein hydrolases that lead to the destruction of the cell wall and subsequent cell lysis are known as autolysins.
It is widely believed that murein hydrolases are required for making precise cuts in the peptidoglycan that allow for the insertion of new muropeptide strands. However, the cell wall acts as a load-bearing molecule that counteracts the internal osmotic pressure of the cell. Therefore, how are these breaches in the cell tolerated without causing lysis? One possibility involves the “three-for-one” model proposed by Höltje (112), whereby three new muropeptide strands are covalently attached underneath an existing load-bearing muropeptide strand. After the removal of the old muropeptide strand, the newly inserted strands are automatically forced into place by the internal pressure of the cell, thus adding a total of two new muropeptide strands to the growing sacculus. This model predicts that the PBPs are working in tandem with murein hydrolases during cell wall synthesis, forming a “murein replicase holoenzyme” (112). Indeed, evidence for this multienzyme complex in E. coli was generated by demonstrating specific protein-protein interactions between purified murein hydrolases (Slt70 and MltA) and PBPs (219, 262). Other studies of Haemophilus influenzae used the chemical cross-linker cyanogen to identify membrane-bound PBPs in close association with each other (3). The results suggest the presence of two multienzyme complexes, which were predicted to be involved in cell elongation and septum formation, respectively. Furthermore, the molecular weight of the complex was greater than the sum of the PBPs identified, suggesting that other proteins (e.g., murein hydrolases) are associated with these complexes. Although this holoenzyme model is attractive and supported by solid evidence, the striking reality is that not a single bona fide murein hydrolase, including Slt70 and MltA mentioned above, has been shown to have a dramatic impact on bacterial growth. In fact, the disruption of multiple murein hydrolase genes in a single strain of Bacillus subtilis also had little effect on bacterial growth, although additional murein hydrolase activities remained (25). An exception to this may be the essential PcsB protein from S. pneumoniae, which is believed to function as a murein hydrolase since it contains a CHAP (cysteine- and histidine-dependent amidohydrolase/peptidase) domain found in many of these enzymes (177). Thus, due to the inability to conclusively demonstrate the essential nature of these enzymes, it is likely that redundant murein hydrolase activities are associated with the proposed murein replicase holoenzyme or that these activities are intrinsic to the PBPs themselves.
What is clear is that many murein hydrolases are required for daughter cell separation after the completion of the newly formed septum. There are multiple examples of mutations in murein hydrolase genes in both gram-positive and gram-negative bacteria that result in the inability of the cells to separate (25, 38, 51, 65, 70, 71, 108, 118, 244, 249). Based on the frequency with which murein hydrolase mutations are associated with this phenotype, the primary functions of these enzymes in daughter cell separation are unambiguous. However, this does not exclude the possibility that they are involved in other essential functions such as that related to a putative murein replicase holoenzyme. For a more detailed description of bacterial murein hydrolases and their impact on cell wall metabolism, the reader is referred to a variety of reviews on this subject (111, 230, 241).
The specific regulation of murein hydrolase activity in gram-negative bacteria has been an active area of investigation and has also been the subject of review (111). Posttranslational regulatory mechanisms of these enzymes have been proposed to include (i) the activation of enzyme activity as a result of substrate modification (90, 255), (ii) sequestration within lipid membranes (102, 114), (iii) controlled transport across the cytoplasmic membrane (21), and (iv) topographical control within the peptidoglycan. The latter relies on evidence suggesting a multilayered arrangement of the peptidoglycan (113, 204, 225) in which the outside, unstressed layers would be susceptible to murein hydrolase activity, leaving the inner, load-bearing layers intact. Regulation of murein hydrolase activity by a variety of cofactors including phospholipids (258), coenzyme A-glutathione disulfide (115), magnesium (147), EDTA (147), and DNA (120, 141, 252) has also been demonstrated. Although the precise mechanism by which murein hydrolases are controlled in gram-negative bacteria has yet to be elucidated, likely attributable to the complexity of this system, this mechanism is undoubtably specific to these organisms due to the unique environment afforded by the membrane-enclosed periplasmic space.
Teichoic Acids
Since gram-positive bacteria lack an outer cell membrane, the peptidoglycan is exposed to the external environment. This poses a significant problem to these organisms, as many of the enzymatic reactions essential for normal cell wall metabolism must be performed without interference from external factors. Although it is widely believed that the gram-positive cell wall is a mesh-like network that is completely permeable to its surrounding environment, several studies suggested that the cell wall provides a unique compartment analogous to the periplasmic space of gram-negative bacteria. One of the most important components of the gram-positive cell wall that helps to define this compartment is a family of carbohydrates referred to as teichoic acids. As detailed extensively in an outstanding review of these molecules by Neuhaus and Baddiley (176), teichoic acids serve a critical yet ill-defined function in the overall physiology of the gram-positive cell wall. There are two basic forms of these molecules; wall teichoic acids, which are covalently attached to the peptidoglycan, and lipoteichoic acids, which are anchored in the cytoplasmic membrane. Along with peptidoglycan, the teichoic acids form a polyanionic gel that has broad-reaching roles ranging from maintaining metal cation homeostasis to functioning as a gatekeeper for the flow of ions, nutrients, and proteins to and from the cytoplasmic membrane. Thus, these molecules appear to be important for the formation of a buffer zone between the external environment and the cytoplasmic membrane, much like the periplasmic space of gram-negative bacteria.
Although gram-positive bacteria clearly do not have a membrane-enclosed compartment like a periplasm, several studies have suggested the presence of a distinct environment associated with the cell walls of these organisms. Using a cryotransmission electron microscopy technique to better preserve the ultrastructure of the bacterial cell wall, the presence of a clearly visible tripartite (or bipartite, depending on the technique used) wall in both B. subtilis and S. aureus has been demonstrated (160, 161). As shown in Fig. 2, a low-density inner wall zone (IWZ), thought to be analogous to a periplasmic space, is surrounded by a high-density outer wall zone (OWZ) containing peptidoglycan and teichoic acid. The IWZ would presumably contain lipoteichoic acids extending into the OWZ and would possibly provide the “scaffolding” (along with other lipoproteins) needed to withstand the internal osmotic forces that would be pressing the cell membrane against the peptidoglycan layer (161). On the outer surface is a fibrous layer that extends into the surrounding medium that is most prominently visualized using a freeze-substituted electron microscopic technique (93, 160, 161).
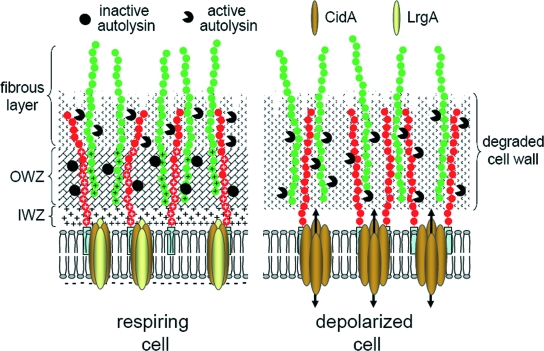
FIG. 2.
Model for the control of murein hydrolase activity. As hypothesized previously (35), the respiring cell produces a proton gradient outside of the cytoplasmic membrane that causes a localized reduction in the pH within the cell wall. This acidic pH suppresses the activity of murein hydrolases associated with the teichoic acids (green) and lipoteichoic acids (red) by promoting the protonation of the d-Ala ester linkages (+ signs). Upon dissipation of the membrane potential, the pH in the cell wall increases and destabilizes or deprotonates the d-Ala ester linkages, thus derepressing the murein hydrolases. The IWZ and OWZ, identified by electron microscopy, are proposed to correspond with the periplasmic space and intact peptidoglycan layer, respectively (160, 161). The fibrous layer is proposed to be composed of partially degraded peptidoglycan on the outer surface of the cell wall, where peptidoglycan turnover is thought to take place. In a depolarized cell, the murein hydrolases are activated and the intact peptidoglycan is degraded, compromising the structural integrity of the cell wall and resulting in cell lysis. Also shown is the CidA protein, whose proposed holin activity would dissipate the membrane potential and trigger cell lysis. As an antiholin, LrgA would inhibit CidA activity and prevent cell lysis.
As intrinsically polyanionic molecules, teichoic acids are likely targets for the plethora of cationic peptide antibiotics produced by a variety of immune cells and multiple bacterial species (176). However, their primary function appears to be in the intrinsic control of murein hydrolases produced during normal cell metabolism (63, 176). This is achieved by acting as both targets of murein hydrolase binding as well as regulators of their activity. Targeting is best studied in S. pneumoniae, where the binding of murein hydrolases is mediated by repeat elements called choline-binding domains that specifically associate with choline-substituted teichoic acids in the cell wall (85, 221). The primary murein hydrolase of S. pneumoniae, LytA, is made in an inactive form and is activated in vitro by association with choline-substituted teichoic acid in a process known as “conversion” (250). Unfortunately, little is understood regarding the molecular mechanisms involved in conversion in vivo other than the speculation that it is associated with minor structural modifications of LytA upon interactions with choline (220). However, it is clear that the mere presence of choline in vivo is insufficient to induce LytA activity during active growth. Thus, the question remains as to what prevents LytA from digesting the pneumococcal cell wall in a metabolically active cell.
Although the process of conversion and the use of choline as a binding substrate appear to be unique to S. pneumoniae and closely related species, many of the murein hydrolases produced by other bacteria also contain repeat elements important for targeting to the cell wall (174). For example, the S. aureus Atl murein hydrolase is proteolytically cleaved to generate amidase and glucosaminidase enzymes, both containing repeat elements (187). These repeat elements (designated R1, R2, and R3) have been shown to target these murein hydrolases to the equatorial surface rings (marking the sites of future cell division) of S. aureus (11, 279), consistent with the role of atl in daughter cell separation (65, 242). Presumably, these repeat elements also target these enzymes to the teichoic acids, although definitive evidence for this has not been reported. Interestingly, the repeat elements were shown to be necessary and sufficient for targeting proteins to the equatorial rings, leading to the proposal that the receptor was positioned at these sites (11). Thus, either the receptor is distinct from teichoic acids or the teichoic acids associated with the future division sites are specifically modified to attract murein hydrolases to these regions of the cell wall.
Indeed, the modification of teichoic acids by the addition of d-alanine (d-Ala) ester linkages has been shown to be critical to their function. The complex biochemical reactions required for d-alanylation of teichoic acids have been thoroughly examined (176) and thus will not be elaborated on in detail here. However, of particular interest to the current review is the demonstration by several laboratories that the dlt genes required for d-alanylation of teichoic acids in many gram-positive bacteria play a vital role in modulating surface charge and controlling murein hydrolase activity. For example, dlt mutations in B. subtilis resulted in enhanced autolysis and increased susceptibility to methicillin (270, 271). Furthermore, the cell walls of the mutants were more negatively charged, as indicated by the increased binding of the cationic protein cytochrome c. Effects of d-Ala ester content on autolysis were also observed in a variety of other bacterial species including S. aureus (64, 140, 181, 197), Lactococcus lactis (234), and Lactobacillus plantarum (188). Other consequences of d-Ala depletion include increased susceptibility to a variety of different antimicrobial compounds (61, 132, 136, 137, 196, 197), decreased survival in neutrophils (49, 137), decreased virulence (61, 138), decreased epithelial cell invasion (137), and decreased biofilm formation (61). All of these effects are likely to be a result of the more positive surface charge of the bacteria as a consequence of the reduced d-alanylation of the teichoic acids within the cell wall.
The modification of teichoic acids within the cell wall for the purpose of controlling murein hydrolase activity is likely to be a common theme in gram-positive bacteria. In addition to d-Ala and choline modifications, the teichoic acids of Streptomyces roseoflavus have been reported to be substituted with l-lysine (176). Studies with Bacillus anthracis have also demonstrated that an unspecified carbohydrate (perhaps teichoic acid) within the cell wall is subject to pyruvate modification, which is important for the recognition and binding of S-layer proteins (162). Like murein hydrolases, these proteins contain repeat elements referred to in this case as S-layer homology (SLH) domains, which are required for cell wall targeting (149, 185, 217). Interestingly, many of the other proteins found to contain SLH domains are murein hydrolases, suggesting that they also recognize and bind these pyruvylated carbohydrates (162). Furthermore, disruption of the B. anthracis csaAB genes involved in pyruvylation exhibits defects in cell division and autolysis, consistent with a role for pyruvylation in the control of murein hydrolase activity (162).
Possibly as a result of the absence of a membrane-enclosed periplasmic space that could provide a stable environment for enzymatic function, a unique and sophisticated strategy to regulate the murein hydrolases produced by gram-positive bacteria has evolved. Previous work (34, 35, 117) suggested that this mechanism may involve the charged state of the membrane (proton motive force [PMF]) of B. subtilis and the subsequent protonation of the cell wall, presumably including the alanines associated with teichoic acid (176). Those studies also suggested the presence of a “compartment” in which this protonation could be maintained despite the essentially infinite buffering capacity of the external medium. As protonation is thought to stabilize the d-alanyl ester linkage within the teichoic acids (176), changes in PMF could alter the degree of d-alanylation and, in turn, the association and/or activity of murein hydrolases. Moreover, a similar mechanism with cell walls modified with pyruvate, choline, and lysine could exist. We envision a complex mechanism that functions in the cell wall to control murein hydrolase activity in a respiring cell. At the heart of this model are the teichoic acids, which inhibit murein hydrolase activity depending on the presence or absence of protonated d-alanines. In turn, the inhibitory activity of d-alanylation is controlled by a pH gradient that is established outside of the respiring cell (121, 127). As the distance from the cell membrane increases, the pH would increase, depending on the pH of the surrounding environment (near neutrality under physiological conditions). Consequences of this pH change would be a gradient of destabilized d-alanyl ester linkages and/or deprotonation of the d-alanyl moieties, both of which would release the inhibitory effects on the associated murein hydrolases. Thus, as depicted in Fig. 2, cell wall-associated murein hydrolases positioned farther away from the cell membrane would be more active than those located proximal to the cell membrane, accounting for the increased peptidoglycan turnover in the outer layers of the cell wall. This model is supported by studies using both cryoelectron and freeze substitution electron microscopy, revealing the presence of distinct “compartments” within the cell wall of B. subtilis as described above. More importantly, these imaging techniques revealed a decreasing gradient of cell wall density, which is also consistent with the idea that cell wall turnover occurs within the outer layers. Furthermore, this model is supported by the observation that murein hydrolase activity and autolysis are inhibited by growth at low pH in several bacterial species (89). It is also consistent with studies demonstrating the importance of PMF for controlling murein hydrolase activity and autolysis in B. subtilis (34, 35, 117). Interestingly, S. aureus did not exhibit a gradient of cell wall density, suggesting that cell wall turnover occurs in distinct sites, possibly within the septa (161). Consistent with this is the observation that staphylococcal murein hydrolases specifically localize to the septa of a dividing cell (11, 279). The activation of murein hydrolase activity and disruption of the cell wall in the presence of penicillin were also localized to the division planes in S. aureus (82, 83), suggesting that murein hydrolase activity in this pathogen is not homogeneously distributed.
REGULATED DEATH AND LYSIS
Bacteriophage Holins
The best-characterized example of regulated bacterial death and lysis comes from studies of the control of the lytic cycle during a bacteriophage infection. As originally demonstrated by Wang and colleagues, there exists a sophisticated bacteriophage-encoded mechanism that controls the timing of lysis during the lytic cycle such that the bacteriophage particles accumulating within the cytoplasm can be released into the surrounding environment at a time that maximizes the reproductive potential of the bacteriophage population (265). This timing has been carefully fine-tuned by evolution to achieve a balance between the consequences of early termination of bacteriophage replication within a host cell, with the benefits of potentially achieving logarithmic amplification by subsequent infection of and replication within additional host cells. The mechanism controlling the timing of bacteriophage-induced lysis is simple and elegant, involving a “holin” and an “endolysin” (a murein hydrolase) that are both necessary and sufficient to induce precisely timed and rapid cell lysis (285). The timing of cell lysis is dictated by the holin, which controls the activity of the endolysin and is achieved using one of two proposed mechanisms. The first mechanism, which is utilized by holins encoded by lambda- and T4-like bacteriophages, involves the control of murein hydrolase transport across the membrane (Fig. 3), where it has access to its substrate, peptidoglycan. Murein hydrolases transported in this fashion lack signal peptides and thus harmlessly accumulate in the cytoplasm until the holin allows passage across the cytoplasmic membrane (285). Although the mechanism by which these holins mediate the passage of murein hydrolases is unclear, it is thought to involve their oligomerization in the membrane to a point that causes membrane permeabilization, passively allowing the murein hydrolase to cross into the periplasm (99, 266, 283). The other mechanism, utilized by bacteriophage P1, involves bacteriophage-encoded murein hydrolases containing “signal-arrest-release” (SAR) domains (277). Like signal sequences, SAR domains target their cargo to the Sec machinery but anchor the protein in the outer face of the membrane in an inactive form until the holin releases it (Fig. 3). Importantly, the release of these proteins is also accompanied by a disulfide isomerization event, converting them to active murein hydrolases (276).
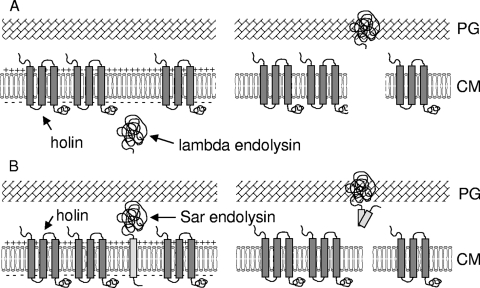
FIG. 3.
Models of holin function. (A) Within a respiring cell with an energized cytoplasmic membrane (CM), endolysins, such as that produced by bacteriophage lambda, accumulate in an active form in the cytoplasm. Upon holin activation, the membrane becomes leaky and allows the escape of the endolysin to the peptidoglycan (PG). (B) SAR-type endolysins, such as that produced by bacteriophage P1, are transported in a Sec-dependent manner to the outer face of the inner membrane, where they are held in an inactive form via the N-terminal “SAR” domain (light gray bar). After holin activation, the membrane becomes de-energized, causing the release and activation of the endolysin. (Adapted from reference 277 with permission of the publisher. Copyright 2005 National Academy of Sciences, U.S.A.)
In early studies of lambda holin and endolysin function, plasmid constructs were generated such that the holin (designated S) and endolysin (designated R) were independently expressed using an inducible promoter (72). What is clear from those studies is that the induction of S and R together caused a rapid loss of cell viability, which was followed by cell lysis 30 min later. Interestingly, the expression of S alone was sufficient to induce a loss of cell viability but not lysis, while the expression of the endolysin alone had no effect on the cells, as these proteins harmlessly accumulated in the cytoplasm. Those authors concluded that the accumulation of the S protein in the cytoplasmic membrane caused membrane lesions resulting in the collapse of the PMF and the subsequent loss of cell viability, release of the endolysin, and, ultimately, cell lysis. Based on these experiments, it was proposed that the timing of lysis was a function of a gradual decrease in PMF until a critical level was reached (72, 285). Several years later, this model was rejected by an elegant experiment in which the PMF of the cell was monitored in real time by tethering the flagellum to a microscope slide and recording the velocity of bacterial rotation (an indirect measure of PMF) at various time points after the induction of S and R (100). The results demonstrated that rather than achieving a gradual decline in PMF, the individual cells maintained their PMF for approximately 20 min after induction until the rotation abruptly stopped several seconds before lysis. Thus, it appears that bacteriophage-induced lysis is controlled in such a way that maintains the integrity of the cell for viral replication. Once the cell is de-energized, it rapidly lyses so that the viral particles can quickly enter the surrounding environment in search of new host cells.
So what dictates when a holin triggers cell lysis? The answer to this appears to be “programmed” into the structure of holins, since single-amino-acid changes within holins can either increase or decrease the timing of lysis (99). While the amino acid sequences of holins are quite divergent, most contain distinct structural features, including a relatively small size (60 to 145 amino acids), two or more putative membrane-spanning domains often separated by a predicted beta-turn linker region, a hydrophilic N terminus, and a highly polar, charge-rich C-terminal domain (284). Interestingly, holin genes often contain a so-called “dual-start motif” that produces two different protein products. The shorter form of the protein functions as the holin, while the longer version, which is often different by the addition of only a few amino acids to the N terminus, functions as the antiholin (26, 206). Despite the minor difference in protein length, the presence of these amino acids at the N terminus has a drastic effect on the function of these proteins. This effect is due to the fact that these additional amino acids almost invariably include lysine or arginine, whose positive charge has a dramatic impact on the orientation of the N terminus within the membrane. Using a series of experiments in which the N termini of the S105 holin and S107 antiholin of bacteriophage lambda were fused to a signal sequence, and then using signal sequence cleavage as a measure of translocation, Graschopf and Blasi (94) demonstrated that the presence of the additional positive charge at the N terminus of S107 orients the N terminus of this protein to the cytoplasmic face of the inner membrane. In contrast, the N terminus of the active S105 holin, which lacks this N-terminal positive charge, is localized to the periplasmic face of the membrane. The conclusion of this study was that the presence of an N-terminal-inside topology of S107 in the presence of an energized membrane confers an inhibitory effect to S105. As the holins accumulate within the membrane, a gradual dissipation of the proton gradient occurs, reaching a point that causes the N terminus of the antiholin to flip to the periplasmic face of the membrane. Once this occurs, the antiholin functions as a holin, and the complete and rapid de-energization of the membrane is achieved (26). Thus, two mechanisms appear to control the “lysis clock”: one that is programmed into the primary structure of the holin protein, presumably affecting the rate of proton leakage, and the other that involves alterations in the ratio between the holin and antiholin. Once the holin timer has gone off, exactly how it mediates the transport of the endolysin (in the case of λ and T4) is unclear. However, studies demonstrate that it likely does not involve the formation of a pore of a specific size. This conclusion was based on the observation that an endolysin fused to β-galactosidase still traversed the membrane in a holin-dependent fashion and retained both endolysin and β-galactosidase activities (266). An alternative model in which the holins accumulate into higher-order oligomers coalescing into lipid-excluding “rafts” was proposed (266). These rafts are believed to grow to a size that can no longer provide an effective barrier to the outside, thus resulting in the leakage of the cytoplasmic contents (including the cognate endolysin) and the eventual lysis of the cell.
Interestingly, not all bacteriophages utilize a holin-endolysin system. Bernhardt et al. (19, 20) demonstrated that bacteriophage φX174 encodes a “protein antibiotic” that targets translocase I (MraY), inhibiting the formation of the first lipid-linked intermediate in cell wall biosynthesis. Thus, this bacteriophage induces cell lysis using a mechanism similar to those of antibiotics like penicillin that target the cell wall synthesis machinery.
The Cid/Lrg Regulatory System
The cid and lrg operons.
The discovery and characterization of the cid and lrg operons evolved from the initial identification in 1996 of a novel two-component regulatory system from S. aureus, termed LytSR, that affected murein hydrolase activity and autolysis (31). As that study was conducted before the sequence of the staphylococcal genome was available, the identification of the lytSR operon was the fortuitous result of a random molecular-based search for novel two-component regulatory systems (16). That approach made use of degenerate oligonucleotide primers specific for the DNA sequences encoding the conserved regions of sensor histidine kinases with the goal of amplifying and cloning DNA fragments originating from sensor histidine kinase genes. Once isolated, the DNA fragments were used to generate mutations in the corresponding genes by homologous recombination. The idea was to then screen each of these mutants for phenotypes that were of interest (e.g., changes in virulence gene expression or altered antibiotic susceptibility). One of the genes identified, originally termed kin1, produced an interesting phenotype when disrupted. Upon growth in liquid culture, the kin1 mutant exhibited increased lysis and altered murein hydrolase activity relative to the parental strain (31), leading to the proposal that this gene was a novel regulator of autolysis. Analysis of the DNA sequences flanking kin1 revealed the presence of another gene immediately downstream encoding its cognate response regulator. Based on the phenotype of the kin1 mutation, this putative two-component regulatory system was renamed LytS and LytR. In a search for genes regulated by LytSR, we then examined the effect of the lytSR mutation on the expression of two downstream genes (32). That study revealed that these genes (designated lrgA and lrgB for LytSR-regulated genes A and B, respectively) were cotranscribed and that their expression was dependent on an intact copy of the lytSR operon.
Analysis of the lrgAB gene products demonstrated that they are both predicted to be extremely hydrophobic and likely to be integral membrane proteins (32). The amino acid sequence of the lrgA gene product (LrgA) contains 148 amino acids and has a deduced molecular mass of 16.3 kDa. Immediately adjacent to lrgA is another open reading frame, termed lrgB, encoding a 233-amino-acid protein (LrgB) with a molecular mass of 25.1 kDa. A clue to the function of LrgA was that it shares sequence characteristics in common with holins (32, 265). Interestingly, it had been speculated that holin-like proteins involved in the transport of bacterial murein hydrolases might exist, based on the observation that some murein hydrolases lack N-terminal signal sequences required for transport across the cytoplasmic membrane (230). The LrgA protein contains four putative membrane-spanning domains, two potential linker regions, and a charged rich amino-terminal domain. The function of lrgB was also addressed by generating an lrgB mutant and examining its phenotypic characteristics (32). Although no effect of this mutation on growth or autolysis was observed, zymographic analysis of this strain revealed the absence of a 25- to 30-kDa murein hydrolase, leading to the speculation that it encodes either a murein hydrolase or a regulator of murein hydrolase activity (32). However, given the predicted hydrophobic nature of the lrgA and lrgB gene products (96), it is unlikely that either of these proteins possess murein hydrolase activity. Consistent with this is the inability to demonstrate increased murein hydrolase activity in E. coli or B. subtilis strains expressing either the lrgA or the lrgB gene (unpublished results). The hypothesis that lrgA encodes an antiholin-like protein was supported by the finding that an lrgAB mutation resulted in increased murein hydrolase activity produced by the bacteria (96). This result is in contrast to the deletion of lrgB alone, as described above, which resulted in the loss of a specific murein hydrolase band by zymography. Deletion of the lrgAB operon also conferred decreased tolerance to penicillin but only in cells that were nearing stationary phase. However, the overexpression of lrgAB caused increased penicillin tolerance in both the lrgAB mutant and parental strains in early exponential phase, consistent with the observation that lrgAB transcription was only minimally expressed during the early exponential phase of growth (96). Based on these results, it was hypothesized that the lrgAB operon encodes an antiholin, although a detailed analysis of the individual genes remains to be conducted. Furthermore, due to the absence of a dual-start motif within the lrgA and/or lrgB gene, it was predicted that the holin protein component of this system would be encoded by another gene within the S. aureus chromosome.
Once the sequence of the S. aureus genome became available, this prediction was supported by the identification and study of lrgAB homologues that were designated cidA and cidB (213). The cidA gene product (CidA) shares 23% amino acid sequence identity with LrgA, whereas the cidB product (CidB) shares 31% amino acid sequence identity with LrgB. The putative CidA protein contains 131 amino acids and has a deduced molecular mass of 14.7 kDa, while CidB contains 229 amino acids and has a molecular mass of 25.0 kDa. Like LrgA and LrgB, the CidA and CidB proteins contain multiple predicted membrane-spanning domains. The putative function of the cid operon as an effector of murein hydrolase activity was demonstrated by generating a cidA mutant and showing that this strain produced decreased murein hydrolase activity relative to that of its parental strain (213). Furthermore, this mutation was also shown to confer tolerance to a variety of antibiotics including penicillin, rifampin, and vancomycin (213, 215). Overall, these results are consistent with the hypothesis that the cid operon encodes a holin and the lrg operon encodes an antiholin. One important problem with further genetic analysis of this operon is the inability to complement the cidA mutation (214, 215). Despite multiple attempts with a variety of vectors, the inability to complement the cidA mutation is unlikely to be due to the presence of a secondary-site mutation since similar cidA mutations in different S. aureus genetic backgrounds gave similar phenotypes. This phenotype was also not due to a polar effect on the downstream cid genes, since isogenic cidBC and cidC mutants grown under the same conditions resulted in a different phenotype (191; unpublished data). Thus, the reason for the inability to complement the cidA mutation remains unknown.
Based on the putative functions of the cidA and lrgA gene products as holins and antiholins, respectively, a model for their roles in murein hydrolase regulation is proposed. As depicted in Fig. 2, the fully energized membrane of a respiring cell produces a gradient of protons that reduces the pH of the environment within the IWZ and OWZ. As described above, the effect of membrane depolarization would be to destabilize the d-Ala ester linkages on the teichoic acids or to deprotonate the d-Ala ester residues, the consequence of which is predicted to relieve the repression of murein hydrolase activity. The activation of the CidA proteins via a holin-like mechanism is hypothesized to dissipate the membrane potential, thus triggering murein hydrolase activity and lysis. The presence of LrgA is thought to inhibit the activity of the CidA holin in a way analogous to the inhibitory effect of an antiholin. This model is consistent with the observation that most gram-positive murein hydrolases possess signal sequences but lack SAR domains that might attach them to the outer leaflet of the cytoplasmic membrane.
A major advance in the study of the cid and lrg operons was the understanding of the importance of using low-passage clinical isolates. This realization came after performing a careful analysis of cid transcription (216). In that study, it was found that the cid operon contained a third gene, designated cidC, encoding a pyruvate oxidase homologue. Unexpectedly, the cidB and cidC genes were found to be cotranscribed on a distinct transcript whose expression was dependent on the alternative sigma factor σB. A separate cidABC transcript was also detected but at much lower levels under the conditions tested. Given that the standard laboratory isolates used all contain a mutation affecting σB function in S. aureus, the clinical osteomyelitis isolate UAMS-1, which contains a functional σB, was employed for all subsequent studies. Interestingly, the generation of the identical cidA mutation in UAMS-1 resulted in a much more pronounced impact on murein hydrolase activity, reducing the amount of activity released into the extracellular environment to nearly undetectable levels (215). Similar to other mutations affecting murein hydrolase activity, this strain also formed multicellular aggregates when grown in liquid culture due to the inability of the cells to separate completely during growth. Furthermore, unlike the laboratory isolates containing the cidA mutation, lysis of the UAMS-1 cidA mutant in stationary phase was dramatically reduced, as determined by using both measurements of optical density of the cultures (191) and enzymatic assays of released cytoplasmic proteins (214). The dependence of cidBC expression on σB and the dramatic differences seen with the cidA mutation in UAMS-1 prompted speculation that the mechanism controlling cell death in S. aureus was a hotspot for the accumulation of mutations, since growth under laboratory culture conditions would likely select against genes promoting cell death (216). However, this remains to be tested by comparing this system in a variety of clinical and laboratory backgrounds.
Another important advance in this research was the recent discovery that the cid operon plays a significant role during biofilm development (214). This is evident in both static and flow-cell biofilm assays. Overall, the biofilm produced by the cidA mutant is more loosely compacted and is less adherent to the substrate. Several studies demonstrated the importance of cell death and lysis in biofilm development in other organisms (5, 157, 167, 198, 214, 233, 235, 268, 273). Most notable is the study by Allesen-Holm et al. (5), which shows that the specific lysis of Pseudomonas aeruginosa cells and release of DNA in a biofilm occur in an ordered pattern. This observation provided strong evidence for the programmed death and lysis of cells as a function of their spatial orientation within the biofilm, not unlike the role of apoptosis in development of more complex eukaryotic organisms. Those studies also revealed an important role for extracellular DNA (eDNA) as a matrix molecule, contributing to the overall structural stability of the biofilm. In support of a structural role for eDNA in a staphylococcal biofilm, treatment of wild-type S. aureus biofilm with DNase resulted in its destabilization, while a similar treatment of the lysis-defective cidA mutant had a minimal effect (214). Furthermore, treatment of the growing biofilm with the murein hydrolase inhibitor polyanethole sulfonate also reduced the adherence of the biofilm. These results indicate that the DNA released as a result of the cidA-mediated lysis of a subset of the bacterial population is an important cohesive component of the biofilm structure. Finally, additional experiments revealed that the cidA mutation caused reduced biofilm formation in animal models of biofilm development (214), indicating that the cidA-mediated control of cell death and lysis has clinical relevance.
The CidR and LytSR regulators.
Although much remains to be learned about the functions of the cid and lrg genes and their potential roles in bacterial PCD, considerable progress toward an understanding of the way in which these operons are regulated has recently been made. Specifically, studies have demonstrated that the transcription of both cidABC and lrgAB was induced by growth in the presence of excess glucose (Fig. 4), an effect that was shown to be a result of the metabolism of this carbohydrate and the subsequent generation of acetic acid (215). Interestingly, the cidC gene was found to encode a pyruvate oxidase that could contribute to the acetate (and acetic acid) accumulation in the culture medium during growth in excess glucose (Fig. 4) (191). Furthermore, cells containing a cidC mutation maintained a much higher level of cell viability in stationary phase than did the parental strain when grown in the presence of excess glucose (191). Besides providing a functional role for the cidC gene, these results demonstrated that growth in the presence of excess glucose enhanced the rate of cell death induced by antibiotics (215). Interestingly, recent studies of the pyruvate oxidase produced by Streptococcus pneumoniae have also revealed an important role for this enzyme in cell death during stationary phase (211). S. pneumoniae cells containing a mutation in the spxB gene encoding this pyruvate oxidase exhibited increased viability in stationary phase due to the absence of the reactive oxygen species (ROS) hydrogen peroxide, generated as a product of this enzyme's activity. Furthermore, the death process induced by hydrogen peroxide exhibited features similar to those of apoptosis in eukaryotic organisms, including alterations in membrane characteristics and increased degradation of DNA. More recently, Kohanski et al. (128) demonstrated that the treatment of both gram-positive and gram-negative bacteria with three major classes of bactericidal antibiotics, each with different cellular targets (cell wall biosynthesis, translation, and DNA replication), induced a common cell death response involving the generation of ROS. Importantly, bacteriostatic antibiotics did not induce this response. Although no studies have examined the effect of the cid and lrg operons on ROS-induced cell death, the presence of a gene encoding a pyruvate oxidase (cidC) within the cid operon makes this an intriguing possibility. Although ROS are thought to function by directly inducing cellular damage, the role of these molecules as signals in the induction of apoptosis (37, 145, 228, 256) indicates that a similar role in bacterial cell death may also exist.
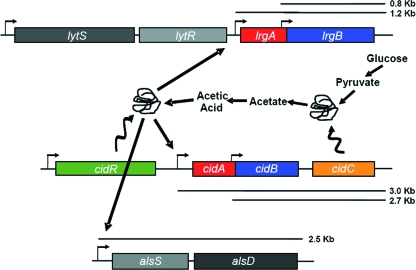
FIG. 4.
CidR-mediated regulation. The cidA and lrgA genes encode homologous hydrophobic proteins believed to function as a holin and an antiholin, respectively. The cidB and lrgB genes also encode homologous hydrophobic proteins (32) whose functions are unknown. The CidR protein, a LysR-type transcription regulator, enhances the expression of cidABC, lrgAB, and alsSD (encoding acetolactate synthase and acetolactate decarboxylase) in response to carbohydrate metabolism (280, 281). The cidC gene encodes pyruvate oxidase (191). The transcripts associated with each operon are indicated by black bars.
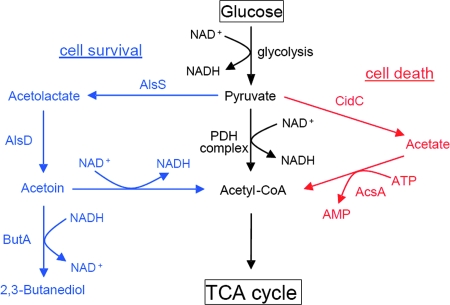
Studies investigating the mechanism by which cidABC and lrgAB transcription is induced by acetic acid led to the identification of the cidR gene (Fig. 4), encoding a putative LysR-type transcription regulator. The disruption of cidR resulted in the loss of both glucose- and acetic acid-induced cidABC and lrgAB transcription (281). The cidR mutation also caused reduced murein hydrolase activity and decreased survival in stationary phase (281). Although the reduction in murein hydrolase activity observed was not as dramatic as that observed with the cidA mutant, this is likely a result of the fact that baseline cidA transcription was still detectable (using RT-PCR) in the cidR mutant. To identify other CidR-regulated genes, a microarray approach was utilized to compare the transcription profiles of the cidR mutant and parental strains grown under inducing conditions. As expected, the cidABC and lrgAB operons were identified by this analysis. Surprisingly, the only other genes identified were the alsSD operon, whose products (acetolactate synthase and acetolactate decarboxylase) (Fig. 4) are required for acetoin production (280). Like the cidC-encoded pyruvate oxidase, the alsSD gene products function to convert the pyruvate formed under conditions of excess glucose to a form that could be utilized at a later time. However, unlike pyruvate oxidase, the alsSD gene products generate the neutral compound acetoin.
Two notable findings from the study of the alsSD mutant were the observation that this strain exhibited a rapid cell death (RCD) phenotype in stationary phase (280) and that the transcription of cid and lrg was not inducible by glucose or acetic acid in a strain harboring the alsSD mutation. The latter observation suggests that the pathway in which pyruvate is converted to acetoin produces a metabolite required for the CidR-mediated induction of cidABC and lrgAB transcription. Inspection of the biochemical pathways responsible for the conversion of glucose to acetyl coenzyme A (acetyl-CoA) reveals an apparent balance between the AlsSD and CidC pathways that impacts cell viability (Fig. 5). Although one pathway in which pyruvate is metabolized likely involves the multienzyme pyruvate dehydrogenase complex (261), the AlsSD and CidC pathways provide alternative routes that are activated in response to ill-defined physiological signals. The former appears to be important for converting NAD+ to NADH (via the conversion of acetoin to acetyl-CoA) consumed in the glycolytic pathway, as the alsSD mutant exhibits a dramatically increased NAD+/NADH ratio compared to the parental and complemented strains (unpublished data). Whether or not these changes in the NAD+/NADH ratio (or some other aspect of the redox state of the cell, including ROS levels) are important in CidR signaling and/or the induction of cell death is currently under investigation. Interestingly, mitochondria are thought to integrate a variety of cellular signals including NAD+/NADH in the control of apoptosis (139). Perhaps a careful look at these signals will provide clues to additional signaling molecules that control Cid/Lrg-mediated cell death and lysis.
FIG. 5.
Conversion of pyruvate to acetyl-CoA in S. aureus. A major pathway involved in the conversion of pyruvate to acetyl-CoA (shown in black) in bacteria requires the pyruvate dehydrogenase (PDH) complex. Other pathways involved include the AlsSD pathway (blue) and the CidC pathway (red), which appear to promote cell survival and death, respectively. Enzymes contributing to these pathways include acetolactate synthase (AlsS), acetolactate decarboxylase (AlsD), pyruvate oxidase (CidC), and acetyl-CoA synthetase (AcsA). Also shown is the conversion of acetoin to 2,3-butanediol, requiring the enzyme acetoin reductase (ButA).
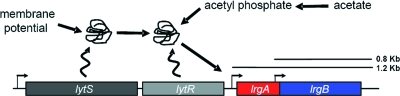
As described above, the control of lrgAB expression has been shown long ago to involve the S. aureus LytSR two-component regulatory system (32) (Fig. 6). Despite the fact that this system was first described over a decade ago (31), the specific signal to which it responds has only recently been investigated. The extensive hydrophobic nature of the LytS sensor domain (31), along with the role that PMF has in the regulation of autolysis in B. subtilis (described above), suggests that the LytS protein responds to a membrane-associated signal. In support of this hypothesis, our studies have shown that agents known to dissipate the membrane potential (Δψ) strongly induce lrgAB but not cidABC transcription (192). The induction of lrgAB expression did not correlate with intracellular ATP levels, indicating that an indirect effect of altered Δψ on ATP synthesis did not affect the expression of this operon. Furthermore, the effect of membrane potential on lrgAB expression was shown to be dependent on lytSR and independent of cidR, clearly demonstrating the presence of two independent regulatory pathways controlling cid and lrg expression. Recent studies demonstrated that the induction of lrgAB expression by acetic acid is dependent on LytR (but not LytS) (unpublished data), suggesting that small-molecule phosphodonors (perhaps acetyl phosphate) (125) directly activate this response regulator as an alternative signaling mechanism involving the LytSR pathway (Fig. 6). Although the biological function of the LytSR regulatory system is unknown, one possibility is that this system provides a means by which the cell can sense its overall metabolic state (192). If the cell is damaged in some way that reduces its ability to maintain a normal energy balance due to the reduction in Δψ, LytSR might function to sense these changes and induce the cell death pathway so that the cellular components can be recycled (14, 152).
FIG. 6.
LytSR-mediated regulation. The LytSR two-component regulatory system is hypothesized to sense decreases in membrane potential and respond by inducing lrgAB transcription (192). Recent evidence also suggests that small-molecule phosphodonors can phosphorylate LytR and induce lrgAB transcription. The overlapping transcripts associated with the lrgAB operon are indicated by black bars.
Similarities between the Cid/Lrg regulatory system and apoptosis.
As previously described, the cid and lrg regulatory system is widely conserved in bacteria (15), suggesting that this mechanism controlling cell death is a common feature in bacterial physiology. Interestingly, this mechanism also shares remarkable similarity with the control strategies mediated by the Bcl-2 family of proteins in the regulation of apoptosis (14). The Bcl-2 proteins are a large family of proteins that are well conserved in eukaryotic organisms. Similar to bacteriophage-encoded holins, Bax can cause membrane permeabilization in a process requiring protein oligomerization (7, 77, 286). Again, similar to holins, the molecular details of this process are obscure, but given the size of the proteins that are released (142) and the biophysical impact of Bax (and other proapoptotic proteins) on membranes (13), it is likely that “pore” formation involves lipid destabilization. The consequences of permeabilization include the release of cytochrome c and other proteins, but whether or not this also involves the depolarization of the mitochondrial inner membrane is hotly debated (7, 139). The release of cytochrome c subsequently triggers the caspase cascade, the central effector of the execution/degradation phase of apoptosis. This phase has been described as being a postmortem process involving cellular disassembly that is independent of the mitochondrial pathway involved in apoptosis induction (139). Indeed, the mitochondrial pathway (also known as the “intrinsic” pathway) can induce death in the absence of caspases (40, 139). Like antiholins, Bcl-2 (and related antiapoptotic proteins) can interact with Bax to inhibit the induction of cell death, although the mechanism by which this occurs is also controversial (139). The remarkable similarities between Bax/Bcl-2-mediated control of apoptosis and the holin/antiholin-mediated control of bacterial death and lysis have recently led to the hypothesis that the events leading to bacterial autolysis are analogous, both biochemically and physically, to the events leading to the disruption of mitochondria during the initial stages of apoptosis (15). Moreover, the steps leading to tumor formation (oncogenesis) are not unlike the steps involved in the development of antibiotic resistance. Initial mutations and transcriptional alterations of the bacteria result in tolerance to antibiotics in which the cells can sustain viability in the presence of normally lethal antibiotic doses (165, 171). Interestingly, an increased expression of the B. subtilis lrgAB homologues (ysbAB) has been observed after treatment of these bacteria with antibiotic compounds (153). This change presumably buys time for the acquisition of mutations that lead to resistance or the ability to grow in the presence of these levels of antibiotics. In contrast, Renzoni et al. (212) detected increased cidABC expression and decreased lrgAB expression in S. aureus cells that had acquired resistance to teicoplanin. During the development of a tumor, initial changes often result in elevated levels of Bcl-2, which prevents cell death under conditions that would normally induce apoptosis. Like the secondary mutations in bacteria leading to resistance (171), cancer cells ultimately acquire mutations, such as within the c-myc gene, that result in the uncontrolled replication of the cell (264) despite the presence of cellular factors that would normally prevent cell proliferation.
Cell death and its well-established role during viral infection of eukaryotic cells (274) could also provide an advantage to bacteria as a means to protect against bacteriophage infection. Indeed, it was the characterization of the adenovirus E1B 19K protein, a Bcl-2 homologue (274), that played an important role in defining the function of Bcl-2 as an inhibitor of apoptosis. This was discovered by observing that in contrast to wild-type adenovirus, infection with E1B 19K mutant virus caused the degradation of the host cell DNA (202, 239, 245, 275), a classic sign of apoptosis. Thus, those studies suggested that the host defends itself against viral infection by inducing PCD in infected cells to limit infection to neighboring cells. Furthermore, those studies also indicated that the viruses counter this strategy by producing a protein that inhibits apoptosis during infection. Based on this precedence, we propose that a similar strategy is utilized by bacteria to protect against bacteriophages and that one of the functions of the LytSR system is to sense infection. Studies have shown that upon bacteriophage attachment, a transient depolarization of the membrane occurs (143, 263). As a sensor of decreases in Δψ (192), the LytSR regulatory system could sense bacteriophage-induced membrane depolarization and respond by inducing the transcription of the lrgAB operon. As an inhibitor of cell lysis, the products of lrgAB might allow bacteriophage replication and assembly but would prevent cell lysis and dissemination to other cells within the population. A similar hypothesis was proposed for the role of the MazEF toxin-antitoxin system in the defense against bacteriophage P1 infection of E. coli (105). Interestingly, some bacteriophages induce a form of altruistic death of their own hosts to prevent exogenous bacteriophages from infecting and spreading to other lysogenized host cells. These bacteriophage exclusion systems (231) are quite diverse and are fairly well characterized. The best known of these, the RexAB system of bacteriophage lambda, becomes activated upon infection of a lambda lysogen. Once activated, RexA interacts and activates RexB, which targets and depolarizes the cytoplasmic membrane, leading to cell death.
Another similarity between the Cid/Lrg system and apoptosis is the role that carbohydrate metabolism has in these processes. As described above, the metabolism of glucose appears to play a central role in the control of cid/lrg expression (Fig. 4) and in cell death (175, 215). Interestingly, the role of glucose metabolism in the control of apoptosis has also been demonstrated. This line of research originated back in the 1930s with the observation that tumor cells metabolize high levels of glucose by glycolysis despite the presence of an adequate oxygen supply. In fact, it is now widely believed that most, if not all, cancer cells exhibit increased glucose uptake and metabolism (74). This observation led to the hypothesis that tumor cells contain dysfunctional mitochondria, leading to “aerobic glycolysis” or the “Warburg effect” (124). Recent studies indicate, however, that rather than being dysfunctional, the mitochondria in tumor cells undergo a physiological “remodeling” that promotes aerobic glycolysis (27). It is thought that the utilization of glycolysis is essential early in the transformation of a cell, which typically occurs in a hypoxic environment prior to vascularization (74). Interestingly, it appears that this glycolytic phenotype is associated with the suppression of apoptosis and resistance to the acidosis produced as a consequence of increased lactic acid generation (74, 203). Indeed, it is thought that during carcinogenesis, tumor cells “evolve” phenotypic adaptations to the toxic effects of acidosis, culminating in resistance to apoptosis (73). Moreover, the continued use of glycolysis after vascularization is thought to be advantageous to the tumor cells for tissue invasion, angiogenesis, and metastasis (74) as well as for supplying the building blocks needed for macromolecular synthesis during rapid growth. It is of interest that many bacteria also utilize “aerobic glycolysis” to supply the building blocks needed for macromolecular synthesis during exponential growth (232). These bacteria metabolize glucose in the presence of oxygen via glycolysis and inhibit the tricarboxylic acid cycle, resulting in the secretion of large amounts of acetate into the growth medium. Only after glucose is depleted is the acetate taken up and catabolized via the tricarboxylic acid cycle. As shown Fig. 5, the fate of pyruvate in bacteria (whether it is converted to acetic acid or acetoin) appears to be a key determinant in the decision between life and death, similar to the role that the pyruvate dehydrogenase complex has in controlling the commitment to apoptosis (27, 203). Thus, in both prokaryotes and eukaryotes, evidence suggests that rapid growth is fueled by glycolysis and that pyruvate metabolism plays a critical role in the control of cell death.
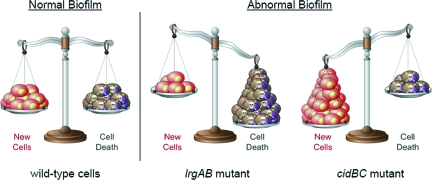
Finally, it is noteworthy that disrupting the balance between life and death in eukaryotic cells can lead to a variety of diseases (109, 264). For example, tipping the balance toward cell death by the overexpression of Bax or the underexpression of Bcl-2 has been shown to be associated with the development of neurodegenerative disorders (12, 135, 246) and heart disease (158, 173, 186). In contrast, the inability to induce cell death when needed leads to the uncontrolled proliferation of cells and cancer (264). A similar balance between the expression of the S. aureus cid and lrg appears to be important for the formation of normal biofilm. Preliminary studies revealed that biofilms produced by the cid and lrg mutants produce opposing biofilm phenotypes based on their effects on cell viability and lysis during development. Whereas the cidA mutant produces a biofilm that accumulates dead cells due to the absence of cell lysis, an lrgAB mutant produces a biofilm that exhibits increased lysis (unpublished data). Both of these mutations had a dramatic effect on biofilm morphology, producing an amorphous mass of cells lacking the distinct three-dimensional tower structures that are characteristic of the wild-type parental strain. Although similar effects on cell death and lysis were produced by disruption of the Pseudomonas aeruginosa cid and lrg homologues, the impact of these mutations on tower development was less dramatic (unpublished data). Thus, similar to the balance between life and death that is afforded by the control of apoptosis in the maintenance of tissue homeostasis (264), a similar balance is observed in the development of a bacterial biofilm (Fig. 7). Moreover, the control of this balance via the differential expression of cid and lrg has recently been proposed to contribute to the tolerance of biofilm cells to antibiotic treatment, analogous to the Bax/Bcl-2-mediated tolerance of tumor cells to chemotherapeutic agents (15).
FIG. 7.
Balance between life and death within a biofilm. Bacterial PCD maintains biofilm homeostasis by balancing cell death and viability during development. Disruptions of this balance, for example, by introducing mutations within the cid and lrg operons, result in defective biofilm formation. (Adapted from reference 264 by permission from Macmillan Publishers Ltd.)
Bacterial Caspases
Thus far, we have argued that the Cid/Lrg regulatory system is functionally analogous to the eukaryotic Bax/Bcl-2 system that lies at the heart of the control of apoptosis. Since bacteria lack the complexity of a eukaryotic cell, it was speculated that the apoptosis machinery outside of the mitochondria (e.g., the caspase pathway) was unique to eukaryotic organisms (15). After all, the caspase pathway is responsible for the cellular disassembly that occurs during apoptosis, including DNA fragmentation, chromatin condensation, membrane blebbing, cell shrinkage, and disassembly into membrane-enclosed vesicles (247). Thus, it comes as a surprise that a few examples of caspase-like enzymes produced by bacteria might exist. In the plant pathogen Xanthomonas campestris, RCD during nutrition stress in the postexponential phase was shown to correlate with the production of a protein that reacts with antibodies generated against human caspase 3 (75). This RCD phenotype was associated with membrane changes and DNA fragmentation, both commonly observed features of apoptosis in eukaryotic organisms. Furthermore, RCD and the production of the caspase-like protein were shown to be inhibited by the presence of starch or by incubation at 4°C (75, 76). Subsequently, it was shown that the RCD phenotype was associated with the intracellular accumulation of pyruvate and citrate and was inhibited by the hydrolytic products of starch, dextrin, and maltose (207). Although these findings demonstrate once again that central carbohydrate metabolism is involved in the control of cell death, similar to the role of pyruvate and acetic acid accumulation in S. aureus (191), acetic acid did not induce the RCD phenotype in X. campestris (207). More recently, studies demonstrated that poly(ADP-ribose) polymerase (PARP) activity, another hallmark of apoptosis, is associated with the production of the X. campestris caspase (208). This activity was found to be responsible for the depletion of NAD+ during the induction of RCD and that the addition of compounds known to specifically inhibit PARP activity prevented NAD+ depletion and RCD. Furthermore, PARP and caspase-specific antibodies cross-reacted with what appeared to be the same protein as well as a cloned and expressed X. campestris protein (polysaccharide deacetylase) containing a “caspase-like” domain. The conclusive identification of this protein as a participant in the induction of RCD X. campestris remains to be established.
Another example of caspase activity and “autocatalyzed PCD” in prokaryotic organisms is provided by studies of marine phytoplankton (22), particularly the cyanobacterium Trichodesmium sp., which demonstrates rapid growth in the ocean and laboratory settings, followed by abrupt cell lysis and biomass degradation (184). This process was shown to function in aging cultures and in response to conditions associated with the marine environment (18) and was accompanied by distinct morphological changes including the degradation of intracellular compartments (thylakoids, carboxysomes, gas vacuoles, and cyanophycin granules). These cells also exhibited signs of DNA degradation and cell shrinkage but retained normal cell membrane integrity. The entry into the death phase was also associated with the production of a protein that cross-reacted with polyclonal antibodies to human caspase-3 as well as with the cleavage of a caspase-specific substrate. Importantly, caspase activity was inhibited by a specific inhibitor of caspase activity, providing evidence that a true caspase was identified. A functional role of PCD was demonstrated by establishing that cells exposed to high light irradiation (as commonly occurs in the marine environment) exhibited increased caspase activity, enhanced sinking (presumably due to the loss of gas vesicles), increased vacuolization, and the selective removal of cells with high caspase activity. Thus, it was speculated that this process facilitates biogeochemical cycling through the transfer of organic and inorganic matter to heterotrophic microbial communities. Overall, these findings were interpreted as support for the hypothesis that this system provided the origins for the evolution of caspase-dependent PCD in higher plants and animals (22). However, follow-up experiments to identify a caspase-like gene as well as a demonstration that this gene is involved in the death of this organism remain to be performed. If a gene encoding a caspase-like enzyme can be identified in this organism, it may represent the exception rather than the rule.
Fratricide
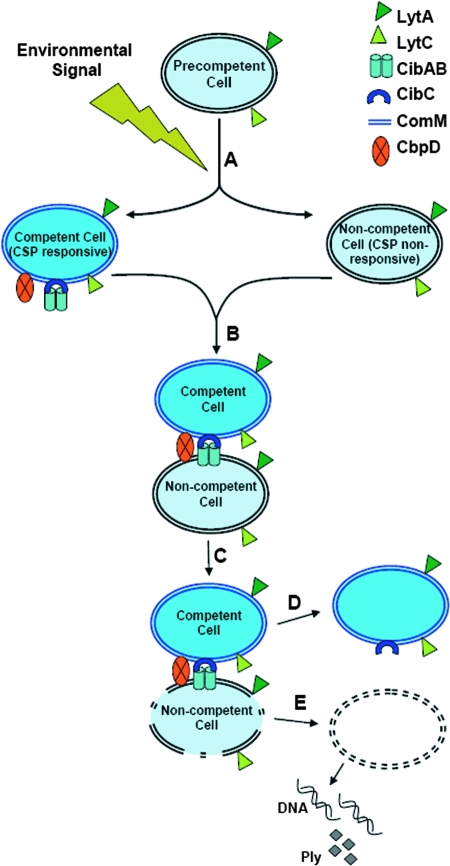
In addition to the examples of altruistic cell suicide (whereby the death of a bacterial cell is self-inflicted for the benefit of the community) discussed above, bacterial populations can also undergo cell death when certain cells in the population kill other sibling cells. One recently identified example of this kind of cell death is the phenomenon of fratricide during competence development of S. pneumoniae. The ability of a population of S. pneumoniae cells to become competent (reviewed in references 45, 46, 47, and 48) is regulated by the ComDE two-component regulatory system. The ComD membrane-bound histidine kinase senses the accumulation of a peptide pheromone called competence-stimulating peptide (CSP) (encoded by the comC gene), which is secreted by the growing S. pneumoniae culture. When the extracellular concentration of CSP reaches a threshold level, it binds to ComD and triggers its autophosphorylation. The phosphoryl group is then transferred to the cognate response regulator ComE, which in turn upregulates the expression of the “early” com genes. One of these early com genes encodes sigma factor X (ComX), which subsequently regulates the expression of the “late” com genes, including the genes necessary for DNA binding, uptake, and recombination. Although this developmental process has been well studied for many decades, relatively little was known about how donor DNA was made available during competence development in the environment. A breakthrough in this field was made when it was shown, by measuring the release of either β-galactosidase (236, 237), pneumolysin (Ply) (an intracellular β-hemolysin) (101), or chromosomal DNA (166, 236, 237) into the culture supernatant, that a lysing subpopulation of cells appeared during natural competence development in S. pneumoniae. The emergence of this lysing subpopulation is dependent on the ComCDE regulatory system (166, 236, 237) and results in the release of chromosomal DNA that could be used as a source of donor DNA for natural transformation (236). It was also shown by cocultivation experiments using mutants deficient in various components of the ComCDE system that two populations of cells are present during competence development: one population of competent, nonlysing cells that lyse the second population of noncompetent cells (101, 237). In other words, during competence development, donor DNA is provided by heterolysis/allolysis (lysis of one bacterial cell that is caused by another cell) as opposed to autolysis (lysis of self) (101, 237). Furthermore, the classic observation that competent S. pneumoniae cells tend to form aggregates when treated with mild acid (251) was found to be dependent upon the presence of a mixture of competent and noncompetent cells as well as the release of extracellular DNA (103). The phenomenon of competence-induced cell lysis was subsequently named “pneumococcal fratricide,” defined as the intraspecies-specific killing of cells that occurs during the development of competence in S. pneumoniae (47, 103).
Two important questions that needed to be addressed in the study of fratricide were (i) what factor(s) produced by the competent cells is responsible for killing their noncompetent siblings, and (ii) how do the competent cells protect themselves from these killing factors? So far, four such “killing” factors have been implicated in fratricide. LytA is a cell wall-associated murein hydrolase (91, 154) that is upregulated in response to the ComDE two-component regulatory system (36, 199, 218). Its role in fratricide was demonstrated by the fact that the competence-induced release of β-galactosidase and chromosomal DNA was significantly reduced in a lytA mutant (166, 236, 237). LytC is another cell wall-associated murein hydrolase that appears to play a role in fratricide; the inactivation of lytC resulted in reduced lysis during competence development, as measured by chromosomal DNA release (166). Additionally, a lytA lytC double mutant displayed a near-complete abolishment of chromosomal DNA (166) and Ply (101) release. Furthermore, the LytA and LytC required during allolysis can be provided by either the competent nonlysing cells or the noncompetent cells that are targeted for killing (101, 103). A third novel murein hydrolase, named choline-binding protein D (CbpD), has also been shown to be necessary for fratricide to occur during competence development (101, 119). The expression of cbpD is highly upregulated during competence induction (119, 200, 218), and both competence-induced cell lysis and DNA release were strongly reduced in a cbpD mutant (119). Furthermore, allolysis (as measured by the release of Ply) was completely abolished in a lytA lytC cbpD triple mutant (101). Finally, the cibAB genes encode a predicted two-peptide bacteriocin whose expression is upregulated by competence induction (199, 218). CibAB has also been shown to be involved in fratricide, as a cibAB mutant was impaired in its ability to release Ply from noncompetent cells targeted for killing (101). Based on these results, a model of fratricide was proposed (Fig. 8) whereby CibAB from the competent cell population acts as a trigger factor for allolysis by killing the noncompetent cells, with their subsequent cell lysis occurring as a secondary event (47, 101). Although the exact mechanism of CibAB function has not been directly demonstrated, it is believed that cell-to-cell contacts between competent and noncompetent cells are necessary for the lethal action of CibAB (101). It was also speculated that CibAB may function by inserting into the cell membrane of target cells and dissipating their membrane potential, similar to the action of other peptide bacteriocins (101, 257). Thus, one obvious mechanism for CibAB function could involve the sensitization of the cell wall to murein hydrolase activity via the model depicted in Fig. 2.
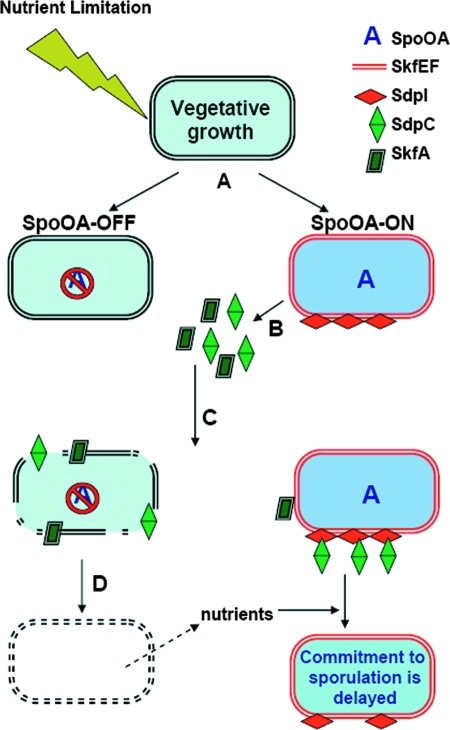
FIG. 8.
Fratricide during competence development in S. pneumoniae. (A) An environmental signal(s) leads to the emergence of two subpopulations, competent cells (CSP responsive) and noncompetent cells (CSP nonresponsive), presumably via a bistable regulatory mechanism. Competent cells express the lytic factor CbpD, putative two-peptide bacteriocin CibAB, and immunity proteins ComM and CibC, whereas the murein hydrolases LytA and LytC are expressed by both competent and noncompetent cells. (B) Cell-to-cell contacts between competent and noncompetent cells allow access of CibAB and CbpD to the noncompetent cells. (C) CibAB triggers the lytic action of CbpD, LytA, and LytC. (D) Competent cells are protected from the actions of these enzymes by the expression of immunity factors CibC and ComM. (E) Noncompetent cells lack these immunity proteins and undergo allolysis, releasing DNA (used for genetic transformation of competent cells) and virulence factors (Ply). (Adapted from reference 45 by permission from Macmillan Publishers Ltd.)
To prevent the lysis of the competent cell population during fratricide, factors that confer protection against the action of the killing factors produced by these cells must be present. To date, two such immunity factors have been identified. The cibAB operon contains a third gene, cibC, which encodes a predicted 65-amino-acid protein with two transmembrane-spanning domains (101). It was found that the expression of the full-length cibABC transcript occurred only in competent cells, and competent cells defective in cibC became sensitive to allolysis in a mixed culture, suggesting that CibC confers immunity to CibAB in competent cells (101). It was also observed that competent cells lacking a functional ComX (and therefore unable to induce the expression of the late com genes) were still resistant to lysis during the development of competence, suggesting that one or more of the early com genes also encode an immunity factor (103). This “early” immunity factor was subsequently identified as ComM, a predicted integral membrane protein (103, 126). The inactivation of comM rendered competent cells sensitive to lysis during fratricide, and comM mutant cultures displayed a drop in optical density relative to that of the parental strain after CSP induction, suggesting that ComM confers immunity against the action of the cells' lytic machinery (103). Despite the progress made in understanding the functions of CibC and ComM, the exact mechanism by which these proteins protect competent cells from killing and/or lysis remains to be determined.
Although many of the molecular components involved in fratricide have been defined, the mechanism governing which cells will become competent and which cells are destined for death in a genetically identical cell population remains to be demonstrated. However, this mechanism is likely similar to the bistable regulatory switch that controls the expression of comK, which encodes the master regulator of competence development in B. subtilis (10, 156, 240). In addition to providing a source of donor DNA during competence development, it is also possible that fratricide fulfills other functions necessary for S. pneumoniae physiology and development. These functions include mediating the release of virulence factors such as pneumolysin (101) during infection of the host or perhaps the release of DNA during biofilm development. eDNA has been shown to be an important component of the biofilm matrix of several microorganisms (5, 167, 198, 214, 233, 235, 273), and it has recently been shown that the release of eDNA is important for biofilm development in S. pneumoniae (167).
Cannibalism
Another example of fratricidal cell killing within a population is cannibalism during B. subtilis sporulation. Sporulation (reviewed in references 45, 110, and 201) is a differentiation process whereby a dormant cell (the endospore) that is able to survive harsh environmental conditions until such time that growth conditions improve is produced. In B. subtilis, sporulation normally occurs as a last-resort response to nutritional stress, but entry into sporulation is also regulated by other signals such as cell density and the cellular redox state (reviewed in reference 201). The various signals and inputs required for entry into sporulation feed into the Spo0A protein, the master regulator of sporulation (201). In a given population of genetically identical cells, only a subset of these cells actually initiates sporulation (44), and this developmental process is irreversible once the asymmetrically positioned division septum (separating the prespore and mother cell) is formed (190). The “decision” by an individual cell to enter into sporulation is regulated by a bistable switch that controls the phosphorylated state of Spo0A and accounts for why some cells contain active Spo0A and others do not (44, 54, 259, 260). Since this developmental process is very time- and energy-consuming, cells need a means to delay spore formation for as long as possible in case the nutritional stress is only a short-term event. For example, if favorable growth conditions were to resume, cells that have irreversibly committed to sporulation would be at a growth disadvantage relative to vegetative cells that could rapidly reinitiate cell division (88). A mechanism by which this is accomplished in B. subtilis, whereby cells that have entered the sporulation pathway (but have not yet passed the irreversible stage of sporulation) are able to block sibling cells from sporulating and kill these cells in order to feed on their nutrients, thereby delaying a commitment to sporulation, has recently been identified (88). This mechanism was dubbed cannibalism (88) and is similar to fratricide in that two groups with distinct fates (“killer” cells verses “victim” cells) arise from a genetically identical population of cells (Fig. 9).
FIG. 9.
Cannibalism during B. subtilis sporulation. (A) Nutrient limitation leads to the emergence of two subpopulations, SpoOA-ON (cells that have entered the sporulation pathway but have not yet passed the irreversible stages of sporulation) and SpoOA-OFF (nonsporulating cells), via a bistable regulatory switch. (B) SpoOA-ON cells express the killing factors SkfA and SdpC as well as their cognate immunity proteins SkfEF and SdpI. (C) SpoOA-OFF cells lack the immunity proteins SkfEF and SdpI and therefore are susceptible to the lethal action of SkfA and SdpC; SpoOA-ON cells are protected from killing by the immunity proteins SkfEF and SdpI. (D) SpoOA-ON cells are able to delay their commitment to sporulation by feeding on the nutrients released from their dead siblings (SpoOA-OFF cells). (Adapted from reference 45 by permission from Macmillan Publishers Ltd.)
Cannibalism was first described by Gonzalez-Pastor et al. (88), who, in generating a mutant library of genes under the control of Spo0A, discovered two loci that are strongly expressed in cells that have entered the sporulation pathway (referred to herein as “Spo0A-ON” cells), and mutations in these loci accelerated sporulation. One of these was the eight-gene skf (sporulation killing factor) (skfABCDEFGH) operon, whose predicted gene products display homology to proteins involved in the production of peptide antibiotics (131). The cocultivation of wild-type and skfABCDEF deletion mutant (ΔskfABCDEF) strains revealed that the mutant was more sensitive to killing after the onset of sporulation. Furthermore, when grown in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside), cells expressing the skf operon under the control of an IPTG-inducible promoter were able to kill wild-type cells (presumably the Spo0A-OFF cells) as well as ΔskfABCDEF mutant cells (88). Those results suggest that the skf operon, expressed in Spo0A-ON cells, encodes both a killing factor that targets nonsporulating cells as well as an immunity protein that protects the Spo0A-ON cells from the action of the killing factor. Based on sequence similarity to a bacteriocin-like protein, it was proposed that skfA encodes the killing factor (88). Also, the expression of the skfEF genes, which are homologous to ABC-type transporters, in a ΔskfABCDEF mutant rendered the mutant cells less susceptible to killing by Spo0A-ON wild-type cells, suggesting that SkfEF provides immunity to Spo0A-ON cells expressing the skf operon (88). It was also recently shown that a separate skfA-specific transcript is highly expressed in response to phosphate starvation via regulation by PhoP, whereas the induction of the full-length skf transcript occurred to a lesser degree (4). The presence of a potential stem-loop structure between the skfA and skfB genes was thought to account for the apparent stability of the skfA transcript, allowing the production of an excess amount of the SkfA killing factor in relation to the rest of the products of the skf operon (4). It was speculated that this regulation coordinates the expression of all the genes in the skf operon while allowing the preferential transcription and translation of skfA mRNA (4).
The second locus identified by Gonzalez-Pastor et al. is the three-gene sdpABC (sporulation delay protein) operon. A mutation within this operon resulted in rapid sporulation and reduced expression of the sdpRI operon adjacent to sdpABC (56, 88). The factor responsible for this regulation is a 5-kDa protein originating from the sdpC gene that acts as an intercellular effector of sdpRI expression as well as a toxin against Spo0A-OFF cells (56, 88). SdpI is a predicted membrane protein, and SdpR is a potential transcriptional regulator (88). Mutational analysis of SdpI revealed that this protein acts as both a signaling molecule as well as an immunity protein against the toxic nature of SdpC (56). Based on those studies, it was proposed that SdpR, SdpI, and SdpC comprise a three-component signal transduction pathway: SdpC binds to the receptor-signal transduction membrane protein SdpI, inducing a conformational change that allows SdpI to bind and sequester SdpR, which is thought to have an autorepressive effect on the transcription of sdpRI. The binding of SdpR by SdpI-SdpC would thus allow the expression of sdpRI in Spo0A-ON cells and confer protection against the toxic action of SdpC (56). Since the expression of sdpRI is under the indirect control of Spo0A, the Spo0A-OFF population is not able to synthesize the immunity protein SdpI and therefore is susceptible to killing by SdpC (56).
One puzzling aspect of cannibalism is explaining how the expression of the skf and sdp operons is temporally controlled in Spo0A-ON cells such that they are expressed during the early, reversible stage of sporulation. This has recently been addressed by the observation that genes not directly contributing to sporulation, such as those involved in cannibalism, are actually activated earlier and at lower concentrations of phosphorylated Spo0A than genes that are directly involved in spore formation (67). Another interesting twist to the cannibalism story is the recent observation that when grown under conditions of nutrient limitation in the presence of either E. coli or other bacteria, B. subtilis preferentially kills the second bacterial species before resorting to cannibalism (172). The killing of E. coli in these cocultivation experiments appeared to be dependent on Spo0A but independent of SkfA, suggesting the presence of additional killing factors that aid in the predation of E. coli by B. subtilis (172).
Toxin-Antitoxin Systems
The term “toxin-antitoxin (TA) module” is used to describe a family of gene pairs whereby one gene encodes an unstable antitoxin that inhibits the potentially lethal action of its cognate toxin encoded by the second gene. These genes were originally identified on extrachromosomal DNA such as low-copy plasmids (30, 116, 183, 254) and prophages (148) and function to ensure the propagation of these otherwise nonessential genetic elements by killing cells that no longer harbor them. When cells are cured of these genetic elements, for example, during cell division, the unstable antitoxin is rapidly degraded, usually by a specific cytosolic protease, and thus allows its relatively stable cognate toxin to kill the cell in the absence of the de novo expression of the antitoxin. These TA genes have also been dubbed “addiction modules” since their lethal action causes the bacterial host to be “addicted” to the ongoing presence of these extrachromosomal elements (148, 282).
More recently, inspection of sequenced microbial genomes has revealed the presence of TA modules on the chromosomes of many bacteria (2, 57, 97, 164, 189). The number of TA loci varies from none (as in the case of many obligate host-associated bacteria) to as many as 45 found in the Nitrosomonas europaea genome (189). The chromosomal TA modules of E. coli have thus far been the most extensively characterized, but note that chromosomal TA modules in other organisms including Streptococcus mutans (150), Bacillus anthracis (1), S. aureus (66), and S. pneumoniae (123, 178) have recently been studied. Several reviews detailing the advances in chromosomal TA module research have been published (33, 57-60, 80); therefore, this review will focus on two well-characterized E. coli TA modules, MazEF and RelBE, as well as on the E. coli TA-like module HipBA, highlighting the evidence for and evidence against their controversial roles in eliciting bacterial cell death.
MazEF.
The E. coli mazEF genes were first identified by Metzger et al. (163) as being located immediately downstream of relA, whose gene product synthesizes 3′,5′-bispyrophosphate (ppGpp) during the stringent response (reviewed in references 29 and 39). Sequence analysis revealed that the predicted protein sequences of MazE and MazF were homologous to the antitoxin and toxin, respectively, encoded by the pemI/pemK plasmid addiction module (159). Aizenman et al. (2) subsequently demonstrated that MazEF displays many characteristics of TA modules: mazEF or mazE expressed alone from an inducible promoter did not affect the viability of E. coli cells, whereas cells expressing mazF alone experienced a 4-log decrease in cell viability. That study also showed that the MazE protein is sensitive to proteolysis by ClpPA and was unstable (30-min half-life), whereas MazF was stable for at least 4 h under the same experimental conditions (2). Collectively, those results demonstrated that mazE and mazF encode the antitoxin and toxin components, respectively, of this chromosomal TA module. Interestingly, they also found that mazEF transcription was negatively regulated by ppGpp accumulation, either due to the artificial overproduction of ppGpp or by inducing amino acid starvation, and that both mazEF and clpP mutants displayed increased cell survival relative to that of the parental strain when cultures were induced with ppGpp (2). Those results led the authors of that study to propose a model of MazEF-dependent PCD (2): the production of ppGpp by RelA (in response to amino acid starvation) inhibits the expression of the mazEF transcript and decreases the amount of the unstable MazE antitoxin in the cell. This, combined with proteolytic degradation of MazE by ClpAP, allows the relatively stable MazF toxin to exert its lethal effect on the cell.
Subsequent studies reported that MazEF-dependent cell death could be triggered by a variety of different stresses. When wild-type E. coli and its isogenic mazEF and clpP mutants were grown in minimal medium to mid-logarithmic phase and exposed to antibiotics that inhibit transcription or translation (rifampin, chloramphenicol, or spectinomycin) for 10 min, both the mazEF and clpP mutants displayed nearly a 100% survival rate, whereas the survival rate of the parental strain was less than 20% in each case (222). However, the survival rates of the mazEF and clpP mutants were comparable to that of the parental strain when they were treated with ampicillin, an inhibitor of cell wall synthesis (222). Therefore, it was suggested that treatment with transcription and/or translation inhibitors causes a corresponding reduction in cellular levels of MazE and subsequent MazF-induced toxicity (222). Further support for the ability of MazEF to cause cell death in response to the inhibition of transcription and/or translation was provided by the observation that the toxin component of the plasmid prophage P1 phd-doc addiction module (a general inhibitor of translation) was also able to induce mazEF-dependent cell death (107).
Interestingly, DNA damage induced by a variety of treatments, including UV irradiation (106), nalidixic acid (106), mitomycin C (106), and thymine starvation (223), was also found to induce MazEF-dependent cell death. In each case, the mazEF mutant displayed increased survival in response to DNA damage relative to the parental strain (106, 223). It was postulated that MazEF-induced cell death in response to DNA damage is a consequence of reduced mazEF transcription, thus preventing the continuous production of the MazE antitoxin (223). Indeed, it was observed that mazEF transcription was reduced in thymine-starved cells (223). This could be caused by mutations generated within the mazEF promoter region, by the induction of ppGpp synthesis, or by an as-yet-unknown protein(s) that may sense replication errors (223). Other stressful conditions that were found to trigger MazEF-dependent cell death include exposure to high temperatures and oxidative stress (106).
As mentioned above, the labile antitoxin MazE is rapidly degraded by ClpPA protease, and in the absence of a constant source of MazE, the MazF toxin is free to exert its lethal effect on the cell (2). Another protein involved in this system is MazG, which was originally identified in a yeast two-hybrid screen for proteins interacting with Era (an essential E. coli GTPase) (287). As its name implies, mazG is located immediately downstream from mazEF (287) and is cotranscribed with mazEF (98). While the inactivation of mazG alone did not affect the viability of E. coli under standard growth conditions (287), the ectopic overexpression of mazG in a mazEFG mutant severely inhibited cell growth (98). Recombinant MazG protein was found to display nucleoside triphosphate pyrophosphohydrolase activity (with a preference for deoxynucleotides) (287), and the MazE-MazF protein complex was able to inhibit the activity of MazG (98). Furthermore, the overexpression of MazG was shown to decrease the accumulation of ppGpp (98). Based on those results, it was proposed that MazG functions to protect the cells against the toxic action of MazF during nutritional stress by counteracting ppGpp accumulation either directly, by hydrolyzing ppGpp itself, or indirectly, by hydrolyzing GTP and/or ATP (which are required by RelA for ppGpp synthesis) (98). This decrease in ppGpp would restore the expression of mazEF, thereby producing more MazE antitoxin and providing a window of time in which cell death is delayed in case the growth conditions are improved (98).
Based on the studies mentioned above, it appears that signals resulting in a decrease in the amount of cellular MazE antitoxin lead to cell death via the toxic action of MazF. The toxic nature of the MazF protein has been identified as its ability to inhibit translation by cleavage of mRNAs (43, 170, 288, 289), and MazF-dependent mRNA cleavage is inhibited in the presence of the MazE antitoxin (170, 289). The ability of MazF to cleave mRNAs was first reported by Christensen et al. (43), who demonstrated that the overexpression of MazF inhibited translation by cleaving actively translated mRNAs. This was strengthened by the observation that the overproduction of tmRNA, a specialized RNA molecule that participates in the recognition and rescue of stalled ribosomes (55), was able to reduce the inhibitory effect of overexpressing MazF (43). It was subsequently shown that recombinant MazF protein acts as a sequence-specific (ACA) endoribonuclease that cleaves single-stranded RNA (289). However, that study reported that MazF activity was ribosome independent (289). This specificity for mRNAs containing the ACA sequence was also demonstrated in an E. coli single-protein production system, where the simultaneous expression of MazF and a target gene engineered to encode an ACA-less mRNA resulted in the high-level expression of the target gene and a dramatic overall decrease in cellular protein synthesis (243). Munoz-Gomez et al. (170) found that purified native MazF protein was capable of cleaving both single-stranded and double-stranded mRNA but displayed a preference for 5′-NAC-3′ sites contained in single-stranded mRNA.
RelBE.
Study of the E. coli RelBE TA system originated with the characterization of a class of mutants, designated RelB mutants, which conferred a “delayed-relaxed” phenotype during amino acid starvation. In cells with an intact stringent response, rRNA and tRNA synthesis is shut down in response to the accumulation of ppGpp. However, the “delayed-relaxed” phenotype observed in RelB mutants is characterized by the production of stable RNAs that resumes after a 10-min lag period following amino acid starvation (52, 53). The locus responsible for the delayed-relaxed phenotype was sequenced and identified by Bech et al. (17) and was found to contain a three-gene operon comprised of the relB, relE, and relF genes. The relF open reading frame displayed both functional and structural similarity to the hok gene of plasmid R1, whose gene product is responsible for the postsegregational killing of cells that lose plasmid R1 during cell division (79, 81). In that study, cell death caused by the induction of relF expression shared several features with Hok-mediated cell death, including the collapse of membrane potential, inhibition of cellular respiration, and changes in cellular morphology (79).
The relB and relE genes were subsequently found to encode the antitoxin and toxin components, respectively, of a TA module (92). The function of each protein was demonstrated by the observation that the ectopic expression of relE from a low-copy plasmid resulted in a 600-fold decrease in viable cell counts, whereas the simultaneous expression of relB from a high-copy plasmid was able to prevent the RelE-mediated loss of viability (92). Other evidence supporting the function of RelBE as a TA module included the ability of relBE to confer stability to a mini-R1 test plasmid (92), the negative regulation of relBE expression by the RelB antitoxin (92), the direct protein-protein interaction of the RelB and RelE proteins as demonstrated in a yeast two-hybrid assay (68), and the sensitivity of the RelB antitoxin to degradation by Lon protease (42).
The RelE toxin was shown to function as a global inhibitor of translation: the overexpression of RelE severely inhibited translation, and a relBE mutant achieved a higher steady-state level of translation after amino acid starvation than did the parental strain (42). The ability of RelE to inhibit translation was also implicated by its ability to bind ribosomes in vitro (68). Likewise, the expression of the relBE transcript was shown to be induced under conditions of either amino acid or glucose starvation as well as in the presence of chloramphenicol, a inhibitor of translation (42). Those results suggested that RelE functions as a global inhibitor of translation during nutritional stress (42). The induction of relBE expression during amino acid starvation was independent of ppGpp accumulation but was dependent on the presence of Lon protease, suggesting that relBE expression is derepressed as a consequence of RelB degradation (42, 43).
It was also found that RelE-dependent inhibition of translation during amino acid starvation did not interfere with cell viability (42). This was confirmed by Pedersen et al. (193), who demonstrated that cultures overexpressing the RelE toxin experienced a decrease in global translation and also lost the ability to form colonies on plates. However, both these conditions could be rescued by the subsequent overproduction of the RelB antitoxin, suggesting that RelE induces a reversible bacteriostatic condition and not cell death (193).
The activity of E. coli RelE has been suggested to be an endoribonuclease that cleaves specific codons of mRNA when they are located in the ribosomal A site (194). Specifically, the stop codon UAG as well as the sense codons UCG and CAG were found to be the most efficiently cleaved in this in vitro system (194). The specificity of RelE to cleave actively translated mRNA was also demonstrated in vivo: the induction of relE expression had no effect on the cleavage pattern of mRNA lacking a functional ATG start codon, and RelE-induced cleavage patterns were observed in the coding region of intact mRNA but not in the untranslated leader region (41). The RelE-dependent cleavage of translated mRNAs was also induced by amino acid starvation, and tmRNA was able to alleviate RelE toxicity, presumably by rescuing stalled ribosomes present on mRNAs damaged by RelE (41, 194). Since the cleavage of mRNA codons at the ribosomal A site appears to be an intrinsic characteristic of the ribosome during its pausing, RelE may simply stimulate this cleavage at the A site rather than having intrinsic ribonucleolytic activity (104).
HipBA.
The hip locus was originally identified by Moyed and Bertrand (168) in a screen of E. coli mutants for a high frequency of persister cell formation (Hip mutants). Persister cell formation, or bacterial persistence, refers to the emergence of a small population of cells that survive antibiotic treatment yet are genetically identical to the original culture (recently reviewed in reference 151). Persister cells that are regrown and exposed to antibiotic demonstrate the same frequency of persister cell formation as the original population, and it has been suggested that this phenomenon may explain the recalcitrance of certain bacterial infections to antibiotic treatment, such as those caused by biofilms (151). The study of Moyed and Bertrand (168) led to the identification of two Hip mutants that displayed increased persister cell formation (1 in 100 cells) relative to the parental strain (1 in 1,000,000 cells), hipA7 and hipA9, which both mapped at 33.8 min of the E. coli chromosome. This two-gene operon, designated hipBA, was subsequently cloned and sequenced (24, 169).
The regulation of the hipBA operon and the function of its gene products are similar to those of the family of TA modules. The expression of hipBA is negatively autoregulated by the HipB protein, which encodes a small DNA-binding protein that binds to the promoter region of hipBA (23, 24). HipB interacts with the HipA protein, and the HipA protein appears to be toxic in the absence of HipB, suggesting that hipA encodes the toxin and hipB encodes the antitoxin of this TA-like module (23, 169). Clues regarding the potential function of HipA have been gleaned from studies of the hipA7 allele, which confers increased persister cell formation as described above. In addition to increased persistence in response to treatment with inhibitors of cell wall synthesis, the hipA7 allele was also found to confer increased survival in response to inhibitors of DNA synthesis (224). Subsequent characterization of the hipA7 allele revealed that hipA7 produces a nontoxic protein whose ability to confer high-level persistence is independent of the antitoxin HipB (133). The highly persistent phenotype of hipA7 mutants can be lost when relA is inactivated in a hipA7 mutant, suggesting a model whereby HipA7 somehow increases the basal level production of ppGpp, which in turn alters the expression of as-yet-unidentified genes required to enter the persistent state (133).
More recently, it has been shown that the in vivo expression of either the wild-type hipA allele or the hipA7 allele in excess of chromosomal hipB each confers a similar level of persistence (134). Furthermore, the expression of hipA under these conditions was able to inhibit DNA replication, RNA transcription, and protein synthesis, but the expression of the mutant hipA7 gene did not noticeably inhibit protein synthesis, suggesting that the ability of HipA to generate persisters may be distinct from its ability to block macromolecular synthesis (134). The expression of hipA is also able to confer tolerance and increase persister cell formation in response to antibiotic treatment when expressed at low levels (62, 134). Although the exact cellular target of HipA remains to be defined, recent evidence demonstrated that HipA belongs to a family of phosphatidylinositide and protein kinases and is capable of autophosphorylation (50). Furthermore, HipA mutant proteins that lack kinase activity were unable to confer antibiotic tolerance, whereas the expression of intact HipA effectively protected cells from antibiotic-induced killing (50).
It should be noted that recent reports have implicated a role for other TA modules in regulating persistence. Gene expression profiling of persister cells formed in the E. coli strain containing the hipA7 allele in response to ampicillin treatment revealed that a number of chromosomal TA genes were upregulated in persister cells, including dinJ/yafQ, yefM, relBE, and mazEF (122). However, mutations in either mazEF or relBE did not affect the ability to form persisters, while the deletion of hipBA significantly reduced persister formation (122). Furthermore, dormant cells (displaying the same phenotypic characteristics of persister cells) isolated from exponential-phase wild-type E. coli cultures displayed an upregulated expression of several TA genes (227). Collectively, these results suggest that TA genes, in addition to hipBA, may contribute to the distinct physiology of persister cells.
Role(s) of TA systems in bacterial physiology.
While it is apparent that chromosomal TA modules are ubiquitous among bacteria, the actual function that they serve in bacterial physiology has been a lively source of debate. Some groups proposed that these TA modules function as part of the bacterium's cell death machinery, killing the cells in response to various sources of stress (57, 60, 106, 222), while others believe that these TA modules have a reversible, bacteriostatic function that aids in surviving nutritional stress (78, 80, 253). The main indication that MazF, like the RelE toxin, induces bacteriostasis in response to stress stems from reports that the loss of cell viability due to the ectopic expression of MazF can be restored by the subsequent expression of the MazE antitoxin (193) and that the activation of MazF in response to amino acid starvation does not induce cell death (43). Furthermore, amino acid starvation was also shown in that study to stimulate the expression of the mazEF transcript in a Lon protease-dependent, ppGpp-independent manner, leading those authors to conclude that the modulation of translation by toxin-antitoxin pairs during amino acid starvation represents a ppGpp-independent branch of the stringent response (43). Those results contradict the earlier reports that ppGpp inhibits mazEF expression (2).
In response to those findings, Engelberg-Kulka and coworkers reported studies in which they confirmed that the loss of cell viability due to the ectopic overexpression of MazF can be rescued by the subsequent induction of MazE expression (6). However, they also found that the effect of MazF on cell viability was not reversible by MazE after the cells were exposed to MazF beyond a certain period of time (6). This was also demonstrated by subjecting wild-type E. coli cultures to conditions previously reported to inhibit chromosomal mazEF expression, such as treatment with DNA-damaging agents or inhibitors of translation (129). In these treated cultures, the restoration of cell viability by overexpressing MazE could occur only within a certain period of time posttreatment (129). Those results (6, 129) suggested a “point-of-no-return” model for MazF-induced cell death, whereby the initial inhibition of translation by MazF can be reversed by the antagonistic effect of MazE, but if this process is not stopped in time, cell death becomes unavoidable and irreversible. It was suggested that this is possibly due to the selective synthesis of cell death proteins encoded by mRNAs that are resistant to cleavage by MazF (6, 59, 129). Thus, in this model, the MazE and MazF proteins would function as intermediaries in the pathway to cell death. Support for this theory was also provided by a study by Godoy and colleagues (87), who demonstrated that E. coli cell death induced by treatment with the intracellular deoxynucleoside triphosphate-depleting agent hydroxyurea involved mazEF and relBE.
A recent publication also reported testing of the involvement of TA modules in eliciting cell death or bacteriostasis (253). In that study, the authors identified the presence of two relA mutations in the strains used in previous studies that may have affected the interpretation of the results (253). When they compared a “true” wild-type strain with its isogenic derivative lacking five chromosomal TA systems (including relBE and mazEF), TA-dependent cell death was not detectable under any of the stress conditions tested (253). Those results were suggested to corroborate data from previous work showing that single deletions of either mazEF or relBE did not affect the frequency of persister cell formation (122). It should be noted that persister cell formation via the HipBA system was still considered to be valid by those authors, since, unlike the deletion of other single TA modules, the deletion of hipBA significantly reduced persister formation (122, 253). Furthermore, the presence of these TA modules did not appear to confer a competitive advantage in the recovery from various stress conditions (amino acid starvation, high temperature, and antibiotics) or in cell growth under nutrient-limited conditions (253).
As described above, a very subtle balance (decision) between life and death may exist in bacteria, similar to that observed in eukaryotic organisms. Therefore, the use of different experimental conditions and/or strains in the studies of the TA modules discussed above may account for the conflicting results obtained in different laboratories. The importance of the use of proper physiological conditions, such as the density of the bacterial culture, is supported by a very recent report (130) showing that mazEF-mediated cell death is a population phenomenon requiring a quorum-sensing factor called extracellular death factor. Extracellular death factor was characterized to be a linear pentapeptide, with each of the five amino acids shown to be important for its mazEF-mediated killing activity. That study supports the idea that a bacterial culture can behave as a multicellular organism. When challenged by stressful conditions that trigger mazEF-mediated cell death, the bacterial population can respond like a multicellular organism in which a subpopulation of cells dies and permits the survival of the bacterial population as a whole (130). Thus, the specific functions of the toxin-antitoxin systems remain an intriguing area of research.
CONCLUSIONS
Based on the examples highlighted in this review, it is obvious that the traditional view of bacterial cell death and lysis as a passive and insipid process is being rapidly replaced by a vision of cell death and lysis as a complex, highly regulated, and important component of bacterial physiology. This concept is mirrored by our ever-evolving view and study of bacteria from selfish, single-celled organisms to complex multicellular communities of cells that display sophisticated social behaviors and developmental processes. Because we have been slow to appreciate the multicellular nature of bacterial communities, our understanding of the importance and control of cell death and apoptosis in the more “highly evolved” eukaryotic organisms is much more advanced compared to the study of the relatively “simple” prokaryotes. Perhaps current and future studies of bacterial cell death and lysis can glean additional clues from the well-studied and defined mechanisms of eukaryotic apoptosis in developmental processes, cellular defense, and cancer. Although we are a long way from completely understanding the complex regulatory signals and pathways that dictate whether individual bacterial cells live or die, further study in this exciting field of research is bound to yield insights into these processes. Furthermore, these studies should also provide new insight into the potential evolutionary link between control of cell death in bacteria and PCD in eukaryotes.
Acknowledgments
We thank members of the Bayles laboratory, past and present, for helpful discussions regarding the cid and lrg operons. Special thanks go to Xu Luo, Alexander Tomasz, Fritz Gotz, Kim Lewis, Ry Young, and Frank Neuhaus for providing support and guidance in the preparation of the manuscript. We also thank Dan Wozniak for providing preliminary data related to cidAB and lrgAB function in P. aeruginosa.
This project was funded by NIH grant R01AI038901 and DOD grant DAAD 19-03-1-0191 to K.W.B.
REFERENCES
- 1.Agarwal, S., S. Agarwal, and R. Bhatnagar. 2007. Identification and characterization of a novel toxin-antitoxin module from Bacillus anthracis. FEBS Lett. 5811727-1734. [DOI] [PubMed] [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 936059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alaedini, A., and R. A. Day. 1999. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem. Biophys. Res. Commun. 264191-195. [DOI] [PubMed] [Google Scholar]
- 4.Allenby, N. E., C. A. Watts, G. Homuth, Z. Pragai, A. Wipat, A. C. Ward, and C. R. Harwood. 2006. Phosphate starvation induces the sporulation killing factor of Bacillus subtilis. J. Bacteriol. 1885299-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. [DOI] [PubMed] [Google Scholar]
- 6.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 1868295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonsson, B., S. Montessuit, S. Lauper, R. Eskes, and J. C. Martinou. 2000. Bax oligomerization is required for channel-forming activity in liposomes and to trigger cytochrome c release from mitochondria. Biochem. J. 345271-278. [PMC free article] [PubMed] [Google Scholar]
- 8.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 9.Avery, O. T., and G. E. Cullen. 1923. Studies on the enzymes of pneumococcus. Part IV. Bacteriolytic enzyme. J. Exp. Med. 38199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avery, S. V. 2005. Cell individuality: the bistability of competence development. Trends Microbiol. 13459-462. [DOI] [PubMed] [Google Scholar]
- 11.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 174639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baliga, B. C., and S. Kumar. 2002. Role of Bcl-2 family of proteins in malignancy. Hematol. Oncol. 2063-74. [DOI] [PubMed] [Google Scholar]
- 13.Basanez, G., J. C. Sharpe, J. Galanis, T. B. Brandt, J. M. Hardwick, and J. Zimmerberg. 2002. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J. Biol. Chem. 27749360-49365. [DOI] [PubMed] [Google Scholar]
- 14.Bayles, K. W. 2003. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 11306-311. [DOI] [PubMed] [Google Scholar]
- 15.Bayles, K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5721-726. [DOI] [PubMed] [Google Scholar]
- 16.Bayles, K. W. 1993. The use of degenerate, sensor gene-specific, oligodeoxyribonucleotide primers to amplify DNA fragments from Staphylococcus aureus. Gene 12399-103. [DOI] [PubMed] [Google Scholar]
- 17.Bech, F. W., S. T. Jorgensen, B. Diderichsen, and O. H. Karlstrom. 1985. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 41059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman-Frank, I., and K. D. Bidle. 2004. The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol. Oceanogr. 49997-1005. [Google Scholar]
- 19.Bernhardt, T. G., W. D. Roof, and R. Young. 2000. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. Proc. Natl. Acad. Sci. USA 974297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhardt, T. G., D. K. Struck, and R. Young. 2001. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 2766093-6097. [DOI] [PubMed] [Google Scholar]
- 21.Betzner, A. S., and W. Keck. 1989. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol. Gen. Genet. 219489-491. [DOI] [PubMed] [Google Scholar]
- 22.Bidle, K. D., and P. G. Falkowski. 2004. Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2643-655. [DOI] [PubMed] [Google Scholar]
- 23.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1764081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1735732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 14473-82. [DOI] [PubMed] [Google Scholar]
- 26.Blasi, U., and R. Young. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21675-682. [DOI] [PubMed] [Google Scholar]
- 27.Bonnet, S., S. L. Archer, J. Allalunis-Turner, A. Haromy, C. Beaulieu, R. Thompson, C. T. Lee, G. D. Lopaschuk, L. Puttagunta, S. Bonnet, G. Harry, K. Hashimoto, C. J. Porter, M. A. Andrade, B. Thebaud, and E. D. Michelakis. 2007. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 1137-51. [DOI] [PubMed] [Google Scholar]
- 28.Bowman, B. U., Jr., and W. B. Redmond. 1956. The effects of glucose and of oxygen on autolysis of Mycobacterium tuberculosis. Am. Rev. Tuberc. 73907-916. [DOI] [PubMed] [Google Scholar]
- 29.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 1445-54. [DOI] [PubMed] [Google Scholar]
- 30.Bravo, A., G. de Torrontegui, and R. Diaz. 1987. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 210101-110. [DOI] [PubMed] [Google Scholar]
- 31.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1785810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30672-679. [DOI] [PubMed] [Google Scholar]
- 34.Calamita, H. G., and R. J. Doyle. 2002. Regulation of autolysins in teichuronic acid-containing Bacillus subtilis cells. Mol. Microbiol. 44601-606. [DOI] [PubMed] [Google Scholar]
- 35.Calamita, H. G., W. D. Ehringer, A. L. Koch, and R. J. Doyle. 2001. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc. Natl. Acad. Sci. USA 9815260-15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cannell, H., C. Kerawala, A. M. Sefton, J. P. Maskell, A. Seymour, Z. M. Sun, and J. D. Williams. 1991. Failure of two macrolide antibiotics to prevent extraction bacteraemia. Br. Dent. J. 171170-173. [DOI] [PubMed] [Google Scholar]
- 37.Carmody, R. J., and T. G. Cotter. 2001. Signalling apoptosis: a radical approach. Redox Rep. 677-90. [DOI] [PubMed] [Google Scholar]
- 38.Carroll, S. A., T. Hain, U. Technow, A. Darji, P. Pashalidis, S. W. Joseph, and T. Chakraborty. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J. Bacteriol. 1856801-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4160-165. [DOI] [PubMed] [Google Scholar]
- 40.Chipuk, J. E., and D. R. Green. 2005. Do inducers of apoptosis trigger caspase-independent cell death? Nat. Rev. Mol. Cell Biol. 6268-275. [DOI] [PubMed] [Google Scholar]
- 41.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 481389-1400. [DOI] [PubMed] [Google Scholar]
- 42.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 9814328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332809-819. [DOI] [PubMed] [Google Scholar]
- 44.Chung, J. D., G. Stephanopoulos, K. Ireton, and A. D. Grossman. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 1761977-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claverys, J. P., and L. S. Havarstein. 2007. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 5219-229. [DOI] [PubMed] [Google Scholar]
- 46.Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 47.Claverys, J. P., B. Martin, and L. S. Havarstein. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 641423-1433. [DOI] [PubMed] [Google Scholar]
- 48.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60451-475. [DOI] [PubMed] [Google Scholar]
- 49.Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Gotz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186214-219. [DOI] [PubMed] [Google Scholar]
- 50.Correia, F. F., A. D'Onofrio, T. Rejtar, L. Li, B. L. Karger, K. Makarova, E. V. Koonin, and K. Lewis. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 1888360-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 1844988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diderichsen, B., and L. Desmarez. 1980. Variations in phenotype of relB mutants of Escherichia coli and the effect of pus and sup mutations. Mol. Gen. Genet. 180429-437. [DOI] [PubMed] [Google Scholar]
- 53.Diderichsen, B., N. P. Fiil, and R. Lavalle. 1977. Genetics of the relB locus in Escherichia coli. J. Bacteriol. 13130-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubnau, D., and R. Losick. 2006. Bistability in bacteria. Mol. Microbiol. 61564-572. [DOI] [PubMed] [Google Scholar]
- 55.Dulebohn, D., J. Choy, T. Sundermeier, N. Okan, and A. W. Karzai. 2007. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 464681-4693. [DOI] [PubMed] [Google Scholar]
- 56.Ellermeier, C. D., E. C. Hobbs, J. E. Gonzalez-Pastor, and R. Losick. 2006. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124549-559. [DOI] [PubMed] [Google Scholar]
- 57.Engelberg-Kulka, H., S. Amitai, I. Kolodkin-Gal, and R. Hazan. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 5343-70. [DOI] [PubMed] [Google Scholar]
- 59.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 1184327-4332. [DOI] [PubMed] [Google Scholar]
- 60.Engelberg-Kulka, H., B. Sat, M. Reches, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 1266-71. [DOI] [PubMed] [Google Scholar]
- 61.Fabretti, F., C. Theilacker, L. Baldassarri, Z. Kaczynski, A. Kropec, O. Holst, and J. Huebner. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 744164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falla, T. J., and I. Chopra. 1998. Joint tolerance to β-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 423282-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fedtke, I., D. Mader, T. Kohler, H. Moll, G. Nicholson, R. Biswas, K. Henseler, F. Gotz, U. Zahringer, and A. Peschel. 2007. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol. Microbiol. 651078-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer, W., P. Rosel, and H. U. Koch. 1981. Effect of alanine ester substitution and other structural features of lipoteichoic acids on their inhibitory activity against autolysins of Staphylococcus aureus. J. Bacteriol. 146467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 1775723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu, Z., N. P. Donegan, G. Memmi, and A. L. Cheung. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 1898871-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujita, M., J. E. Gonzalez-Pastor, and R. Losick. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 1871357-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galvani, C., J. Terry, and E. E. Ishiguro. 2001. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J. Bacteriol. 1832700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Bustos, J. F., B. T. Chait, and A. Tomasz. 1987. Structure of the peptide network of pneumococcal peptidoglycan. J. Biol. Chem. 26215400-15405. [PubMed] [Google Scholar]
- 70.Garcia, D. L., and J. P. Dillard. 2006. AmiC functions as an N-acetylmuramyl-l-alanine amidase necessary for cell separation and can promote autolysis in Neisseria gonorrhoeae. J. Bacteriol. 1887211-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 311275-1281. [DOI] [PubMed] [Google Scholar]
- 72.Garrett, J. M., and R. Young. 1982. Lethal action of bacteriophage λ S gene. J. Virol. 44886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gatenby, R. A., E. T. Gawlinski, A. F. Gmitro, B. Kaylor, and R. J. Gillies. 2006. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 665216-5223. [DOI] [PubMed] [Google Scholar]
- 74.Gatenby, R. A., and R. J. Gillies. 2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4891-899. [DOI] [PubMed] [Google Scholar]
- 75.Gautam, S., and A. Sharma. 2002. Involvement of caspase-3-like protein in rapid cell death of Xanthomonas. Mol. Microbiol. 44393-401. [DOI] [PubMed] [Google Scholar]
- 76.Gautam, S., and A. Sharma. 2002. Rapid cell death in Xanthomonas campestris pv. glycines. J. Gen. Appl. Microbiol. 4867-76. [DOI] [PubMed] [Google Scholar]
- 77.George, N. M., J. J. Evans, and X. Luo. 2007. A three-helix homo-oligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev. 211937-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerdes, K., F. W. Bech, S. T. Jorgensen, A. Lobner-Olesen, P. B. Rasmussen, T. Atlung, L. Boe, O. Karlstrom, S. Molin, and K. von Meyenburg. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 52023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3371-382. [DOI] [PubMed] [Google Scholar]
- 81.Gerdes, K., and S. Molin. 1986. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol. 190269-279. [DOI] [PubMed] [Google Scholar]
- 82.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 621371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giesbrecht, P., T. Kersten, and J. Wecke. 1992. Fan-shaped ejections of regularly arranged murosomes involved in penicillin-induced death of staphylococci. J. Bacteriol. 1742241-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gilpin, R. W., A. N. Chatterjee, and F. E. Young. 1972. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J. Bacteriol. 111272-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giudicelli, S., and A. Tomasz. 1984. Attachment of pneumococcal autolysin to wall teichoic acids, an essential step in enzymatic wall degradation. J. Bacteriol. 1581188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 26310088-10095. [PubMed] [Google Scholar]
- 87.Godoy, V. G., D. F. Jarosz, F. L. Walker, L. A. Simmons, and G. C. Walker. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 25868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301510-513. [DOI] [PubMed] [Google Scholar]
- 89.Goodell, E. W., R. Lopez, and A. Tomasz. 1976. Suppression of lytic effect of beta lactams on Escherichia coli and other bacteria. Proc. Natl. Acad. Sci. USA 733293-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodell, W., and A. Tomasz. 1980. Alteration of Escherichia coli murein during amino acid starvation. J. Bacteriol. 1441009-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 685690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 291065-1076. [DOI] [PubMed] [Google Scholar]
- 93.Graham, L. L., and T. J. Beveridge. 1994. Structural differentiation of the Bacillus subtilis 168 cell wall. J. Bacteriol. 1761413-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Graschopf, A., and U. Blasi. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33569-582. [DOI] [PubMed] [Google Scholar]
- 95.Greenberg, R. A., and H. O. Halvorson. 1954. Studies on an autolytic substance produced by an aerobic sporeforming bacterium. J. Bacteriol. 6945-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 1821794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gronlund, H., and K. Gerdes. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 2851401-1415. [DOI] [PubMed] [Google Scholar]
- 98.Gross, M., I. Marianovsky, and G. Glaser. 2006. MazG—a regulator of programmed cell death in Escherichia coli. Mol. Microbiol. 59590-601. [DOI] [PubMed] [Google Scholar]
- 99.Gründling, A., U. Bläsi, and R. Y. Young. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 1826082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grundling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 989348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guiral, S., T. J. Mitchell, B. Martin, and J. P. Claverys. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. USA 1028710-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hakenbeck, R., E. W. Goodell, and U. Schwarz. 1974. Compartmentalization of murein hydrolases in the envelope of Escherichia coli. FEBS Lett. 40261-264. [DOI] [PubMed] [Google Scholar]
- 103.Havarstein, L. S., B. Martin, O. Johnsborg, C. Granadel, and J. P. Claverys. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 591297-1307. [DOI] [PubMed] [Google Scholar]
- 104.Hayes, C. S., and R. T. Sauer. 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12903-911. [DOI] [PubMed] [Google Scholar]
- 105.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272227-234. [DOI] [PubMed] [Google Scholar]
- 106.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 1863663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 1832046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Holtje. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41167-178. [DOI] [PubMed] [Google Scholar]
- 109.Hetts, S. W. 1998. To die or not to die: an overview of apoptosis and its role in disease. JAMA 279300-307. [DOI] [PubMed] [Google Scholar]
- 110.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holtje, J.-V., and E. I. Tuomanen. 1991. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J. Gen. Microbiol. 137441-454. [DOI] [PubMed] [Google Scholar]
- 112.Höltje, J. V. 1996. Molecular interplay of murein synthases and murein hydrolases in Escherichia coli. Microb. Drug Resist. 299-103. [DOI] [PubMed] [Google Scholar]
- 113.Holtje, J. V., and B. Glauner. 1990. Structure and metabolism of the murein sacculus. Res. Microbiol. 14175-89. [DOI] [PubMed] [Google Scholar]
- 114.Holtje, J. V., and W. Keck. 1988. Organization of the major autolysin in the envelope of Escherichia coli, p. 181-188. In P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (ed.), Antibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, DC.
- 115.Holtje, J. V., and U. Schwarz. 1983. Coenzyme A-glutathione disulfide:an endogenous inhibitor of the lytic enzymes in Escherichia coli, p. 185-190. In R. Hakenbeck, J. V. Holtje, and H. Labischinski (ed.), The target of penicillin. Walter de Gruyter, Berlin, Germany.
- 116.Jaffe, A., T. Ogura, and S. Hiraga. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25753-763. [DOI] [PubMed] [Google Scholar]
- 118.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 581087-1101. [DOI] [PubMed] [Google Scholar]
- 119.Kausmally, L., O. Johnsborg, M. Lunde, E. Knutsen, and L. S. Havarstein. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 1874338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keck, W., and U. Schwarz. 1979. Escherichia coli murein-DD-endopeptidase insensitive to beta-lactam antibiotics. J. Bacteriol. 139770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kemper, M. A., M. M. Urrutia, T. J. Beveridge, A. L. Koch, and R. J. Doyle. 1993. Proton motive force may regulate cell wall-associated enzymes of Bacillus subtilis. J. Bacteriol. 1755690-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 1868172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khoo, S. K., B. Loll, W. T. Chan, R. L. Shoeman, L. Ngoo, C. C. Yeo, and A. Meinhart. 2007. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J. Biol. Chem. 28219606-19618. [DOI] [PubMed] [Google Scholar]
- 124.Kim, J. W., and C. V. Dang. 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 668927-8930. [DOI] [PubMed] [Google Scholar]
- 125.Klein, A. H., A. Shulla, S. A. Reimann, D. H. Keating, and A. J. Wolfe. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 1895574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Knutsen, E., O. Ween, and L. S. Havarstein. 2004. Two separate quorum-sensing systems upregulate transcription of the same ABC transporter in Streptococcus pneumoniae. J. Bacteriol. 1863078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koch, A. L. 1986. The pH in the neighborhood of membranes generating a protonmotive force. J. Theor. Biol. 12073-84. [DOI] [PubMed] [Google Scholar]
- 128.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130797-810. [DOI] [PubMed] [Google Scholar]
- 129.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 1883420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kolodkin-Gal, I., R. Hazan, A. Gaathon, S. Carmeli, and H. Engelberg-Kulka. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318652-655. [DOI] [PubMed] [Google Scholar]
- 131.Kolter, R., and F. Moreno. 1992. Genetics of ribosomally synthesized peptide antibiotics. Annu. Rev. Microbiol. 46141-163. [DOI] [PubMed] [Google Scholar]
- 132.Koprivnjak, T., A. Peschel, M. H. Gelb, N. S. Liang, and J. P. Weiss. 2002. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J. Biol. Chem. 27747636-47644. [DOI] [PubMed] [Google Scholar]
- 133.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 501199-1213. [DOI] [PubMed] [Google Scholar]
- 134.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 1883826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kostic, V., V. Jackson-Lewis, F. de Bilbao, M. Dubois-Dauphin, and S. Przedborski. 1997. Bcl-2: prolonging life in a transgenic mouse model of familial amyotrophic lateral sclerosis. Science 277559-562. [DOI] [PubMed] [Google Scholar]
- 136.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 1885797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 1876719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kristian, S. A., X. Lauth, V. Nizet, F. Goetz, B. Neumeister, A. Peschel, and R. Landmann. 2003. Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J. Infect. Dis. 188414-423. [DOI] [PubMed] [Google Scholar]
- 139.Kroemer, G., L. Galluzzi, and C. Brenner. 2007. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 8799-163. [DOI] [PubMed] [Google Scholar]
- 140.Kullik, I., R. Jenni, and B. Berger-Bachi. 1998. Sequence of the putative alanine racemase operon in Staphylococcus aureus: insertional interruption of this operon reduces D-alanine substitution of lipoteichoic acid and autolysis. Gene 2199-17. [DOI] [PubMed] [Google Scholar]
- 141.Kusser, W., and U. Schwarz. 1980. Escherichia coli murein transglycosylase. Purification by affinity chromatography and interaction with polynucleotides. Eur. J. Biochem. 103277-281. [DOI] [PubMed] [Google Scholar]
- 142.Kuwana, T., M. R. Mackey, G. Perkins, M. H. Ellisman, M. Latterich, R. Schneiter, D. R. Green, and D. D. Newmeyer. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111331-342. [DOI] [PubMed] [Google Scholar]
- 143.Labedan, B., and L. Letellier. 1981. Membrane potential changes during the first steps of coliphage infection. Proc. Natl. Acad. Sci. USA 78215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Labischinski, H., and H. Maidhof. 1994. Microbial peptidoglycan (murein) hydrolases, p. 23-38. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 145.Le Bras, M., M. V. Clement, S. Pervaiz, and C. Brenner. 2005. Reactive oxygen species and the mitochondrial signaling pathway of cell death. Histol. Histopathol. 20205-219. [DOI] [PubMed] [Google Scholar]
- 146.Leduc, M., C. Frehel, E. Siegel, and J. Van Heijenoort. 1989. Multilayered distribution of peptidoglycan in the periplasmic space of Escherichia coli. J. Gen. Microbiol. 1351243-1254. [DOI] [PubMed] [Google Scholar]
- 147.Leduc, M., R. Kasra, and J. van Heijenoort. 1982. Induction and control of the autolytic system of Escherichia coli. J. Bacteriol. 15226-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lehnherr, H., E. Maguin, S. Jafri, and M. B. Yarmolinsky. 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233414-428. [DOI] [PubMed] [Google Scholar]
- 149.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Beguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 1772451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lemos, J. A., T. A. Brown, Jr., J. Abranches, and R. A. Burne. 2005. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol. Lett. 253251-257. [DOI] [PubMed] [Google Scholar]
- 151.Lewis, K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 70267-274. [DOI] [PubMed] [Google Scholar]
- 152.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin, J. T., M. B. Connelly, C. Amolo, S. Otani, and D. S. Yaver. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 491915-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lopez, R., M. P. Gonzalez, E. Garcia, J. L. Garcia, and P. Garcia. 2000. Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151437-443. [DOI] [PubMed] [Google Scholar]
- 155.Lord, F. T., and R. N. Nye. 1919. The relation of the pneumococcus to hydrogen ion concentration, acid death-point, and dissolution of the organism. J. Exp. Med. 30389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maamar, H., and D. Dubnau. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 703232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mallat, Z., A. Tedgui, F. Fontaliran, R. Frank, M. Durigon, and G. Fontaine. 1996. Evidence of apoptosis in arrhythmogenic right ventricular dysplasia. N. Engl. J. Med. 3351190-1196. [DOI] [PubMed] [Google Scholar]
- 159.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 1756850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Matias, V. R., and T. J. Beveridge. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56240-251. [DOI] [PubMed] [Google Scholar]
- 161.Matias, V. R., and T. J. Beveridge. 2006. Native cell wall organization shown by cryoelectron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 1881011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 194473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 26315699-15704. [PubMed] [Google Scholar]
- 164.Mittenhuber, G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1295-302. [PubMed] [Google Scholar]
- 165.Moreillon, P., and A. Tomasz. 1988. Penicillin resistance and defective lysis in clinical isolates of pneumococci: evidence for two kinds of antibiotic pressure operating in the clinical environment. J. Infect. Dis. 1571150-1157. [DOI] [PubMed] [Google Scholar]
- 166.Moscoso, M., and J. P. Claverys. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 54783-794. [DOI] [PubMed] [Google Scholar]
- 167.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 1887785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Munoz-Gomez, A. J., S. Santos-Sierra, A. Berzal-Herranz, M. Lemonnier, and R. Diaz-Orejas. 2004. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567316-320. [DOI] [PubMed] [Google Scholar]
- 171.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 1049451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nandy, S. K., P. M. Bapat, and K. V. Venkatesh. 2007. Sporulating bacteria prefers predation to cannibalism in mixed cultures. FEBS Lett. 581151-156. [DOI] [PubMed] [Google Scholar]
- 173.Narula, J., N. Haider, R. Virmani, T. G. DiSalvo, F. D. Kolodgie, R. J. Hajjar, U. Schmidt, M. J. Semigran, G. W. Dec, and B. A. Khaw. 1996. Apoptosis in myocytes in end-stage heart failure. N. Engl. J. Med. 3351182-1189. [DOI] [PubMed] [Google Scholar]
- 174.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nelson, J. L., K. C. Rice, S. R. Slater, P. M. Fox, G. L. Archer, K. W. Bayles, P. D. Fey, B. N. Kreiswirth, and G. A. Somerville. 2007. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob. Agents Chemother. 51616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 531161-1175. [DOI] [PubMed] [Google Scholar]
- 178.Nieto, C., T. Pellicer, D. Balsa, S. K. Christensen, K. Gerdes, and M. Espinosa. 2006. The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol. Microbiol. 591280-1296. [DOI] [PubMed] [Google Scholar]
- 179.Nomura, M. 1955. The lytic phenomena of Bacillus subtilis. Part 2. Discoveries of the killing factor and autolysine in the lysate and induced lysis of Bac. subtilis with ultraviolet light. J. Agric. Chem. Soc. Jpn. 29678-682. [Google Scholar]
- 180.Nomura, M., and J. Hosoda. 1956. Nature of the primary action of the autolysin of Bacillus subtilis. J. Bacteriol. 72573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.O'Brien, M. J., S. A. Kuhl, and M. J. Starzyk. 1995. Correlation of teichoic acid D-alanyl esterification with the expression of methicillin resistance in Staphylococcus aureus. Microbios 83119-137. [PubMed] [Google Scholar]
- 182.Ochiai, T. 1999. Salt-sensitive growth of Staphylococcus aureus: stimulation of salt-induced autolysis by multiple environmental factors. Microbiol. Immunol. 43705-709. [DOI] [PubMed] [Google Scholar]
- 183.Ogura, T., and S. Hiraga. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 804784-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ohki, K. 1999. A possible role of temperate phage in the regulation of Trichodesmium biomass. Bull. Inst. Oceanogr. Monaco 19287-291. [Google Scholar]
- 185.Olabarria, G., J. L. Carrascosa, M. A. de Pedro, and J. Berenguer. 1996. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J. Bacteriol. 1784765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Olivetti, G., R. Abbi, F. Quaini, J. Kajstura, W. Cheng, J. A. Nitahara, E. Quaini, C. Di Loreto, C. A. Beltrami, S. Krajewski, J. C. Reed, and P. Anversa. 1997. Apoptosis in the failing human heart. N. Engl. J. Med. 3361131-1141. [DOI] [PubMed] [Google Scholar]
- 187.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Palumbo, E., M. Deghorain, P. S. Cocconcelli, M. Kleerebezem, A. Geyer, T. Hartung, S. Morath, and P. Hols. 2006. d-Alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin. J. Bacteriol. 1883709-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Parker, G. F., R. A. Daniel, and J. Errington. 1996. Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology 1423445-3452. [DOI] [PubMed] [Google Scholar]
- 191.Patton, T. G., K. C. Rice, M. K. Foster, and K. W. Bayles. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 561664-1674. [DOI] [PubMed] [Google Scholar]
- 192.Patton, T. G., S. J. Yang, and K. W. Bayles. 2006. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 591395-1404. [DOI] [PubMed] [Google Scholar]
- 193.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45501-510. [DOI] [PubMed] [Google Scholar]
- 194.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112131-140. [DOI] [PubMed] [Google Scholar]
- 195.Perkins, H. R. 1980. The bacterial autolysins, p. 437-456. In H. J. Rogers, H. R. Perkins, and J. B. Ward (ed.), Microbial cell walls. Chapman and Hall, London, United Kingdom.
- 196.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 2748405-8410. [DOI] [PubMed] [Google Scholar]
- 197.Peschel, A., C. Vuong, M. Otto, and F. Gotz. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 442845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 1874392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 1826192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 511051-1070. [DOI] [PubMed] [Google Scholar]
- 201.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7579-586. [DOI] [PubMed] [Google Scholar]
- 202.Pilder, S., J. Logan, and T. Shenk. 1984. Deletion of the gene encoding the adenovirus 5 early region 1b 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J. Virol. 52664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Plas, D. R., and C. B. Thompson. 2002. Cell metabolism in the regulation of programmed cell death. Trends Endocrinol. Metab. 1375-78. [DOI] [PubMed] [Google Scholar]
- 204.Prats, R., and M. A. de Pedro. 1989. Normal growth and division of Escherichia coli with a reduced amount of murein. J. Bacteriol. 1713740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qoronfleh, M. W., J. E. Gustafson, and B. J. Wilkinson. 1998. Conditions that induce Staphylococcus aureus heat shock proteins also inhibit autolysis. FEMS Microbiol. Lett. 166103-107. [DOI] [PubMed] [Google Scholar]
- 206.Raab, R., G. Neal, C. Sohaskey, J. Smith, and R. Young. 1988. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 19995-105. [DOI] [PubMed] [Google Scholar]
- 207.Raju, K. K., S. Gautam, and A. Sharma. 2006. Molecules involved in the modulation of rapid cell death in Xanthomonas. J. Bacteriol. 1885408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Raju, K. K., H. S. Misra, and A. Sharma. 2007. Xanthomonas caspase displays an inherent PARP-like activity. FEMS Microbiol. Lett. 272259-268. [DOI] [PubMed] [Google Scholar]
- 209.Redmond, W. B. 1957. Studies on autolysis of mycobacteria. II. Autolysis of various pathogenic and nonpathogenic strains grown in a medium deficient in nitrogen. J. Bacteriol. 73279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Redmond, W. B., and B. U. Bowman, Jr. 1955. Studies on autolysis of mycobacteria, the effect of temperature change. J. Bacteriol. 69293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, M. Lipsitch, and R. Malley. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 1896532-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Renzoni, A., C. Barras, P. Francois, Y. Charbonnier, E. Huggler, C. Garzoni, W. L. Kelley, P. Majcherczyk, J. Schrenzel, D. P. Lew, and P. Vaudaux. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 503048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in the regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 1852635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rice, K. C., J. B. Nelson, T. G. Patton, S. J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 1863029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ries, W., C. Hotzy, I. Schocher, U. B. Sleytr, and M. Sara. 1997. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J. Bacteriol. 1793892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 361279-1292. [DOI] [PubMed] [Google Scholar]
- 219.Romeis, T., and J. V. Holtje. 1994. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J. Biol. Chem. 26921603-21607. [PubMed] [Google Scholar]
- 220.Romero, P., R. Lopez, and E. Garcia. 2007. Key role of amino acid residues in the dimerization and catalytic activation of the autolysin LytA, an important virulence factor in Streptococcus pneumoniae. J. Biol. Chem. 28217729-17737. [DOI] [PubMed] [Google Scholar]
- 221.Sanchez-Puelles, J. M., J. M. Sanz, J. L. Garcia, and E. Garcia. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 8969-75. [DOI] [PubMed] [Google Scholar]
- 222.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 1832041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 1851803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 1703321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Schwarz, U., and B. Glauner. 1988. Murein structure data and their relevance for the understanding of murein metabolism in Escherichia coli, p. 33-40. In P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (ed.), Anitibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, DC.
- 226.Severin, A., and A. Tomasz. 1996. Naturally occurring peptidoglycan variants of Streptococcus pneumoniae. J. Bacteriol. 178168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shah, D., Z. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shen, H. M., and S. Pervaiz. 2006. TNF receptor superfamily-induced cell death: redox-dependent execution. FASEB J. 201589-1598. [DOI] [PubMed] [Google Scholar]
- 229.Shockman, G. D., and J. F. Barrett. 1983. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu. Rev. Microbiol. 37501-527. [DOI] [PubMed] [Google Scholar]
- 230.Shockman, G. D., and J.-V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 231.Snyder, L. 1995. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 15415-420. [DOI] [PubMed] [Google Scholar]
- 232.Somerville, G. A., B. Said-Salim, J. M. Wickman, S. J. Raffel, B. N. Kreiswirth, and J. M. Musser. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 714724-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9133-137. [DOI] [PubMed] [Google Scholar]
- 234.Steen, A., E. Palumbo, M. Deghorain, P. S. Cocconcelli, J. Delcour, O. P. Kuipers, J. Kok, G. Buist, and P. Hols. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187114-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 715404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 997681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Steinmoen, H., A. Teigen, and L. S. Havarstein. 2003. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J. Bacteriol. 1857176-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sturges, W. S., and L. F. Rettger. 1922. Bacterial autolysis. J. Bacteriol. 7551-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Subramanian, T., M. Kuppuswamy, J. Gysbers, S. Mak, and G. Chinnadurai. 1984. 19-kDa tumor antigen coded by early region E1b of adenovirus 2 is required for efficient synthesis and for protection of viral DNA. J. Biol. Chem. 25911777-11783. [PubMed] [Google Scholar]
- 240.Suel, G. M., J. Garcia-Ojalvo, L. M. Liberman, and M. B. Elowitz. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440545-550. [DOI] [PubMed] [Google Scholar]
- 241.Sugai, M. 1997. Peptidoglycan hydrolases of the staphylococci. J. Infect. Chemother. 3113-127. [Google Scholar]
- 242.Sugai, M., H. Komatsuzawa, T. Akiyama, Y. Hong, T. Oshida, Y. Miyake, T. Yamaguchi, and H. Suginaka. 1995. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 1771491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Suzuki, M., J. Zhang, M. Liu, N. A. Woychik, and M. Inouye. 2005. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 18253-261. [DOI] [PubMed] [Google Scholar]
- 244.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46601-612. [DOI] [PubMed] [Google Scholar]
- 245.Takemori, N., C. Cladaras, B. Bhat, A. J. Conley, and W. S. Wold. 1984. cyt gene of adenoviruses 2 and 5 is an oncogene for transforming function in early region E1B and encodes the E1B 19,000-molecular-weight polypeptide. J. Virol. 52793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Thompson, C. B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science 2671456-1462. [DOI] [PubMed] [Google Scholar]
- 247.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 2811312-1316. [DOI] [PubMed] [Google Scholar]
- 248.Tobin, P. J., N. Mani, and R. K. Jayaswal. 1994. Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie van Leeuwenhoek 6571-78. [DOI] [PubMed] [Google Scholar]
- 249.Tomasz, A., P. Moreillon, and G. Pozzi. 1988. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J. Bacteriol. 1705931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tomasz, A., and M. Westphal. 1971. Abnormal autolytic enzyme in a pneumococus with altered teichoic acid composition. Proc. Natl. Acad. Sci. USA 682627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tomasz, A., and E. Zanati. 1971. Appearance of a protein “agglutinin” on the spheroplast membrane of pneumococci during induction of competence. J. Bacteriol. 1051213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tomioka, S., and M. Matsuhashi. 1978. Purification of penicillin-insensitive DD-endopeptidase, a new cell wall peptidoglycan-hydrolyzing enzyme in Escherichia coli, and its inhibition by deoxyribonucleic acids. Biochem. Biophys. Res. Commun. 84978-984. [DOI] [PubMed] [Google Scholar]
- 253.Tsilibaris, V., G. Maenhaut-Michel, N. Mine, and L. Van Melderen. 2007. What is the benefit for E. coli to have multiple toxin-antitoxin systems in their genomes? J. Bacteriol. 1896101-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tsuchimoto, S., H. Ohtsubo, and E. Ohtsubo. 1988. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 1701461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tuomanen, E., Z. Markiewicz, and A. Tomasz. 1988. Autolysis-resistant peptidoglycan of anomalous composition in amino-acid-starved Escherichia coli. J. Bacteriol. 1701373-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ueda, S., H. Masutani, H. Nakamura, T. Tanaka, M. Ueno, and J. Yodoi. 2002. Redox control of cell death. Antioxid. Redox Signal 4405-414. [DOI] [PubMed] [Google Scholar]
- 257.van Belkum, M. J., J. Kok, G. Venema, H. Holo, I. F. Nes, W. N. Konings, and T. Abee. 1991. The bacteriocin lactococcin A specifically increases permeability of lactococcal cytoplasmic membranes in a voltage-independent, protein-mediated manner. J. Bacteriol. 1737934-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Vanderwinkel, E., and M. De Vlieghere. 1985. Modulation of Escherichia coli N-acetylmuramoyl-L-alanine amidase activity by phosphatidylglycerol. Biochim. Biophys. Acta 83854-59. [DOI] [PubMed] [Google Scholar]
- 259.Veening, J. W., L. W. Hamoen, and O. P. Kuipers. 2005. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol. Microbiol. 561481-1494. [DOI] [PubMed] [Google Scholar]
- 260.Veening, J. W., W. K. Smits, L. W. Hamoen, and O. P. Kuipers. 2006. Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J. Appl. Microbiol. 101531-541. [DOI] [PubMed] [Google Scholar]
- 261.Vilhelmsson, O., and K. J. Miller. 2002. Synthesis of pyruvate dehydrogenase in Staphylococcus aureus is stimulated by osmotic stress. Appl. Environ. Microbiol. 682353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Vollmer, W., M. von Rechenberg, and J. V. Holtje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 2746726-6734. [DOI] [PubMed] [Google Scholar]
- 263.Wagner, E. F., H. Ponta, and M. Schweiger. 1980. Development of Escherichia coli virus T1. The role of the proton-motive force. J. Biol. Chem. 255534-539. [PubMed] [Google Scholar]
- 264.Walensky, L. D. 2006. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ. 131339-1350. [DOI] [PubMed] [Google Scholar]
- 265.Wang, I.-N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54799-825. [DOI] [PubMed] [Google Scholar]
- 266.Wang, I. N., J. Deaton, and R. Young. 2003. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J. Bacteriol. 185779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ward, J. B., and R. Williamson. 1985. Bacterial autolysins: specificity and function, p. 159-166. In C. Nombela (ed.), Microbial cell wall synthesis and autolysis. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 268.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wecke, J., M. Lahav, I. Ginsburg, E. Kwa, and P. Giesbrecht. 1986. Inhibition of wall autolysis of staphylococci by sodium polyanethole sulfonate “liquoid.” Arch. Microbiol. 144110-115. [DOI] [PubMed] [Google Scholar]
- 270.Wecke, J., K. Madela, and W. Fischer. 1997. The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology 1432953-2960. [DOI] [PubMed] [Google Scholar]
- 271.Wecke, J., M. Perego, and W. Fischer. 1996. D-alanine deprivation of Bacillus subtilis teichoic acids is without effect on cell growth and morphology but affects the autolytic activity. Microb. Drug Resist. 2123-129. [DOI] [PubMed] [Google Scholar]
- 272.Wells, J. E., and J. B. Russell. 1996. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl. Environ. Microbiol. 621342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 2951487. [DOI] [PubMed] [Google Scholar]
- 274.White, E. 2006. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 131371-1377. [DOI] [PubMed] [Google Scholar]
- 275.White, E., T. Grodzicker, and B. W. Stillman. 1984. Mutations in the gene encoding the adenovirus early region 1B 19,000-molecular-weight tumor antigen cause the degradation of chromosomal DNA. J. Virol. 52410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Xu, M., A. Arulandu, D. K. Struck, S. Swanson, J. C. Sacchettini, and R. Young. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307113-117. [DOI] [PubMed] [Google Scholar]
- 277.Xu, M., D. K. Struck, J. Deaton, I. N. Wang, and R. Young. 2004. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. USA 1016415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Yabu, K., and S. Kaneda. 1995. Salt-induced cell lysis of Staphylococcus aureus. Curr. Microbiol. 30299-303. [DOI] [PubMed] [Google Scholar]
- 279.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 1781565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang, S. J., P. M. Dunman, S. J. Projan, and K. W. Bayles. 2006. Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 60458-468. [DOI] [PubMed] [Google Scholar]
- 281.Yang, S. J., K. C. Rice, R. J. Brown, T. G. Patton, L. E. Liou, Y. H. Park, and K. W. Bayles. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 1875893-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267836-837. [DOI] [PubMed] [Google Scholar]
- 283.Young, I., I. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8120-128. [DOI] [PubMed] [Google Scholar]
- 284.Young, R., and U. Blasi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17191-205. [DOI] [PubMed] [Google Scholar]
- 285.Young, R. Y. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zagotta, M. T., and D. B. Wilson. 1990. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J. Bacteriol. 172912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zhang, J., and M. Inouye. 2002. MazG, a nucleoside triphosphate pyrophosphohydrolase, interacts with Era, an essential GTPase in Escherichia coli. J. Bacteriol. 1845323-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhang, J., Y. Zhang, and M. Inouye. 2003. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 27832300-32306. [DOI] [PubMed] [Google Scholar]
- 289.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]