Abstract
Summary: Many enzymes and other proteins are difficult subjects for bioinformatic analysis because they exhibit variant catalytic, structural, regulatory, and fusion mode features within a protein family whose sequences are not highly conserved. However, such features reflect dynamic and interesting scenarios of evolutionary importance. The value of experimental data obtained from individual organisms is instantly magnified to the extent that given features of the experimental organism can be projected upon related organisms. But how can one decide how far along the similarity scale it is reasonable to go before such inferences become doubtful? How can a credible picture of evolutionary events be deduced within the vertical trace of inheritance in combination with intervening events of lateral gene transfer (LGT)? We present a comprehensive analysis of a dehydrogenase protein family (TyrA) as a prototype example of how these goals can be accomplished through the use of cohesion group analysis. With this approach, the full collection of homologs is sorted into groups by a method that eliminates bias caused by an uneven representation of sequences from organisms whose phylogenetic spacing is not optimal. Each sufficiently populated cohesion group is phylogenetically coherent and defined by an overall congruence with a distinct section of the 16S rRNA gene tree. Exceptions that occasionally are found implicate a clearly defined LGT scenario whereby the recipient lineage is apparent and the donor lineage of the gene transferred is localized to those organisms that define the cohesion group. Systematic procedures to manage and organize otherwise overwhelming amounts of data are demonstrated.
INTRODUCTION
Gene products and the genes encoding them exhibit a wealth of alternative character states (see Table 1 for definitions). This diversity can be equated with a vast repertoire of biochemical and physiological individualities that define the ever-divergent tree of life. For the most intensively studied gene/gene product systems, experimental documentation exists for only a small fraction of the hundreds of finished genomes that are now available. Given the contemporary pace of genome sequencing, this fraction will become increasingly smaller. Any new experimental results with a given gene product in a given organism immediately become of greatly expanded interest to the extent to which the various character states found and described can be extrapolated to related organisms. But how far can one proceed along a scale of diminishing sequence resemblance before confidence in projections of a known character state (e.g., the specificity of a specificity-variable enzyme) to its closest relatives becomes uncertain? How can one achieve an integrated and credible picture of what evolutionary events proceeded within the vertical genealogical trace and what events intervened via lateral gene transfer (LGT)?
TABLE 1.
Definitions of terms used
| Term | Description |
|---|---|
| Finished genomes | Organisms whose genomes have been fully sequenced; also referred to as “complete genomes” |
| LGT | Lateral gene transfer; any transfer of genetic material between cells which do not have a direct parent-offspring relationship; the term “horizontal gene transfer” is also frequently used |
| Character state | Phylogenetic term for a heritable trait (character) that can have different states; thus, prephenate dehydrogenase is a “character” that can have alternative “states” of being present or absent, of being fused with another protein or not fused with another protein, or of having either a lysine or an arginine residue at position 73, etc. |
| Substrate ambiguity | Descriptive of an enzyme having broad substrate specificity, i.e., able to utilize two or more related compounds as alternative substrate reactants |
| Homologs | Genes descended from a common ancestor; three types of homologs include paralogs, which originate in a common cell via gene duplication, orthologs, which originate by speciation, and xenologs, which originate via LGT |
| Phylogenetic tree | Multiple branches extend divergently from the nodes of a phylogenetic tree; if a single branch is used to represent the tree at a node position, it is said to be collapsed; restoration is achieved by expansion of the tree at that node position |
| Lower Gammaproteobacteria | Informal superorder designation for the class Gammaproteobacteria that is based upon many varied character states of aromatic amino acid biosynthesis; so far, this includes the orders Enterobacterales, Pasteurellales, and Vibrionales and all of the Alteromonadales (except for the family Alteromonadaceae) |
| Upper Gammaproteobacteria | Informal superorder designation for the class Gammaproteobacteria that is based upon many varied character states of aromatic amino acid biosynthesis; so far, this includes the orders Chromatiales, Oceanospirillales, Pseudomonadales, and Xanthomonadales and the family Alteromonadaceae of the Alteromonadales |
| Indels | Collective term for insertions or deletions that account for unmatched regions when amino acid sequence alignments are performed |
| Cohesion group | Collection of a given protein from various organisms whose amino acid sequences assemble as a compact cluster on a phylogenetic tree; the protein tree of adequately populated cohesion groups will generally parallel a section of a 16S rRNA phylogenetic tree, thus rigorously supporting a vertical genealogy (derivation from a common ancestor); organisms having occasional cohesion group members that are inconsistent with the 16S rRNA expectations have been the recipient of LGT originating from some organismal source represented by the cohesion group lineage |
| Supercohesion group | For each of many organisms, the sequences of multiple proteins in a biochemical pathway (e.g., the seven Trp pathway proteins) can be joined together in the same order (concatenated) prior to multiple alignment in order to provide a more powerful basis for cohesion group analysis |
| Xenolog intruder | Sequence member of a cohesion group encoded by a gene which arrived in its host organism via LGT; the donor organism can be assumed to be somewhere within the specific lineage defined by the cohesion group members |
| TyrA protein family | Dehydrogenase enzyme family, members of which function almost exclusively for l-tyrosine biosynthesis; the family exhibits widely variable substrate and cofactor specificities |
| TyrAα and TyrAβ | Two assemblages of TyrA cohesion groups which comprise distinct subhomology groups of the global TyrA protein tree |
| TyrAa | TyrA enzyme that is specific for l-arogenate (arogenate dehydrogenase) |
| TyrAp | TyrA enzyme that is specific for prephenate (prephenate dehydrogenase) |
| TyrAc | TyrA enzyme that can accept either prephenate or l-arogenate as substrate (cyclohexadienyl dehydrogenase) |
In this review, we focus upon a dehydrogenase that functions in l-tyrosine biosynthesis as a prototype example of numerous enzymes which are important to understand but which are at the same time “difficult” subjects for bioinformatic analysis due to moderate sequence length, moderate conservation of sequence, and variable catalytic properties (e.g., substrate specificity). We introduce the concept of cohesion group analysis, whereby the available collection of a given protein homolog is sorted into many separate groups of high identity. Each sufficiently populated cohesion group is phylogenetically coherent and defined by an overall congruence with a distinct section of a 16S rRNA tree. Evolutionary progressions can be rigorously ascertained within cohesion groups, and interesting LGT events can be recognized. Because evolution often proceeds in a circuitous fashion, can make “jumps,” and may even reverse course, the evolutionary path is most reliably traced in a continuum of closely related organisms as a beginning step. Cohesion groups are thus rigorous units for making bioinformatic and evolutionary inferences because they represent genealogical segments taken at relatively shallow hierarchical levels. Once the latter foundation is established, the scope of the analysis can be progressively enlarged because the continual availability of sequences from new genomes is expected to result not only in the formulation of new cohesion groups but also with the merging of cohesion groups as phylogenetic gaps are progressively filled. In addition, as exemplified by previous work with the seven proteins of tryptophan biosynthesis (78), concatenation of multiple proteins has been shown to be a next step that confers greatly expanded resolving power. The assembly of such “supercohesion groups,” which correspond to metabolic segments, is envisioned as an advanced step.
The current TyrA assemblage consists of two subhomology groupings designated TyrAα (40 cohesion groups) and TyrAβ (18 cohesion groups). Evidence in support of the thesis that the TyrAβ subhomology grouping consists of TyrA enzymes that interact with either fused domains or complexed domains of other enzymes is presented. Multiple examples of the logic used to make evolutionary conclusions are given, and examples of tentative evolutionary scenarios that are experimentally testable are also given. Motif variations conserved within a cohesion group are discussed as reflections of probable mechanistic variations of an otherwise widely conserved mechanism. How a rationale can be developed to select key organisms that have ideal phylogenetic placements to advance an overall analysis by filling information gaps with experimental data is demonstrated. Systematic procedures to manage and organize otherwise overwhelming amounts of data are described. Web resources are introduced, which are interactive and freely available. A set of character state snapshots that are displayed on a sortable set of cohesion group trees using tools developed at the SEED (http://theseed.uchicago.edu/FIG/Html/tyrASubsystem.html). This includes a viewer link that displays the context of gene organization around tyrA genes within a cohesion group. The approaches herein applied should be easily applicable to other metabolic subsystems.
TyrA AND l-TYROSINE BIOSYNTHESIS
Enzyme Order Alternatives Dictate Substrate Specificity Patterns
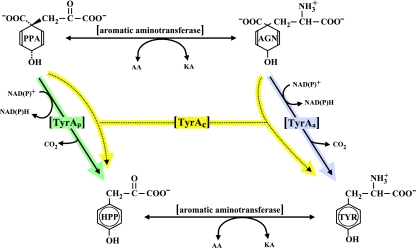
l-Tyrosine biosynthesis almost always deploys a member of the TyrA family, the subject of this review. The alternative flow routes that proceed from prephenate to l-tyrosine are shown in Fig. 1. Detailed visualizations of the alternative flow routes to l-tyrosine portrayed in larger contexts of aromatic amino acid biosynthesis can be found at the AroPath website (http://www.aropath.lanl.gov/Visualizations/index.html). These printable diagrams feature a full suite of clickable enzyme abbreviations that are hyperlinked to a comprehensive table of tyrosine pathway nomenclature. This, in turn, is linked to sequences at the NCBI that exemplify the various nomenclature entries.
FIG. 1.
Alternative flow routes between prephenate and l-tyrosine. The l-arogenate (AGN) flow route to l-tyrosine (TYR) is initiated when prephenate (PPA) is transaminated to produce l-arogenate. A specific and irreversible arogenate dehydrogenase (TyrAa) then converts l-arogenate to l-tyrosine. The 4-hydroxyphenylpyruvate (HPP) flow route to l-tyrosine is initiated when prephenate is utilized by a specific and irreversible prephenate dehydrogenase (TyrAp). An aromatic aminotransferase then transaminates 4-hydroxyphenylpyruvate to produce l-tyrosine. Broad-specificity dehydrogenases that are capable of using both prephenate and 4-hydroxyphenylpyruvate as reaction substrates are known as cyclohexadiencyl dehydrogenases (TyrAc). AA, amino acid; KA, keto acid.
Among the enzymes of amino acid biosynthesis, those of the TyrA family have perhaps been the most widely surveyed in comparative enzymological studies. The TyrA protein family includes enzymes of varied specificities that have in common the catalysis of an oxidative, irreversible reaction in l-tyrosine biosynthesis in all three domains of life. The single known exception to this general physiological role within the homology family is 4-amino-prephenate dehydrogenase, a sparsely distributed enzyme involved in antibiotic synthesis in some species of Streptomyces (7, 81). The universal overall reaction (which includes the latter functional role) involves oxidative decarboxylation and aromatization of one of several possible cyclohexadienyl substrates in the presence of a pyridine nucleotide cosubstrate. Protein families such as the TyrA protein family that can accomplish related but different reactions under the umbrella of a common overall chemistry are herein referred to as pliant proteins. The final two reactions of l-tyrosine biosynthesis consist of an aminotransferase step and the TyrA-mediated dehydrogenase step, which follow from prephenate, an obligatory cyclohexadienyl precursor of l-tyrosine. However, these two steps can occur in either order, a phenomenon that accounts for two mutually exclusive intermediates that may intervene between prephenate and l-tyrosine. If prephenate is first transaminated, then l-arogenate (a cyclohexadienyl amino acid) (82) is generated; if prephenate first undergoes oxidative decarboxylation, then 4-hydroxyphenylpyruvate is generated. Hence, some dehydrogenases of tyrosine biosynthesis are specific for prephenate (prephenate dehydrogenase), whereas others are specific for l-arogenate (arogenate dehydrogenase). A third qualitative category of specificity is one where either of the cyclohexadienyl substrates can be accepted (dual-specificity cyclohexadienyl dehydrogenases). The latter category is probably the most widespread. Cyclohexadienyl dehydrogenases exhibit substantial quantitative variation in that the degree of preference for one substrate or the other varies through a wide range.
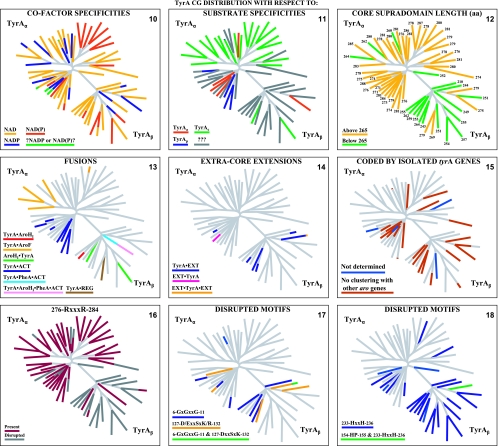
The TyrA family of dehydrogenases also exhibits varied specificities for the pyridine nucleotide substrate that can be accepted. Thus, some are specific for NAD+, some are specific for NADP+, and some will utilize either cofactor (again varying through a wide continuum of preference for the cofactor).
In the following assessment of substrate specificities, it should be noted that various technical pitfalls for working with crude extracts and partially purified enzyme preparations have been recognized over the years. Adequate controls are needed to ensure that prephenate is not contaminated with l-arogenate or prephenyllactate (83), that a phosphatase is not converting NADP+ to NAD+ to give a false-positive result for NADP+ reactivity, that an oxidase is not recycling a reduced cofactor product back to the oxidized form to give unduly low (or null) apparent activities, and that apparent prephenate dehydrogenase activity is not in fact due to the production of l-arogenate via prephenate aminotransferase. Functional complementation of a mutant deficient in a known prephenate-specific dehydrogenase is not proof that the heterologous donor gene specifies a prephenate-specific enzyme because prephenate, accumulated at abnormally high concentrations behind the block, can be anomalously transaminated in vivo to l-arogenate. Indeed, a tyrA mutant of Salmonella enterica serovar Typhimurium, widely used as a source of prephenate, is also the main source of l-arogenate for biochemical preparations (8). Some of these phenomena have been responsible for errors in older literature. Saccharomyces cerevisiae is an example of an organism that has sometimes been assumed to possess a prephenate-specific TyrA dehydrogenase, but we are not aware of rigorous enzymological data in support of this.
Strict specificity for prephenate.
Prephenate-specific dehydrogenases (TyrAp) are thus far limited to two experimental documentations. One is within a large clade of gram-positive bacteria related to Bacillus subtilis, where the most detailed enzymological characterization remains that described previously Champney and Jensen (17). Here, the specificity for prephenate is coupled with specificity for NAD+. The other set of experimental data are from Gluconobacter oxydans, Brevundimonas vesicularis, Brevundimonas diminuta, and species of Acetobacter (13; data not shown). This group couples specificity for prephenate with specificity for NADP+. (All of the latter organisms are also distinctive in the possession of two other character states: an arogenate-specific dehydratase for phenylalanine synthesis and a single 3-deoxy-d-arabino-heptulosonate synthase of a distinctive homology type [AroAII] [38]), which is sensitive to tryptophan-mediated feedback inhibition.) Unfortunately, genomes of species of Brevundimonas (previously named Pseudomonas) have yet to be sequenced. Caulobacter crescentus is inferred to have a prephenate/NADP+-specific dehydrogenase by virtue of its close relationship with Brevundimonas species within the family Caulobacteraceae as well as the motif similarity in the G-rich cofactor discriminator region (see Fig. 4). Gluconobacter and Acetobacter belong in common to the family Acetobacteraceae. By extrapolation, it is possible that the prephenate/NADP+ specificity combination (and perhaps the other two character states) might persist throughout two orders (Caulobacterales and Rhodospirillales) of the Alphaproteobacteria. However, there is a report (51) of specificity for the arogenate/NAD+ substrate combination in Phenylobacterium immobile, which belongs to the family Caulobacteraceae. The sequence of P. immobile is not yet available, and it will be interesting to see whether this unexpected result might be explained by acquisition via LGT.
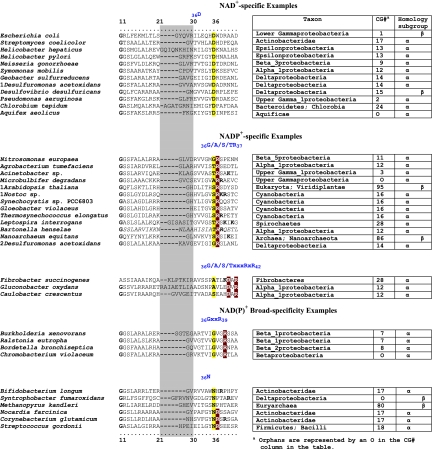
FIG. 4.
Selected examples of motifs in the discriminator region for cofactor binding. N-terminal TyrA sequence patterns that distinguish specificity for NAD+ (top), specificity for NADP+ (middle), and the ability to accept either cofactor [NAD(P)+] (bottom) are shown. Sequences shown begin with the last G (residue 11) of the GxGxxG motif in the Wierenga fingerprint (73). The variable gap of the Wierenga fingerprint is shown as a gray column. Examples of the smallest gap (one residue) and the largest gap (nine residues) are given. Two different patterns are shown for the NADP+ category, and two patterns are shown for the broad-specificity category. Motifs that center around the all-important residue 36 are shown for each of the five groups.
Although TyrA from Escherichia coli is widely referred to as a prephenate dehydrogenase, it is properly designated a cyclohexadienyl dehydrogenase since it exhibits a poor but distinct ability to utilize l-arogenate as an alternative substrate (4, 5). Actually, most of the closely related sister enterics within the lower Gammaproteobacteria, although also exhibiting a clear preference for prephenate, have relatively more dehydrogenase activity with l-arogenate than does E. coli. (4).
Broad specificity.
An early wide-ranging enzymological survey revealed the ubiquity of dual-specificity cyclohexadienyl dehydrogenases (TyrAc) (13). The implication is that an uncertain mixture of both orders of reaction may be ongoing simultaneously in a single organism. Beyond the many subsequent characterizations of partially purified enzymes cited in the following references, detailed studies of purified cyclohexadienyl dehydrogenases include those cloned from Zymomonas mobilis (86), Erwinia herbicola (75), and Pseudomonas stutzeri (77).
Strict specificity for l-arogenate.
l-Arogenate-specific dehydrogenases (TyrAa), also fairly widespread in nature, have been purified and characterized from a cyanobacterium (Synechocystis sp.) (10) and from a higher plant (Arabidopsis thaliana) (64). All photosynthetic bacteria and photosynthetic eukaryotes studied thus far possess l-arogenate-specific, NADP+-specific dehydrogenases. This specificity combination is present in the enzymes from red algae and green algae (9) as well as from Euglena gracilis (14). Coryneform bacteria, other actinomycetes, and Nitrosomonas europeae exemplify bacteria whose possession of l-arogenate-specific dehydrogenases are well documented (see reference 67 and references therein). Although the Nitrosomonas enzyme provides yet another example where specificity for the l-arogenate/NADP+ couple exists, the l-arogenate-specific enzymes from coryneform bacteria will utilize either cofactor, whereas l-arogenate-specific enzymes from most actinomycetes (39, 40) other than coryneform bacteria exhibit NAD+ specificity.
One plausible and interesting selective basis for the enzymatic utilization of l-arogenate and the avoidance of 4-hydroxyphenylpyruvate as an intermediate of l-tyrosine biosynthesis is to prevent cross-pathway complications in cases where 4-hydroxyphenylpyruvate has additional functional roles in metabolism that could lead to futile cycling. For example, the catabolism of l-tyrosine often deploys an initial transamination step that generates 4-hydroxyphenylpyruvate, which could wastefully enter the biosynthetic pathway. An additional example is when 4-hydroxyphenylpyruvate formed from l-tyrosine is utilized as a biosynthetic precursor of plastoquinone and vitamin E, as is uniquely typical of photosynthetic organisms. It is likely no accident that photosynthetic organisms typically utilize l-arogenate as an obligatory intermediate of l-tyrosine biosynthesis, thus avoiding the possibility that 4-hydroxyphenylpyruvate molecules that should be plastoquinone precursors would erroneously enter the l-tyrosine biosynthetic pathway (futile cycling). It is an intriguing example of metabolic plasticity that the latter coupling of biochemical pathways (l-arogenate for l-tyrosine biosynthesis and 4-hydroxyphenylpyruvate for plastoquinone/vitamin E biosynthesis) results in a novel situation where l-arogenate is a precursor of 4-hydroxyphenylpyruvate, with l-tyrosine serving as the intermediate. Thus, in this case, 4-hydroxyphenylpyruvate, rather than being an intermediate of tyrosine biosynthesis, is a following, posttyrosine intermediate of plastoquinone biosynthesis.
Patterns of substrate specificity and regulatory interplay in Tyr/Phe branches.
Organisms such as Bacillus subtilis that deploy a specific prephenate dehydratase and a specific prephenate dehydrogenase at the prephenate branchpoint (the classic pathway configuration) have a regulatory domain known as the ACT domain (49) attached to each of the competitively positioned enzymes to accomplish direct feedback inhibitions that are easily visualized. However, a less straightforward (albeit rather common) pattern for the biosynthesis of l-phenylalanine and l-tyrosine in nature is the utilization of l-arogenate for l-tyrosine synthesis but not for l-phenylalanine synthesis. This occurs in cyanobacteria (69), coryneform bacteria (24-26), and other actinomycetes such as Amycolatopsis methanolica (1). In fact, in the absence of early information that l-arogenate could be a precursor of phenylalanine, l-arogenate was initially named “pretyrosine” (69). With this pathway configuration (consult the figure at http://www.aropath.lanl.gov/Visualizations/TyrPath/TyrPath.htm), the tyrosine branch is unsuited for direct allosteric control. This is because at the branchpoint in this pathway configuration, the prephenate aminotransferase reaction is catalyzed by an aromatic aminotransferase, none of which have ever been found to be subject to allosteric control. It seems likely that catalytic interference caused by the structural overlap of the l-tyrosine end product with the substrates that can be accommodated by aromatic aminotransferases would account for this. On the other hand, the phenylalanine branch is well equipped for allosteric control (since prephenate dehydratase [PheA], which competes with prephenate aminotransferase at the prephenate branchpoint, catalyzes an irreversible initial step of substrate commitment). The ACT domains of cyanobacterial and coryneform PheA proteins mediate a novel mechanism of control to balance flux to both end products. PheA is subject to opposing influences of allosteric activation by l-tyrosine and allosteric feedback inhibition by l-phenylalanine. Starvation for l-phenylalanine enhances the flow of prephenate to l-phenylalanine due to an unrestrained PheA enzyme that is not only transiently free from feedback inhibition by l-phenylalanine but also activated by endogenous l-tyrosine. On the other hand, starvation for l-tyrosine results in the potent inhibition of PheA by endogenous l-phenylalanine, which relieves prephenate aminotransferase from competition with PheA at the branchpoint, thus enhancing flux toward tyrosine. In this manner, l-tyrosine synthesis is indirectly regulated by an enzyme of l-phenylalanine synthesis. It is intriguing that Pseudomonas aeruginosa exhibits a similar pattern whereby flux to l-phenylalanine is regulated directly and flux to l-tyrosine is regulated indirectly. Here, rather than deploying an arogenate dehydrogenase, a cyclohexadienyl dehydrogenase is used. Since the sole chorismate mutase for aromatic biosynthesis is fused to prephenate dehydratase, prephenate is channeled toward l-phenylalanine preferentially. Potent feedback inhibition of prephenate dehydratase by l-phenylalanine allows the release of prephenate from the complex and its utilization for l-tyrosine biosynthesis. This has been described as a channel-shuttle mechanism of regulation (15).
With the background that TyrA proteins that are specific for prephenate are suitable for highly sensitive allosteric control and therefore likely to possess an allosteric domain such as the ACT domain, one might expect that all TyrA proteins that are fused with an ACT domain would be prephenate specific or at least exhibit an overwhelming preference for prephenate. However, TyrA from Streptomyces has an ACT domain but has been reported to be l-arogenate specific (39, 40). This is surprising because the implied inhibition of arogenate dehydrogenase by l-tyrosine could occur, albeit with less refinement, via direct product inhibition without an ACT domain. Moreover, the selective value of this inhibition, however implemented, is questionable because it would cause the accumulation of l-arogenate, which cannot enter the l-phenylalanine pathway directly, requiring back-transamination to prephenate first. One possible mechanism to explain the role of an ACT domain in keeping phenylalanine and tyrosine synthesis balanced would be for l-phenylalanine to activate arogenate dehydrogenase (via the ACT domain) in addition to inhibiting prephenate dehydratase. Another possibility is that Streptomyces might deploy an arogenate dehydratase instead of the much more ubiquitous prephenate dehydratase, thus placing l-arogenate at the metabolic branchpoint (an alternative pathway pattern). If so, backed-up l-arogenate caused by the inhibition of arogenate dehydratase and arogenate dehydrogenase by l-phenylalanine and l-tyrosine, respectively, may in turn feedback inhibit the initial common-pathway step of aromatic biosynthesis (in a pattern of sequential feedback inhibition similar to that discovered in higher plants) (21). This illustrates how an organized basis for desirable experimental inquiries can be driven by detailed analyses that are grounded in phylogenetic context, a point made recently by Osterman (58).
Coexisting Pathway to l-Tyrosine in Some Anaerobic Organisms
It should be noted that in some cases, a second interesting pathway of tyrosine biosynthesis coexists with the chorismate pathway. This second pathway can convert aryl acids to aromatic amino acids and is probably of limited distribution in anaerobes. It has been shown (63) that Methanococcus maripaludis illustrates the ability to scavenge environmental 4-hydroxyphenylacetate produced by the microbial community via peptide catabolism. Following activation to the coenzyme A thioester, reductive carboxylation, and transamination, the l-tyrosine product spares the use of the more expensive de novo pathway derived from chorismate. This aryl acid pathway is well integrated by regulation, such that it is the first-choice option, favored over the coexisting chorismate-derived pathway whenever 4-hydroxyphenylacetate is available.
How Common Is Variation of Substrate Specificity?
Enzymes are so well known for the truly remarkable specificities which often exist that an impression endures that broad-specificity enzymes are not common. However, aside from enzymes such as aminotransferases, which typically possess broadly overlapping substrate specificities (36), many enzymes also carry latent specificity potentials that can be enhanced under positive selective conditions (3, 52). Primordial enzymes with broad substrate specificity are central to the “recruitment hypothesis” (sometimes called the “patchwork hypothesis”) whereby differentially narrowed specificities and regulatory properties were attached to gene products of duplicated genes (34). These genes form paralog families, distinguished by differentially specialized functions but sharing a common catalytic mechanism and united by the ability to regain one or more of the related functions. In contemporary experimental systems, the latter expression of latent catalytic abilities is obtained by the selection of suppressor mutations. Two categories of substrate ambiguity exist: (i) those confined to operation within a pathway where the order of reaction steps can vary (same-pathway ambiguity) and (ii) those where an enzyme is competent for two or more alternative reactions that belong to different pathways (multipathway ambiguity).
Same-pathway ambiguity.
The TyrA family exemplifies same-pathway ambiguity. In most cases, the chemistry needed to build a given molecule dictates a particular order of steps that must be followed. In the case of l-tyrosine biosynthesis, modification of the side chain (via aromatic aminotransferase) and decarboxylation/aromatization (via dehydrogenase) are not interdependent. Thus, the overall conversion of prephenate to l-tyrosine can be accomplished with either order of steps. This is potentially true for any pathway where enzymatic chemistries performed are independent of one another. It would not be surprising if many such ambiguities exist but have not yet been recognized. For example, within the early common aromatic pathway, dehydroquinate proceeds to shikimate in two steps: dehydration (dehydroquinate dehydratase) and reduction (shikimate dehydrogenase). There is no reason a priori that these two steps could not occur in the opposite order, in which case quinate (rather than dehydroshikimate) would be the unique intermediate. Quinate dehydrogenase is widely known as a catabolic enzyme but potentially could perform as a biosynthetic enzyme in some systems.
Multipathway ambiguity.
A fuller modern appreciation of the extent of substrate ambiguity has been greatly accelerated by the contemporary surge in research designed to find and exploit substrate ambiguity for biotechnological objectives. It has become increasingly apparent with modern techniques of metabolite detection that the number of metabolites present in an organism far exceeds the number of genes that would be required if the gene product/enzymes were specific (66). Macchiarulo et al. (50) applied a sophisticated docking algorithm in a computational study that revealed a very high potential for cross-reactivity of endogenous metabolites and enzymes in metabolic reactions. There are two levels of enzymatic promiscuity. In addition to substrate ambiguity (34), it has become clear that surprisingly many enzymes can catalyze seemingly disparate reactions (catalytic promiscuity) that are normally classified as different types of reactions (55). Kurakin (46) made the case that both substrate ambiguity and catalytic promiscuity are in fact expected features in a new paradigm of dynamic and adaptive protein structure. In this paradigm, major and established biochemical pathways operate against a background where many diverse “micrometabolites” are fortuitously generated, a background thought to supply latent evolutionary potential.
Even a minimal sampling of the very recent literature reveals a rapid proliferation of new examples. These include (i) a detailed assessment of the basis for the catalytic promiscuity of E. coli alkaline phosphatase, which can also act as a sulfatase (16); (ii) a new family of lactonases that hydrolyze a variety of lactones, possess low phosphotriesterase activities, and have been shown to be the source of a newly evolved and highly efficient phosphotriesterase (2); (iii) a gentisate dioxygenase that also functions with 1,4-dihydroxy-2-napthoate and salicylate (31); (iv) an ATP-dependent hexokinase from Sulfolobus tokadaii that can phosphorylate glucose, mannose, glucosamine, and N-acetylglucosamine (54); (v) a higher-plant isopropylmalate synthase that not only condenses acetyl coenzyme A (acetyl-CoA) with 2-ketoisovalerate but will also accept 2-oxo acid substrates of two-carbon to six-carbon lengths (19); (vi) a number of variations in the substrate specificities of glutathione synthesis enzymes in comparison to E. coli, Streptococcus agalactiae, and Clostridium acetobutylicum (42); (vii) an amino acid racemase from Pseudomonas putida with an unusual breadth of specificity for amino acids (43); (viii) ATP-forming acetyl-CoA synthetases that accept acetate, propionate, and some longer straight- and branched-chain acyl substrates (32); (ix) an isochorismate pyruvate lyase from Pseudomonas aeruginosa that also has weak chorismate mutase activity (45); and (x) Sulfolobus species that condense pyruvate and aldehydes with two to four carbon atoms (phosphorylated or not) (74). d-2-Hydroxyacid dehydrogenase from Haloferax mediterranei exhibits interesting parallels to the broad-specificity TyrA variants. This d-stereospecific enzyme has broad specificity for alpha-keto carboxylic acids and dual coenzyme specificity (NADH and NADPH) (20). This is striking because most members of this family are NADH dependent. A thorough and scholarly recent review on the subject of enzyme promiscuity was written by Khersonsky et al. (41).
It should be noted that the above-described consideration of same-pathway and multipathway ambiguities is not all-comprehensive with respect to the large topic area of variations that occur in reaction/substrate/cofactor specificity, e.g., phosphorylation in alternative positions of some carbohydrates by the same enzyme and alternative positions of cleavage in the same peptide by protease, etc.
The TyrA Supradomain
The Structural Classification of Proteins (SCOP) database defines a protein domain as an evolutionary unit that can function independently or that can interact with other domains in a multidomain protein to achieve function. TyrA proteins exemplify a case where an N-terminal Rossmann fold and a C-terminal domain comprise a “supradomain” (72), a combination that is essential for catalysis mediated by TyrA. Sun et al. (71) noted that TyrA proteins belong to the “6-phosphogluconate dehydrogenase C-terminal domain-like superfamily” in the SCOP structural classification of protein domains. This superfamily has a ubiquitous N-terminal Rossmann fold joined to a C-terminal extension that is family specific. The latter extension has a common core that is formed around two long antiparallel helices.
A supradomain of about 180 amino acids that is central to TyrA proteins has been identified (10, 77). All TyrA sequences used in this analysis have been trimmed to the boundaries of the supradomain and are available for download (http://theseed.uchicago.edu/FIG/tyra_sequence.cgi). Well-characterized TyrA proteins from Neisseria gonorrhoeae (70), Zymomonas mobilis (86), and Synechocystis sp. (10) as well as the engineered TyrA domain from Pseudomonas stutzeri (77) represent phylogenetically well-spaced proteins (cohesion groups 2, 9, 12, and 16) that exemplify the minimal domain length. It has been suggested (77) that the foregoing four sequences, although of different specificities, define a basic catalytic domain. In this model, it was proposed that the specificity for the side chains of the substrates utilized would parallel the specificity for side chains of inhibitors that are postulated to bind directly to the active site. The only difference between the prephenate and l-arogenate substrate molecules is the side chain, which remains unaltered in the coupled overall reactions of oxidative decarboxylation and aromatization (Fig. 1). Thus, for example, N. gonorrhoeae TyrA has an overwhelming preference for prephenate (pyruvyl side chain) and exhibits classical competitive inhibition by the product 4-hydroxyphenylpyruvate (pyruvyl side chain) but is insensitive to inhibition by l-tyrosine (alanyl side chain).
Cohesion Groups
Rigorous unit of analysis.
Unlike 16S rRNA sequences, which have been famously used to obtain genomic phylogenies, protein sequences are of limited value for making phylogenetic inferences over wide phylogenetic distances, especially if the proteins are neither great in length nor highly conserved. Valid phylogenetic trees for proteins require an adequate continuum of close relatives. Indeed, where genome representation is sufficiently dense in subsections of the overall phylogenetic tree, protein trees can be more informative than 16S rRNA sequences because of the greater resolving power of amino acid variation (84).
Xie et al. (80) assembled trees for the seven individual tryptophan pathway enzymes from then-available prokaryotes in a comprehensive analysis in which divergent paralogs and xenologs engaged in specialized metabolic activities were sorted out from the genes dedicated to primary biosynthesis. Examination of the distribution of gene fusions and gene organization patterns in a context where these distributions were mapped to the 16S rRNA tree elucidated a variety of lineage-specific evolutionary trends. Landmark evolutionary events of operon splitting and rejoining could be reconstructed by following individual divergences in narrow phylogenetic slices and placing these together in a broader phylogenetic context. With avoidance of errors due to ancient paralogy and LGT, one can deduce the most likely character state(s) that represents a given phylogenetic node. The hierarchical placement of each node is determined by the membership of a cohesion group. The more dynamic the evolutionary pace and therefore the greater the divergence, the more narrow (albeit more informationally enriched) the phylogenetic piece captured and therefore the more shallow the position of the node will be. If nodes at the bottom of the phylogenetic tree are sufficiently well represented to deduce any given character state(s) at those nodes, one can hope to apply parsimony principles to deduce the most likely common ancestor at progressively more ancient nodes, thus moving backwards in evolutionary time. It was shown (80) how contexts of flanking genes at relatively shallow hierarchical levels can illuminate which of two evolutionary states is ancestral and which is derived.
Expansion via concatenation: supercohesion groups.
The above-cited work was the basis for a follow-up effort in 2004 (78), which showed that a concatenation of the seven tryptophan pathway proteins yielded protein trees made up of individual sections that, while exhibiting an uncertain connectivity with one another, were each congruent with a portion of the 16S rRNA tree. Ten orphan concatenates were also obtained from genomes with no close relatives among the finished genomes. The seven single-protein tryptophan pathway trees were compared to the concatenate tree. They faintly resembled the concatenate tree but with much weaker support (depending upon highly individualistic degrees of conservation and protein length).
Since the cohesion group approach is fundamental to the thrust of this review, some clarification of terminology is in order. Proteins whose sequences cluster together with high bootstrap values on a phylogenetic tree comprise a cohesion group. Most or all of these proteins are from organisms that also cluster together on a 16S rRNA tree, and this fraction of the cohesion group defines an evolutionary progression of the encoding gene in a vertical genealogy. Genes encoding one or more members of a cohesion group may have been transferred to phylogenetically distant organisms via LGT, and the protein thus will not fit 16S rRNA expectations. Such cohesion group members are called intruder sequences, and the genome possessing it is mosaic with respect to the encoding gene. Cohesion groups that are assembled by the concatenation of two or more proteins of a metabolic pathway are called supercohesion groups. A protein or concatenated protein that is too divergent to share membership in cohesion groups or supercohesion groups is called an orphan sequence and is the sole occupant of an orphan cohesion group or supercohesion group.
Tryptophan pathway congruency groups within the Bacteria were so named because most or all members of a given group were congruent with 16S rRNA expectations. However, some congruency groups contain “intruder” sequences that, due to LGT, are not congruent with 16S rRNA expectations. To avoid semantic confusion, we herein rename these groups “cohesion groups,” since each group is a uniformly cohesive collection of sequences that all originated from a relatively recent ancestor. A given protein member of a cohesion group either is congruent with 16S rRNA expectations and therefore embedded within a vertical genealogy or is an intruder sequence that was translocated to an alien host organism via LGT. LGT of several whole-pathway trp operons and a few partial-pathway trp operons complicated but did not obscure the vertical genealogical trace (78). Indeed, the events of paralogy and xenology could be sorted out because of their demonstrated context within a discernible genealogical trace. The cohesion group approach with the tryptophan pathway subsystem facilitated new and very detailed evolutionary inferences that could be broadly applied to the kingdoms Bacteria and Archaea. In this paper, the cohesion group approach is extended to another branch (TyrA) of aromatic amino acid biosynthesis, with an ultimate objective of extending and integrating the knowledge base to the remainder of this large, multibranched pathway (and indeed with related metabolic subsystems).
TyrA HOMOLOGY ISLANDS: AN ASSEMBLAGE OF COHESION GROUPS
Multimember and Orphan Cohesion Groups
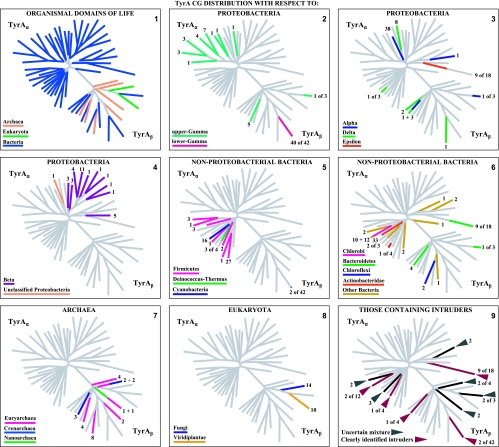
A set of 347 trimmed catalytic core TyrA sequences from all three domains of life were aligned with manual adjustments as needed, particularly at the extreme N-terminal region, where alignment programs consistently yield poor results for the G-rich region that discriminates pyridine nucleotides. The refined alignment was used to obtain a phylogenetic tree. In order to eliminate biases caused by the overrepresentation of relatively large numbers of sequences from closely related organisms, nodes having bootstrap values in excess of a threshold value were collapsed (see below). A single arbitrarily chosen sequence was used to represent each cohesion group at a collapsed node, and these were then used to construct another alignment. Some cohesion “groups” contain a single sequence, and these unnumbered orphans are provisionally designated TyrCG-O. So far, all of the orphan sequences are from the Bacteria. The final alignment, which had received an input of the 18 orphan sequences plus a representative sequence from each of 40 multimember cohesion groups, produced a new tree in which each branch represents a cohesion group. The resulting bifurcated tree, shown in Fig. 2, consists of two subhomology groupings containing branches that all extend from nodes having less-than-threshold bootstrap support. Therefore, the order of branching in each subhomology group is uncertain, and one can describe the tree as TyrA homology islands of uncertain interconnection that are distributed in one (TyrAα) or another (TyrAβ) of two subhomology regions. This tree is used throughout much of this paper as a visually apt way to display various character state features of the cohesion groups. Table 2 provides a list of the organisms containing the sequences chosen to represent the 58 TyrA cohesion groups that are displayed in Fig. 2. The choices were made arbitrarily because any sequence in a cohesion group (even if it is an intruder sequence) is considered to be equally representative of the cohesion group. Table 2 provides the organism abbreviations, the identification numbers in use for sequences at the SEED, and the gi numbers for sequences at the NCBI. The online version of Table 2 (extended table) is hyperlinked to the NCBI taxonomy browser, to the appropriate protein pages at the SEED, and to NCBI gene records. The rightmost column of Table 2 indicates the taxonomic grouping where the cohesion groups are distributed. For example, in TyrCG-1, the TyrA sequence of Erwinia carotovora is representative of multiple orders within the Gammaproteobacteria (but not in all orders of this class). In TyrCG-16, the TyrA sequence of Synechocystis sp. is representative of the entire phylum of Cyanobacteria. The five cohesion groups of the class Betaproteobacteria that are listed are each present at the taxon level of a different family within that class.
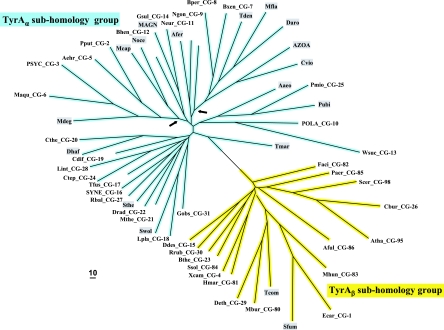
FIG. 2.
Islands of cohesion groups displayed on a phylogenetic tree. Trimmed supradomain sequences, one representing each cohesion group or orphan and aligned as shown in Fig. 3, were used as input into a tree program as described in the Appendix. The resulting radial tree, visualized using TREEVIEW software (62), displays all of the unconnected cohesion groups. Two distinct subhomology groupings are evident: TyrAα (highlighted blue) and TyrAβ (highlighted yellow). See Table 2 for a succinct identification of each cohesion group. A complete, expanded version of Table 2 is available online (http://theseed.uchicago.edu/FIG/Html/TyrAExtended.html). Bootstrap values at all nodes are less than 58%, and therefore, the order of branching shown is not certain. The arrows indicate nodes that are common to TyrA sequences present in most upper Gammaproteobacteria (left arrowhead) or present in most Betaproteobacteria (right arrowhead). See the appendix for a URL for a website at which the organisms indicated by the four-letter codes are identified.
TABLE 2.
Sources and properties of representative sequences of TyrA cohesion groups
| Cohesion groupa | Source of TyrA sequence | AroPath codeb | SEED code | gi no.c | Taxon placement of non-LGT membersd |
|---|---|---|---|---|---|
| TyrCG-1 | Erwinia carotovora | Ecar | fig 218491.3.peg.2590 | 50122273* | Lower Gammaproteobacteria (4 orders) |
| TyrCG-2 | Pseudomonas putida | Pput | fig 160488.1.peg.1756 | 26988501* | Upper Gamma_1proteobacteria (family Pseudomonadaceae) |
| TyrCG-3 | Psychrobacter sp. | PSYC | fig 259536.4.peg.787 | 71038727* | Upper Gamma_2proteobacteria (family Moraxellaceae) |
| TyrCG-4 | Xanthomonas campestris | Xcam | fig 190485.1.peg.1454 | 21230932* | Upper Gamma_3proteobacteria (family Xanthomonadaceae) |
| TyrCG-5 | Alkalilimnicola ehrlichei | Aehr | fig 187272.6.peg.896 | 114320089 | Upper Gamma_4proteobacteria (order Chromatiales) |
| TyrCG-6 | Marinobacter aquaeolei | Maqu | NA | 77952716 | Upper Gamma_5proteobacteria (2 orders) |
| TyrCG-7 | Burkholderia xenovorans | Bxen | fig 36873.1.peg.4890 | 91784814* | Beta_1proteobacteria (family Burkholderiaceae) |
| TyrCG-8 | Bordetella pertussis | Bper-1 | fig 257313.1.peg.827 | 33592104* | Beta_2proteobacteria (family Alcaligenaceae) |
| TyrCG-9 | Neisseria gonorrhoeae | Ngon | fig 242231.4.peg.1532 | 59801853* | Beta_3proteobacteria (family Neisseriaceae) |
| TyrCG-10 | Polaromonas sp. | POLA | fig 296591.1.peg.2815 | 91787673* | Beta_4proteobacteria (family Comamonadaceae) |
| TyrCG-11 | Nitrosomonas europaea | Neur | fig 228410.1.peg.323 | 30248354* | Beta_5proteobacteria (family Nitrosomonadaceae) |
| TyrCG-12 | Bartonella henselae | Bhen | fig 283166.1.peg.1442 | 49476273* | Alpha_1proteobacteria (most orders) |
| TyrCG-13 | Wolinella succinogenes | Wsuc | fig 273121.1.peg.325 | 34482497* | class Flavobacteria |
| TyrCG-14 | Geobacter sulfurreducens | Gsul | fig 243231.1.peg.2590 | 39997701* | Delta_1proteobacteria (order Desulfuromonadales) |
| TyrCG-15 | Desulfovibrio desulfuricans | Ddes | fig 207559.3.peg.3693 | 78358524* | Delta_2proteobacteria (order Desulfovibrionales) |
| TyrCG-16 | Synechocystis sp. | SYNE-3 | fig 1148.1.peg.1391 | 16330562* | Phylum Cyanobacteria |
| TyrCG-17 | Thermobifida fusca | Tfus | fig 269800.4.peg.764 | 72161612* | Subclass Actinobacteridae |
| TyrCG-18 | Lactobacillus plantarum | Lpla | fig 220668.1.peg.1693 | 28271503* | Class Bacilli |
| TyrCG-19 | Clostridium difficile | Cdif | fig 1496.1.peg.2965 | 115250885* | Clostridia_1 (order Clostridiales) |
| TyrCG-20 | Clostridium thermocellum | Cthe-5 | fig 203119.1.peg.2939 | 67874921* | Clostridia_2 (2 orders) |
| TyrCG-21 | Moorella thermoacetica | Mthe-3 | fig 264732.1.peg.2464 | 83590180* | Clostridia_3 (2 orders) |
| TyrCG-22 | Deinococcus radiodurans | Drad | fig 243230.1.peg.1305 | 6458858* | Class Deinococci |
| TyrCG-23 | Bacteroides thetaiotaomicron | Bthe-9 | fig 226186.1.peg.3931 | 29341249* | Class Bacteroidetes |
| TyrCG-24 | Chlorobium tepidum | Ctep | fig 194439.1.peg.84 | 21672925* | Class Chlorobia |
| TyrCG-25 | Petrotoga miotherma | Pmio | NA | NA | Unresolved phylogenetic mixture |
| TyrCG-26 | Coxiella burnetii | Cbur | fig 227377.1.peg.935 | 29654299* | Unresolved phylogenetic mixture |
| TyrCG-27 | Rhodopirellula baltica (Pirellula sp.) | Rbal | fig 243090.1.peg.3009 | 32473675* | Unresolved phylogenetic mixture |
| TyrCG-28 | Leptospira interrogans | Lint-1 | fig 267671.1.peg.2379 | 45658293* | Unresolved phylogenetic mixture |
| TyrCG-29 | Dehalococcoides ethenogenes | Deth | fig 243164.3.peg.722 | 57234714* | Phylum Chloroflexi |
| TyrCG-30 | Rhodospirillum rubrum | 2Rrub-1 | fig 1085.1.peg.3401 | 83592308* | Alpha_2proteobacteria (2 orders) |
| TyrCG-31 | Gemmata obscuriglobus | Gobs | NA | NA | Family Planctomyceteceae |
| TyrCG-O | Acidithiobacillus ferrooxidans | Afer-4 | NA | NA | Upper Gammaproteobacteria orphan |
| TyrCG-O | Aquifex aeolicus | Aaeo | fig 224324.1.peg.1217 | 15606822* | Aquificae orphan |
| TyrCG-O | Azoarcus sp. | AZOA | fig 76114.4.peg.1423 | 56475924* | Betaproteobacteria orphan |
| TyrCG-O | Chromobacterium violaceum | Cvio | fig 243365.1.peg.3407 | 34498862* | Betaproteobacteria orphan |
| TyrCG-O | Dechloromonas aromatica | Daro | fig 159087.4.peg.933 | 71906873* | Betaproteobacteria orphan |
| TyrCG-O | Desulfitobacterium hafniense | Dhaf | fig 49338.1.peg.2227 | 89334457* | Clostridia orphan |
| TyrCG-O | Magnetococcus sp. | MAGN-1 | fig 156889.1.peg.1669 | NA | Unclassified Proteobacteria orphan |
| TyrCG-O | Methylobacillus flagellatus | Mfla-5 | fig 265072.1.peg.206 | 91775427* | Betaproteobacteria orphan |
| TyrCG-O | Methylococcus capsulatus | Mcap-1 | fig 243233.4.peg.782 | 53804254* | Upper Gammaproteobacteria orphan |
| TyrCG-O | Microbulbifer degradans | Mdeg | fig 203122.1.peg.1111 | 90021791* | Upper Gammaproteobacteria orphan |
| TyrCG-O | Nitrosococcus oceani | Noce | fig 323261.3.peg.7 | 77163714* | Upper Gammaproteobacteria orphan |
| TyrCG-O | Pelagibacter ubiquee | Pubi | fig 335992.3.peg.1115 | 71082920* | Alphaproteobacteria orphan |
| TyrCG-O | Symbiobacterium thermophilum | Sthe | fig 292459.1.peg.1361 | 51856245* | Firmicutes orphan |
| TyrCG-O | Syntrophobacter fumaroxidans | Sfum-1 | fig 335543.6.peg.3883 | 71548230* | Deltaproteobacteria orphan |
| TyrCG-O | Syntrophomonas wolfei | Swol-1 | NA | 114566874* | Clostridia orphan |
| TyrCG-O | Thermodesulfobacterium commune | Tcom | NA | NA | Thermodesulfobacteria orphan |
| TyrCG-O | Thermotoga maritima | Tmar | fig 243274.1.peg.339 | 15643112* | Thermotogae orphan |
| TyrCG-O | Thiobacillus denitrificans | Tden | fig 292415.3.peg.543 | 74316971* | Betaproteobacteria orphan |
| TyrCG-80 | Methanococcoides burtonii | Mbur | fig 259564.1.peg.2246 | 91773934* | Euryarchaea (phylum) |
| TyrCG-81 | Haloarcula marismortui | Hmar-2 | fig 272569.1.peg.498 | 55377389* | Halobacteria (class) |
| TyrCG-82 | Ferroplasma acidarmanus | Faci_1 | fig 97393.1.peg.324 | 68141176* | Thermoplasmata (class) |
| TyrCG-83 | Methanospirillum hungatei | Mhun | fig 323259.5.peg.1087 | 88602324* | Methanomicrobia (class) |
| TyrCG-84 | Sulfolobus solfataricus | Ssol | fig 273057.1.peg.273 | 15897245* | Sulfolobales (order) |
| TyrCG-85 | Pyrobaculum aerophilum | Paer-2 | fig 178306.1.peg.1339 | 18312982* | Unresolved phylogenetic mixture |
| TyrCG-86 | Archaeoglobus fulgidus | Aful-1 | fig 224325.1.peg.224 | 11497843* | Unresolved phylogenetic mixture |
| TyrCG-95 | Arabidopsis thaliana | 1Atha | fig 3702.1.peg.1877 | 15218283* | Viridiplantae (kingdom) |
| TyrCG-98 | Saccharomyces cerevisiae | Scer | fig 4932.3.peg.431 | 6319643* | Fungi (kingdom) |
Cohesion groups that belong to the TyrAβ subhomology group, as shown in Fig. 2, are in boldface type.
Organism acronyms consist of four letters: the first letter of the genus name followed by the first three letters of the species name. Any hyphen number designations that follow the acronym proper are used to distinguish potential ambiguities, and multiple TyrA species in a single organism are distinguished by numbers preceding the acronym (as implemented at AroPath [http://www.aropath.lanl.gov/Organisms/Acronyms_sorted_by_species.html]).
Gene identification number. An asterisk indicates that sequencing of the genome is essentially complete. NA, not applicable.
An attempt is made to describe each cohesion group at the hierarchical level at which all organisms having the sequence occupy the same taxon. NCBI's taxonomy page is used as a resource for this. Typically, cohesion groups gather at about the level of family or class, but wide deviations occur in either direction (see the text). In our treatment, the Gammaproteobacteria are clearly divided into two groups, with the lower Gammaproteobacteria and the upper Gammaproteobacteria being the equivalent of “superorder” taxon designations. The organism names and gi numbers are hyperlinked to the taxonomy browser at the NCBI, and the fig/peg numbers are hyperlinked to Protein Pages at the SEED.
Pelagibacter ubique is labeled as “Candidatus Pelagibacter ubique” in many databases. (“Candidatus” refers to an organism that cannot be maintained in a culture collection.)
By design, the orphan sequences used each have as much impact on the alignment (and consequent tree) as do cohesion groups with large numbers of members. The TyrA cohesion groups can be considered to be generally coherent islands in phylogenetic space. As more sequences accumulate, new orphan sequences will emerge, some new sequences will group with previous orphans to yield a multimembered (and newly numbered) cohesion group, and some cohesion groups can be expected to merge as phylogenetic gaps are filled. Eventually, given a sufficient accumulation of new sequences to fill gaps in the phylogenetic space, merged cohesion groups can be expected to yield fewer TyrA cohesion groups that will capture larger phylogenetic slices at deeper hierarchical levels. A complete compilation of the current cohesion group membership (extended table) can be accessed at the SEED (http://theseed.uchicago.edu/FIG/Html/TyrAExtended.html). This is linked to the “Protein Page” at the SEED, which in turn is linked to many popular database resources, including the NCBI (see resources in the Appendix). The extended table is a key interactive resource that displays the source and certain properties of each TyrA sequence. Where it seems clear that a given sequence or group of sequences in a given cohesion group arrived in the host organism by LGT, they are labeled as “intruder sequences.” The taxon level of the organisms possessing the TyrA sequences in a given cohesion group (but excluding intruder sequences) is given in the leftmost column. Organisms with TyrA sequences deemed to be intruder sequences, if present, are listed at the bottom of a given cohesion group. Some cohesion groups are described as being “unresolved phylogenetic mixtures” because one or more of the members appear to be intruder sequences, but it cannot yet be deduced which is the intruder and which is not. Each entry is linked to the NCBI taxonomy browser, to the system used to apply organism acronyms, to the interactive Protein Page at the SEED, and to NCBI gene records. Certain other properties discussed in this review, such as gene fusions, are also tracked in the extended table.
Xenolog Intruders
Multimember cohesion groups are assemblages that are generally congruent with a vertical genealogy, although interesting xenolog intruder sequences were occasionally identified. For example, cohesion group TyrCG-1 contains 40 sequences from a sublineage of Gammaproteobacteria (lower Gammaproteobacteria) that cluster together as expected. Two additional member sequences from several strains of Nostoc (cyanobacteria) are also present as xenolog intruders (that is, a tyrA gene from within the enteric lineage was presumably passed to a common ancestor of Nostoc by LGT). These intruder sequences did not displace the native tyrA genes because Nostoc strains possess a second gene encoding a TyrA sequence which belongs to TyrCG-16, a large cohesive grouping of orthologs present in all 16 finished cyanobacterial genomes available. Thus, the Nostoc tyrA genes in TyrCG-16 are part of an ortholog collection that fits expectations of a vertical genealogy, whereas the Nostoc tyrA genes in TyrCG-1 are not congruent with 16S rRNA expectations (and hence are assumed to be xenolog intruders). The latter xenolog intruders are thought to play a specialized functional role in secondary metabolism (67), and indeed, it has recently been asserted that these genes participate in the provision of l-tyrosine precursor molecules dedicated to the formation of scytonemin, an indole-alkaloid that functions as a sunscreen agent (68).
What is the rationale for the conclusion that the Nostoc genes in the above-described example arrived as intruder sequences rather than the opposite scenario, namely, that the genes from the lower Gammaproteobacteria are LGT intruders derived from Nostoc? Nostoc species are in the same taxon family as species of Anabaena, and Anabaena lacks the intruder sequences. Hence, if Nostoc were the LGT donor, the LGT would have occurred at a relatively recent time after its divergence from the genus Anabaena. In order to account for the possession of the LGT-derived gene by all of the lower Gammaproteobacteria, this fairly recent time would have had to overlap with the more ancient time when the common ancestor of lower Gammaproteobacteria existed, i.e., before divergence to various orders and after divergence from the upper Gammaproteobacteria. These times of Nostoc/Anabaena divergence and upper Gammaproteobacteria/lower Gammaproteobacteria divergence clearly do not overlap, as can be qualitatively assessed by inspection of the appropriate nodes of a 16S rRNA tree. At a hierarchical level of superorder for lower Gammaproteobacteria compared with a level of genus for Nostoc, the lower Gammaproteobacteria lineage is qualitatively older than the Nostoc lineage (even allowing for the uneven hierarchical taxon designations that exist). A gene from a younger lineage cannot have been passed to a common ancestor of the older lineage via LGT because that ancestor would have already diverged very substantially. In short, the common ancestor of lower Gammaproteobacteria could not have been an LGT donor to a Nostoc recipient because the more recent Nostoc lineage had not yet separated at the time when the common ancestor of lower Gammaproteobacteria emerged. Accordingly, it would be feasible for Nostoc to be an LGT donor to only some restricted divergent portion of the lower Gammaproteobacteria membership but not to all of it.
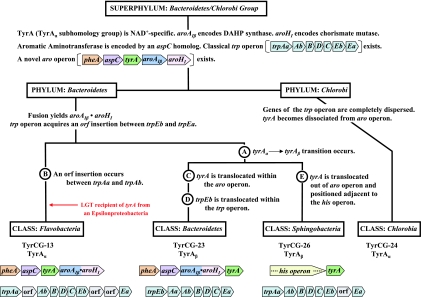
TyrCG-13 is striking because it contains all of the current TyrA sequences from two taxonomic classes (Flavobacteria and Epsilonproteobacteria), each belonging to a different phylum. One set must be derived from a relatively ancient intruder sequence that was acquired from a member of the other set via LGT. The rationale for concluding that TyrA sequences in the class Flavobacteria arose as an intruder that arrived via LGT from an Epsilonproteobacteria source is explained later in this paper, where Fig. 9 is discussed.
FIG. 9.
Tracking milestone evolutionary events in the group Bacteroidetes/Chlorobi. The dendrogram (not drawn to scale) enumerates character states inferred to be present in the common ancestor of the superphylum at the top. Various evolutionary events affecting genes of the ancestral trp and aro operons are indicated at appropriate lineage positions. At the bottom, the gene organizations of the trp operons and the aro operons present in contemporary classes of the phylum Bacteroidetes are shown.
Those cohesion groups labeled in the extended table as an “unresolved phylogenetic mixture” contain one or more xenolog intruders, but it is unclear which one is the donor and which one is the recipient. For example, TyrCG-27 contains three sequences from three different phyla. Since the Anaeromyxobacter and Rhodopirellula organisms are from phyla that have representation in other cohesion groups, an educated (but still uncertain) guess would be that sequences from the latter two organisms are intruder sequences derived from within the phylum Verrucomicrobia. Acquisition of more sequences from appropriate organisms should clarify this.
As a second example, TyrCG-25 contains TyrA sequences from two organisms in different phyla. Petrotoga miotherma is assumed to carry an intruder TyrA sequence derived from a relative of Dictyoglomus miotherma by LGT, and this is based upon the following line of logic. Petrotoga miotherma has a fairly close relative, Thermotoga maritima, whose TyrA sequence is an orphan. Their TyrA sequences would be expected to belong to the same cohesion group because the divergence of TyrA into multiple cohesion groups is usually not seen below the taxon rank of family. Thus, considering the relationship of TyrA sequences from Petrotoga, Thermotoga, and Dictyoglomus, a single LGT event of transfer of TyrA from within the Dictyoglomus lineage to Petrotoga would simultaneously explain why the TyrA sequences from Dictyoglomus and Petrotoga belong to the same cohesion group and why the TyrA sequences from Petrotoga and Thermotoga do not belong to the same cohesion group. Thus, with the information presently available, the former possibility is the most parsimonious inference. Nevertheless, a conservative approach is taken to still label TyrCG-15 as an “unresolved phylogenetic mixture” until the inference made above can be verified or denied with the help of more genome sampling.
Intra-Cohesion-Group Intruders
Even where a set of TyrA cohesion group members are congruent with a 16S rRNA tree, it must be clarified that one cannot assert an absolute absence of LGT events within the lineage. But such LGT events would have been between very close relatives, where LGT can indeed be expected to occur most frequently (47). For example, TyrCG-18 contains 27 sequences from the class Bacilli. As such, these sequences are all congruent with 16S rRNA expectations at the hierarchical level of the class taxon, and we identified no intruders in the current TyrCG-18 membership. However, it is possible, and even likely, that there may have been LGT exchanges within the cohesion group. LGT events at this level will usually not be noticeable, but given a sufficiently large and well-spaced membership, it should be possible to sort out LGT donors and recipients.
Along these lines, it is instructive to revisit the phenomenon whereby the trp operon has been inserted into the middle of a six-member aromatic pathway (aro) operon concomitant with the gain of the regulatory gene mtrB, the loss of trpAb from the trp operon, and the subsequent conscription of pabAb to perform the amidotransferase function for both the tryptophan and p-aminobenzoic acid pathways (80). Note that this constitutes a suite of four different, but interwoven, character states. At the time of the previous study, the organisms known to have these character states were limited to Bacillus subtilis, Bacillus halodurans, and “Bacillus stearothermophilus.” Taxonomic revision has resulted in the placement of “B. stearothermophilus” into a different genus, Geobacillus (53). An additional Geobacillus genome, G. kaustophilus, as well as some additional Bacillus species are now available. The trp operon insertion and the associated character states can now be updated. They are all present in both of the Geobacillus species and in the following clade of Bacillus species: B. clausii, B. subtilis, B. halodurans, and B. licheniformis. Other Bacillus species (B. cereus, B. anthracis, and B. thuringiensis) lack the trp operon insertion and the three associated character states. Thus, in light of these updates, the simplest scenario is that the trp operon insertion into the aro operon, the loss of trpAb, the broadened functional role of pabAb, and the gain of mtrB regulation occurred initially as dynamic innovations in Geobacillus. Subsequently, the supraoperon was transferred via LGT to a common ancestor of the Bacillus clade and was positioned in the aro operon region by displacement via the recombination of flanking homolog genes. The transferred fragment could have been as long as mtrA>mtrB>hepS>menH>hepT>ndk>cheR>aroG>aroB> aroF>trp operon>hisHb>tyrA>aroF>tpr (the supraoperon is shown in boldface type), with recombination perhaps occurring between the mtrA and tpr orthologs (consult Fig. 11 in reference 80 for a view of this conserved gene region). Note that this would have cotransferred the unique trp regulatory gene mtrB, which encodes TRAP (trp RNA binding attenuation protein) (28). The assertion of an intra-cohesion-group LGT that is herein made is amenable to confirmatory follow-up in that protein trees for most or all of the proteins encoded by genes that flank the trp genes should give the same result as that obtained with the TyrA protein tree, namely, that the proteins of one set of Bacillus species are more similar to their counterparts in Geobacillus than to the remaining set of Bacillus species. If so, a significant evolutionary jump (sufficient to define a new trp cohesion group) has occurred in Geobacillus, and the suite of new character states have fairly recently been passed to a common ancestor of a fraction of the Bacillus genus via LGT. Genes flanking the trp operon may not have been much different in comparison of the donor and recipient of LGT. Accordingly, TyrA proteins from all Bacillus species populate the same cohesion group regardless of LGT from Geobacillus or not. Indeed, TyrA proteins from the entire class Bacilli populate a single cohesion group, except for the Symbiobacterium thermophilum orphan. In contrast, the tryptophan subsystem has experienced such dynamic evolutionary changes within Geobacillus that a new trp supercohesion group (based upon the concatenation of Trp proteins) has emerged. This multicharacter set of genes has then exerted quite a profound effect, via LGT, upon a clade of closely related species in a nearby genus. Since Geobacillus strains are comprised of thermophilic species, the above-mentioned proteins in that fraction of Bacillus species that have a Geobacillus origin might tend to have retained the characteristics of high thermotolerance of Geobacillus. This is experimentally testable.
In the near future, when small cohesion groups expand to a better size for analysis, it should be possible to obtain fine-tuned protein trees that will allow inferences of credible LGT events within a given cohesion group. The availability of more genomes representing the genera Bacillus and Geobacillus in particular (as well as the class Bacilli in general) should allow this to be accomplished with the trp/aro multigene system.
Correspondence of Cohesion Groups with Formal Taxon Ranks
The “extended table” at the SEED supplies in the leftmost column the highest-ranking formal taxonomic designations (from the NCBI taxonomy browser) that bound a given cohesion group. Cohesion groups capture their membership at different hierarchical levels, e.g., TyrCG-7 at the level of family, TyrCG-14 at the level of order, TyrCG-17 at the level of subclass, TyrCG-18 at the level of class, and TyrCG-16 at the level of phylum. TyrA sequences from higher plants and fungi populate TyrCG-95 and TyrCG-98 at the hierarchical level of kingdom (but note that the Eukaryota are vastly more subdivided taxonomically than are the Bacteria). We often found that organisms belonging to a formal class contained two or more TyrA cohesion groups that did not match any formal hierarchical subdivisions of that class, such as subclass or order. Names have been provided for many of these subdivided taxons. For example, the Gammaproteobacteria (a formal class) are represented by 10 cohesion groups that carry the following name labels: lower Gammaproteobacteria (TyrCG-1), upper Gamma_1proteobacteria (TyrCG-2), upper Gamma_2proteobacteria (TyrCG-4), upper Gamma_3proteobacteria (TyrCG-5), upper Gamma_4proteobacteria (TyrCG-6), upper Gamma_5proteobacteria (TyrCG-7), and four orphans (Acidithiobacillus ferrooxidans, Methylococcus capsulatus, Microbulbifer degradans, and Nitrosococcus oceani).
A striking list of many divergent character state features of aromatic amino acid biosynthesis points to two distinct subdivisions of the class Gammaproteobacteria. We have termed these the lower Gammaproteobacteria and the upper Gammaproteobacteria. With respect to the multiple character states of aromatic amino acid biosynthesis and regulation, all of the formal Gammaproteobacteria taxon orders (except one) partition cleanly into either the lower Gammaproteobacteria or the upper Gammaproteobacteria. Thus, we treat the Gammaproteobacteria as being comprised of two superorders: (i) the lower Gammaproteobacteria, containing the orders Enterobacterales, Pasteurellales, and Vibrionales and most families within the Alteromonadales, and (ii) the upper Gammaproteobacteria, containing the orders Chromatiales, Oceanospirillales, Pseudomonadales, and Xanthomonadales and part of the Alteromonadales (67). The latter so far consist only of genera within the family Alteromonadaceae, e.g., Marinobacter and Microbulbifer.
The wide variation in the taxon rank delineated by the organisms whose TyrA sequences belong to a particular cohesion group can be attributed to (i) differing evolutionary dynamics in different lineages and (ii) uneven and erratic taxonomic subdivisions in formal nomenclature schemes (i.e., generously sampled and highly studied groupings become subject to more subdividing than do sparsely represented groupings). In general, it is predictable that TyrA sequences from organisms belonging to the same formal taxon up to the level of family will belong to the same cohesion group and will share similar character state properties.
TWO TyrA SUBHOMOLOGY GROUPS
The Master Cohesion Group Alignment
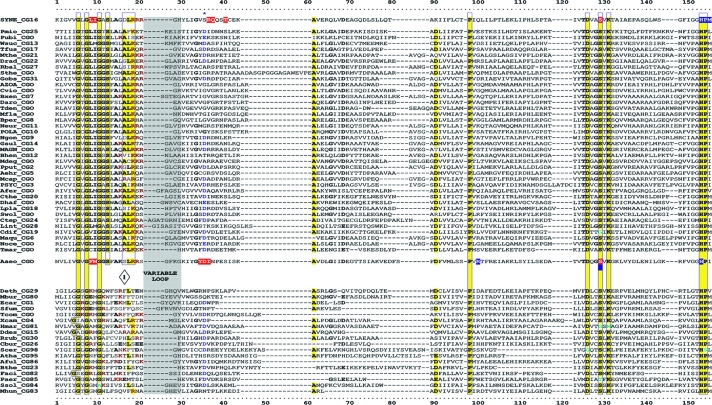
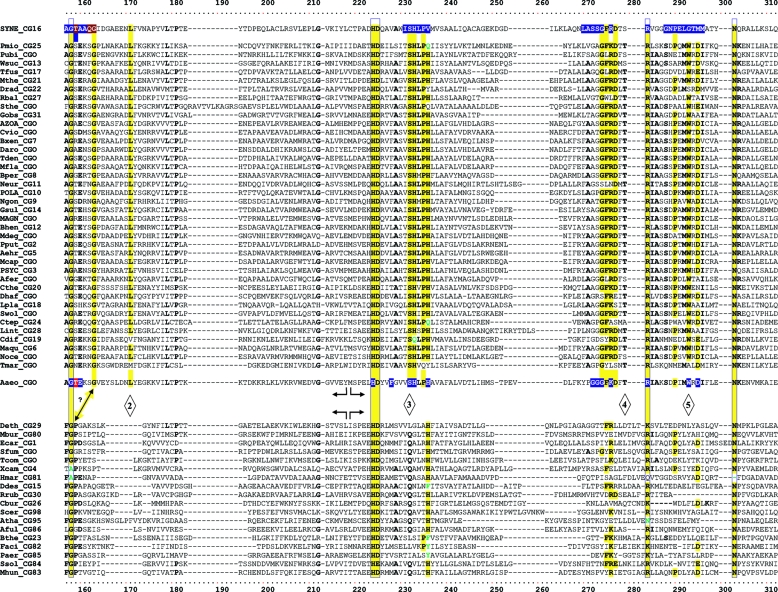
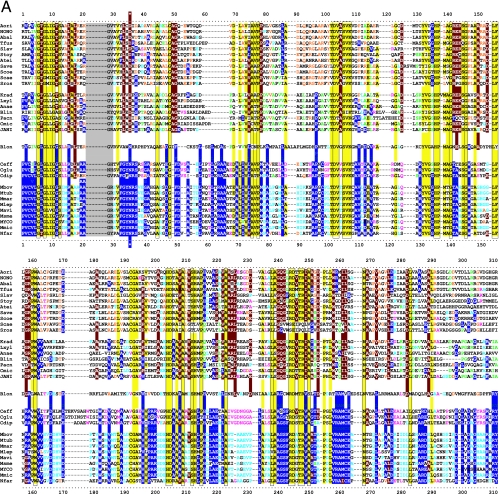
TyrA is a single-homolog assemblage, but the TyrA tree bifurcates into two distinct groupings, labeled in Fig. 2 as the TyrAα and TyrAβ subhomology groups. Although this important bifurcation was not previously recognized, in retrospect, the same split was shown previously (see Fig. 3 in reference 67). Figure 3 shows the master cohesion group alignment that was used to generate the tree portrayed in Fig. 2. Based upon comparisons of TyrA sequences from members of the TyrAα subhomology grouping with the TyrA sequences of E. coli and its closest relatives (which are all TyrAβ members), it was previously concluded (prior to the recognition of a distinct TyrAβ grouping) that the TyrA sequence of E. coli and its close relatives is distinguished from the other sequences by insertion/deletion (indel) structuring (10, 71). Indel structuring refers to a general case where a protein domain makes functionally important contacts with another protein domain to which it is fused. In sequence alignments with homolog counterparts that are not fused and functionally independent of the second protein domain, there are regions of amino acid insertion or deletion that may disrupt conserved and functionally important sequence motifs of the unfused protein. It is envisioned that such important regions are compensated for by a region of the fused protein partner, which exercises an appropriate contact (indel contact). Compensatory indel contacts may operate in both directions for fused proteins, as appears to be the case for the mutually dependent activities of TyrA and chorismate mutase, which are fused in E. coli.
FIG. 3.
Master alignment of cohesion group representatives. The final manual alignment of 58 cohesion group representatives (see the appendix) was imported from the BioEdit alignment editor into the Word program to enhance presentation. TyrAα sequences are shown in the top section bounded at the top and bottom by sequences (Synechocystis sp. and Aquifex aeolicus) for which X-ray crystal structures are available. TyrAβ sequences are shown at the bottom. Amino acid residues shown to be important for NADP+ or for NAD+ in Synechocystis sp. and Aquifex aeolicus, respectively (48, 71), are shown in red with white lettering. Residues modeled in Synechocystis sp. and Aquifex aeolicus to be important for l-arogenate or for prephenate binding, respectively (48, 71), are shown in blue with white lettering. Relative residue position numbers are shown across the top. Invariant or near-invariant anchor residues are enclosed within vertical bars and highlighted yellow. Other highly conserved residues are shown in boldface type and highlighted yellow. Near-invariant residues that differ in a cohesion group representative, but which are nevertheless uniformly different throughout the cohesion group, are shown in boldface green type. The gray vertical band encloses residues in a variable loop (one to nine residues). Divergently pointed arrows at residue positions 216 and 217 mark the boundary between the pyridine nucleotide-binding domain and the catalytic domain. Regions that distinguish TyrAα and TyrAβ, as discussed in the text, are marked with numbers within triangles.
The multiple alignment in Fig. 3 provides a detailed comparison of all 40 TyrAα cohesion group representatives (top) with all 18 TyrAβ cohesion group representatives (bottom). Our collection of trimmed supradomain sequences (10) was used as input into the alignment program. These trimmed sequences (available for download from dropdown boxes activated by cohesion group mouseovers of Fig. 2 online [http://theseed.uchicago.edu/FIG/Html/tyrACGTree.html]) begin with the residues that define the Wierenga fingerprint (73) in the pyridine nucleotide discriminator region at the N terminus of TyrA proteins. Thus, each sequence has been trimmed to begin five residues upstream of the GxGxxG motif (note that three of the cohesion groups within TyrAβ appear to possess an alternative GxxGxxG motif, utilized elsewhere among some other dehydrogenases; these are TyrCG-4, TyrCG-15, and TyrCG-82). For convenience of presentation, the alignment of Fig. 3 does not show about 30 to 35 residues at the C terminus of the supradomain sequences since no patterns of conservation are evident there (however, the complete trimmed supradomain sequences can be obtained at the SEED as described in the Appendix). The vertical gray zone near the N terminus contains from one to nine residues deemed to be within the variable loop of the Wierenga fingerprint. No gaps were allowed prior to position 41 except in the variable loop.
Motif Variations Conserved at the Level of Cohesion Group
Note that some near-invariant residues differ in an occasional cohesion group. Whenever a near-invariant residue differs in a particular cohesion group but nevertheless is conserved in all members of that cohesion group, such deviations are shown in boldface green type in Fig. 3. Although as isolated observations, such deviations might suggest identities as possible pseudogenes, this is quite unlikely when every member of the cohesion group has the same variant residue. For example, the near-invariant DxxSxK motif spanning positions 127 to 132 in Fig. 3 has been shown to be of critical importance in both of the existing X-ray crystal studies (48, 71). Four of the cohesion groups (only one of them in TyrAα) exhibit variations in this motif. It is striking, considering the moderate overall conservation of TyrA sequences, that these DxxSxK motif deviations alone are currently reliable signatures that distinguish the TyrCG-19, TyrCG-81, TyrCG-98, and Tyr-85 cohesion groups. Tyr-98 (containing 14 sequences from fungi) is additionally exceptional at position 155, being the only cohesion group that does possess a proline residue at this highly important position. Such conserved variations undoubtedly correspond to interesting mechanistic variations of an otherwise widely conserved mechanism. As such, these should merit the attention of protein chemists.
Four Regional Sequence Sections That Differentiate TyrAα from TyrAβ
Regions of sequence that clearly differentiate members of TyrAα from members of TyrAβ are indicated by numbers enclosed within diamonds (Fig. 3) as follows. First, The G-rich region of TyrAα is quite orderly, usually being GxGLIGGS and never having adjacent or intervening charged residues. In contrast, the same region of TyrAβ typically has intervening or adjacent R or K residues (shown in red). Occasionally, this region of TyrAβ contains negatively charged D or E residues (shown in blue). Positions 17 to 19 of TyrAα are frequently occupied by the motif ALK/R, with the 19K/R being highly conserved. This motif is altogether absent in TyrAβ. Second, the motif surrounding the highly conserved 158G is 157AGxExxGxxxxxxxL171 in TyrAα, whereas in TyrAβ, the motif is 157FGP159. Note the possibility that the latter motif really corresponds to 162xGx164 of TyrAα. In other words, it may be the G residue at position 163 that is conserved throughout the entire homology family rather than the G residue at position 158. While the former region has been shown to be an important active-site region in both of the X-ray crystal studies done in TyrAα organisms (48, 71), this appears to be a region of indel disruption in TyrAβ. Third, the motif 232SHLPH236 is highly conserved in TyrAα, where it has been shown to be an important active-site region in crystallography studies (48, 71). However, only 236H is conserved in TyrAβ. Fourth, the motif 274GxR/KDxS/TR284, present in TyrAα as an important active-site region (48, 71), is disrupted in TyrAβ, where only the equivalent of the invariant 284R is matched (although here it can be R or K). This motif is discussed in later sections of this review, where, for convenience, it is referred to as the RxxxR motif. Finally, in TyrAα sequences, the motif 290PxMWxDI296 consists of putative active-site residues (48, 71), but this region is totally disrupted in TyrAβ sequences.
COFACTOR DISCRIMINATOR REGION
Specificity Motifs
The pyridine nucleotide-binding domain of TyrA proteins extends to well over the sequence midpoint and abuts the second domain without any linker region (bent, divergent arrows indicate the join points of the two domains in Fig. 3). However, the cofactor specificity [NAD+, NADP+, or NAD(P)+] is determined by a relatively short ADP-binding βαβ discriminator region at the N terminus of TyrA. A negatively charged residue (D or E) at position 36 (Fig. 3) is all-important for hydrogen binding to the diol group of the ribose near the adenine moiety in NAD+-specific enzymes. A negatively charged residue at position 36 absolutely precludes NADP+ utilization. An asparagine residue at position 36 appears to enable the binding of both NAD+ and NADP+ (67, 85).
Most of our curated TyrA sequences can be assigned to one of three specificity classes: specific for NAD+, specific for NADP+, or able to utilize either cofactor. Figure 3 shows an alignment of some representative TyrA discriminator regions in order to illustrate the recognizable patterns. The alignment begins with the third G of the GxGxxG motif; this corresponds to residue 11 of the Wierenga fingerprint (73). Residues that occupy a variable loop (positions 22 to 30) are shaded in Fig. 3 and 4, and gaps are allowed only in this region. The classic Wierenga fingerprint allows for a variable loop containing two to five residues, but our alignment studies suggest that the loop can contain from one to nine residues. Thus, TyrA from Gloeobacter violaceus has only a single residue within the variable loop, whereas the TyrA proteins from Gluconobacter oxydans and from Helicobacter hepaticus contain nine residues within the variable loop. As the name would imply, these variable-loop regions are not always highly conserved within a cohesion group. Thus, Leptospira interrogans and Fibrobacter succinogenes, the two members of TyrCG-28, have variable-loop regions that show few matches (Fig. 4). Such differences in match identities and also in loop lengths can be seen in Fig. 4 for the various Betaproteobacteria and upper Gammaproteobacteria that have been selected. In other cases, though, the variable-loop regions seem to be surprisingly consistent, as exemplified by the cyanobacteria and by the Actinobacteridae.
Figure 4 illustrates selected examples of motifs associated with specificities for NAD+ (top), NADP+ (middle), and NAD(P)+ (bottom). Each specificity category is represented within both the TyrAα and TyrAβ subhomology groups, as indicated on the right. NAD+-specific enzymes possess a D (or occasionally an E) at position 36; this aspartate (or glutamate) residue acts to repel the negatively charged NADP+. The majority of cohesion groups (33 of 58) possess a D residue at position 36, while three cohesion groups (the latter all being in TyrAα) possess an E residue at position 36. TyrA from E. coli and all other members of TyrCG-1 display a DW motif corresponding to positions 36 and 37. This alone is sufficient to distinguish sequence members of TyrCG-1 from any other TyrA sequences. The placement of a D residue at position 36 in the alignment is usually unambiguous. However, Arabidopsis thaliana in the second grouping shown in Fig. 4 illustrates a case where, without the benefit of experimental data, the D residue at position 39 could easily have been aligned to position 36 without creating an abnormally short variable loop. However, rigorous experimental data allow the association of this sequence pattern with NADP+ specificity, and it can be seen that the Arabidopsis thaliana sequence in the cofactor discriminator region aligns well with other experimentally known NADP+-specific enzymes, such as those from Nitrosomonas europaea, Acinetobacter sp., and Synechocystis sp.
NADP+-specific enzymes typically deploy one G/S/T/A residue at position 36, and this is followed most commonly by RS (but sometimes by RR or RK). A second pattern of NADP+ specificity (36G/A/S/TxxxRxR42) was recognized from the sequence from Gluconobacter oxydans, which is known experimentally to be NADP+ specific (and prephenate specific). The pattern from Fibrobacter succinogenes and Caulobacter crescentus matches this quite well. Here, the positively charged R residue, normally located at position 37, is shifted three positions downstream, and the R residue at position 42 may be significant as well.
A broad capability to utilize either of the two cofactors is achieved by one of two variations: a 36GxxR39 motif and a 36N motif. The 36GxxR39 motif, as seen in some of the TyrA sequences from the Betaproteobacteria (70), resembles the 36G/S/T/AR37 motif of NADP+-specific enzymes. From an inspection of Fig. 4, one could envision an evolutionary transition from 36G/S/T/AR37 to 36GxxR39 to have occurred by the insertion of VG (or similar residues) after the G at position 36, displacing the important basic R residue to position 39. Of course, the opposite scenario, whereby two residues are deleted from 36GxxR39, is equally plausible. The presence of asparagine at position 36 correlates with the ability to use both cofactors, as established by experimental data from Corynebacterium glutamicus (24, 25). Interestingly, the 36NRS38 variation correlates with an order-of-magnitude preference for NADP+ in coryneform bacteria, whereas the 36GxxRS40 variation correlates with equal preference for NAD+ or NADP+ in Betaproteobacteria such as Ralstonia. Thus, in cases where 36N is not followed by RS (all in organisms not yet examined experimentally), it would be interesting to know if the preference for NADP+ is lessened, perhaps markedly.
Some cohesion groups have a split membership with respect to cofactor specificity (see Fig. 6, panel 10). One of these, TyrCG-17, is discussed in detail below.
FIG. 6.
Snapshots of character state features. Eighteen panels are shown as mini-semblances of the bifurcated tree of cohesion groups portrayed in Fig. 2. Various character states of interest are displayed on these trees to facilitate comparisons. The organisms in all three domains of life that host the various TyrA cohesion groups are profiled in panels 1 to 8. The numbers at the branch ends in panels 2 to 8 indicate the total number of sequences within the cohesion group. An appropriate fraction of a given branch is color coded if the cohesion group has a “mixed” membership. Thus, in panel 3, the proximal half of the TyrCG-13 branch is color coded for the nine sequences of the Epsilonproteobacteria. In panel 6, the other (distal) half of the branch is color coded to indicate the nine TyrA sequences from the class Flavobacteria (Bacteroidetes). The locations of cohesion groups containing intruder sequences are identified in panel 9, e.g., the Flavobacteria mentioned above. TyrA character states associated with cofactor and cyclohexadienyl substrate specificities are displayed in accord with the color-coded legends (panels 10 and 11). In panel 10, “?NADP or NAD(P)?” means that whether the enzyme is NADP+ specific or whether it can use either cofactor is unknown, but we know that it cannot be NAD+ specific. The amino acid lengths of trimmed core supradomain TyrA sequences are given at the branch ends of panel 12. TyrA enzymes encoded by tyrA genes fused to other genes are depicted in panel 13. TyrA enzymes encoded by tyrA genes which are isolated from other aromatic pathway genes are shown in panel 15. The color-coded legends for panels 17 and 18 show conserved motifs (Fig. 3), which are disrupted or absent in the indicated cohesion groups (or a fraction thereof). These panels can be accessed at http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html, where they can be expanded and sorted in order to facilitate comparisons. The interactive panels are linked to the extended table in order to quickly view the membership of any cohesion group of interest.
Cofactor Specificity Divergence in TyrCG-17
TyrCG-17 is a large cohesion group made up of the Actinobacteridae (subclass rank), whose experimentally studied membership so far possess l-arogenate-specific TyrA proteins. These have, however, diverged with respect to the cofactor-substrate utilized, being either NAD+ specific or broadly NAD(P)+ specific. This divergence of cofactor specificity correlates perfectly with a bifurcation of TyrA sequence clustering, which is evident in both the multiple alignment shown in Fig. 5A and the corresponding protein tree (Fig. 5B). This separation suggests that the sequence divergence has been driven by the alteration of functional specialization for the cofactor. Song et al. (67) in fact found that these two groups (then designated Actinobacteridae_1 and Actinobacteridae_2) were located on apparent separate branches of their TyrA protein tree (see Fig. 3 in reference 67). In retrospect, this was due to bias created by the overrepresentation of each group. Among the nodes collapsed in the process used to formulate cohesion groups were the two where all of the sequences in each of the former Actinobacteridae_1 and Actinobacteridae_2 groups joined with high bootstrap values. Choosing a representative sequence for each collapsed node and then obtaining a new tree resulted in the merging of both chosen representatives into the same cohesion group.
FIG. 5.
Divergence of cofactor specificity within cohesion group TyrCG-17. TyrA sequences from members of nine families of the order Actinomycetales and one (Bifidobacterium longum) from the family Bifidobacteriaceae within the order Bifidobacterales were aligned by entering the appropriate trimmed sequences into ClustalX, carrying out manual adjustments with the aid of the BioEdit alignment editor, and entering the final alignment into the Phylip program. The alignment (A) and the tree visualized with TREEVIEW (B) were imported into Word to enhance presentation. The Bifidobacterium longum sequence is shown in the middle of A for comparison with TyrA sequences from the single family (Corynebacterinceae) members in the bottom block and with members of the remaining families of the Actinomycetales (top block).
Figure 5A shows a multiple alignment of trimmed TyrA supradomains from members of the TyrCG-17 cohesion group. Sequences used begin with the first residue of the Wierenga fingerprint (73) at position 1 and end with the last strongly conserved region at position 310. The outgroup sequence from Bifidobacterium longum, the sole available TyrA sequence from the order Bifidobacteriales, is shown in the middle of the figure in order to facilitate a comparison with the two distinctly divergent sets of sequences from the sister order Actinomycetales. Invariant or near-invariant residues are shown with yellow highlighting. Residues conserved only in the upper grouping are highlighted in red, and residues conserved only in the lower grouping are highlighted in blue. Since the Bifidobacterium longum sequence has an N residue at position 36, it is presumed to possess broad cofactor specificity (Fig. 4), and we suggest that this was the ancestral state prior to the divergence of the two orders. This character state of broad cofactor specificity was conserved in the lower block of sequences, which are TyrA members from a single family within the order Actinomycetales. These divide into the genera Corynebacterium and Mycobacterium. On the other hand, the remainder of the families that populate the Actinomycetales evolved toward narrowed cofactor specificity for NAD+. This scenario is consistent with the much greater similarity of the Bifidobacterium longum sequence with the lower block of sequences than with the upper block of sequences. In the upper block, two subgroupings are apparent: one contains TyrA sequences from the families Pseudocardineae, Streptosporangineae, Streptomycineae, and Micromonosporineae, while the other contains TyrA sequences from the families Frankineae, Micrococcineae, Actinomycineae, and Propionibacterineae. This separation is also conspicuous in the protein tree shown in Fig. 5B.
Members of TyrCG-17 are thought to all be l-arogenate specific, and it is perhaps surprising that the narrowing of cofactor specificity for the upper block of sequences in Fig. 5A is associated with changes throughout the entire TyrA sequence rather than just the N-terminal domain. However, existing X-ray crystal studies have pointed to a substantial functional intercalation of the two domains comprising the TyrA supradomain. In another cohesion group, TyrCG-3, the three member sequences share the character state of being broad-specificity cyclohexadienyl dehydrogenases but differ in cofactor specificities, with two members being NAD+ specific and one being NADP+ specific. Here, where the overall amino acid identity is high (56%), the divergence of cofactor specificity has triggered more sequence divergence in the N-terminal domain (46% identity) than in the C-terminal domain (74% identity) (the join point of the N-terminal and C-terminal domains is marked by divergent arrows in Fig. 3).
SNAPSHOTS OF TyrA CHARACTER STATES IN A PHYLOGENETIC CONTEXT
A Tool To Track Character State Variations
The comparative assessment of various character state features of TyrA proteins is potentially useful for a detailed bioinformatic analysis. The ability to track various features of interest that covary with one another can lead to important insights and to testable hypotheses. Considering the large number of genomes already sequenced, together with the proliferation of new genomes coming online, a systematic way to manage and access data that builds upon a basic store of careful and detailed study is needed. Otherwise, the volume of information is overwhelming. Some questions will be generally applicable to most metabolic subsystems. For example, what are the phylogenetic boundaries of the organisms that in common possess TyrA proteins belonging to a given cohesion group? Which events of LGT can be tracked through the identification of intruder sequences? What gene fusions are present in a given cohesion group (thus implying a common origin)? If gene fusion panels like panel 13 of Fig. 6 were available for multiple subsystems, one could assemble them for comparison across subsystems, e.g., to answer a question such as what are all of the different gene fusions in the Firmicutes, and where are their various hierarchical placements (thus indicating recent or ancient origin)? This would be determined by viewing the panels from other subsystems that correspond to panel 5 (to locate Firmicutes) and panel 13 (to locate gene fusions) of Fig. 6 in parallel. Other features of a subsystem protein will vary with highly individualistic properties. Thus, while many enzymes are highly specific, TyrA enzymes exhibit greatly varied ranges of acceptance for both the cofactor substrate and the cyclohexadienyl substrate. This generates alternative character states of physiological importance that are herein captured as snapshots. Figure 6 contains 18 panels that are a basis for maintaining organized data about features of TyrA proteins that are deemed useful for comparative and evolutionary analyses. Each panel is a semblance of Fig. 2, and the overall effort is oriented to a comparison of how the TyrAα and TyrAβ subhomology groups differ from one another.
Online at the SEED (http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html), clicking the “compare TyrA panels” option allows a choice of up to three panels for side-by-side comparison. The individual panels are expandable with a built-in magnifier, and links are provided at the top for navigation to the extended table.
Phylogenetic Boundaries
Panel 1 of Fig. 6 shows the distribution of cohesion groups among the three domains of life. All TyrA sequences from the Archaea and Eukaryota reside in the TyrAβ subhomology grouping, whereas most (but not all) TyrA sequences from Bacteria are located in the TyrAα subhomology grouping. The allocation of cohesion groups among the major taxa within the superkingdoms of Archaea and Eukaryota are shown in panels 7 and 8, respectively, of Fig. 6. Numbers at the ends of branches (panels 2 to 8) indicate the numbers of sequences contained within a cohesion group. Hence, branches labeled with a “1” indicate an orphan sequence. In panels 5 and 6, the presence of cohesion groups is displayed at the level of phylum, except for the Actinobacteridae (subclass) in panel 6. Panels 2 to 4 show the distribution of cohesion groups present in the various class divisions of the phylum Proteobacteria. The Gammaproteobacteria have been separated into the lower Gammaproteobacteria “superorder” and the upper Gammaproteobacteria “superorder” because extraordinarily dynamic evolutionary jumps in the lower Gammaproteobacteria have created qualitatively significant distinctions. Indeed, features of aromatic biosynthesis in the upper Gammaproteobacteria and the Betaproteobacteria are much more similar to one another than is the case when upper Gammaproteobacteria and lower Gammaproteobacteria are compared (67, 80). It is interesting that a significant change of state of the histidine operon, whereby a gene fusion is embedded in a compact operon, occurs uniquely in exactly the same organisms that we refer to as the lower Gammaproteobacteria (23). The gene organization of the histidine pathway for the upper Gammaproteobacteria differs sharply from that of the lower Gammaproteobacteria.
The phylum Proteobacteria exhibits relatively great overall divergences with respect to TyrA sequences such that cohesion groups usually parallel a formal order or a collection of orders. Only TyrA sequences from the Epsilonproteobacteria are represented at the class taxon level as members of a single cohesion group (Fig. 6, panel 3). The lower Gammaproteobacteria (consisting of the orders Enterobacteriales, Pasteurellales, Alteromonadales, and Vibrionales) possess TyrA sequences that populate TyrCG-1 in the TyrAβ subhomology group. Among the upper Gammaproteobacteria, members of the order Pseudomonadales possess TyrA sequences that fall into TyrCG-2 and TyrCG-3. Members of the order Xanthomonadales possess TyrA sequences that belong to TyrCG-4 in the TyrAβ subhomology grouping. Members of the order Chromatiales possess TyrA sequences that belong to TyrCG-5, except for Nitrosococcus oceani, whose TyrA sequence is an orphan. Members of the order Oceanospirillales possess TyrA sequences that belong to TyrCG-6.
One member of TyrCG-6 (Marinobacter aquaeolei) as well as one orphan of the upper Gammaproteobacteria (Microbulbifer degradans, recently reclassified as Saccharophagus degradans) are classified at the NCBI as belonging to the Alteromonadales. However, TyrA members present in the Alteromonadales are otherwise housed by lower Gammaproteobacteria. M. aquaeolei and M. degradans clearly seem to have multiple properties characteristic of upper Gammaproteobacteria. They lack many evolved characteristics of lower Gammaproteobacteria. For example, a member of the latter superorder (exemplified by species of Shewanella within the Alteromonadales) possesses TyrAβ, an aroHI-tyrA fusion, a tyr operon containing a newly emerged paralog encoding a third regulatory isoenzyme of 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase, a tyrR regulatory gene, and a complete trp operon including a trpD-trpC fusion. These are all newly evolved character states that typify lower Gammaproteobacteria (more detail can be found in Fig. 7 and Table 6 in reference 67). In striking contrast, all upper Gammaproteobacteria (including the above-mentioned Marinobacter aquaeolei and Microbulbifer degradans) possess the TyrAα subhomology type of TyrA, they lack the aroHI-tyrA fusion, they lack a tyr operon containing a gene encoding a regulatory isoenzyme of DAHP synthase, they lack a tyrR repressor gene, tyrA is in fact within a supraoperon containing other aromatic pathway genes, and a “split” trp operon (80) is present. In short, organisms currently contained within the Alteromonadales are a mixture of lower Gammaproteobacteria and upper Gammaproteobacteria that are sharply distinguished by a suite of differing character states.
FIG. 7.
Independent tyrA-aroF fusions in proteobacterial amino acid sequences of TyrA-AroF fusions from the upper Gammaproteobacteria and the Betaproteobacteria were aligned with TyrA and AroF concatenates from other members of these proteobacterial divisions where these genes are unfused. The alignment was used to obtain the Phylip tree shown. Values of bootstrap support are indicated at nodes. Proteins encoded by tyrA-aroF fusions are enclosed within the orange patterning.
Two other TyrA sequences from the upper Gammaproteobacteria are orphans, with one (Methylococcus capsulatus) being from the order Methylococcales and the other (Acidithiobacillus ferrooxidans) being from the order Acidithiobacillales. New TyrA sequences from incoming genomes belonging to these orders will very likely join the orphans, producing new cohesion groups. The distribution of cohesion groups in the various nonproteobacterial taxa are covered in panels 5 and 6 of Fig. 6, and cohesion groups hosted by Archaea and Eukaryota are illustrated in panels 7 and 8.
Xenolog Intruders
The presence of xenologs in some cohesion groups is portrayed in panel 9 of Fig. 6. Note that the “intrusion” event does not refer to the cohesion group but rather refers to the genome that hosts the intruder sequence. In some cases, a mixture of phylogenetically incoherent sequences coexists such that while it is obvious that one or the other is an intruder, it is unclear which is and which is not. The latter cohesion groups are referred to as an “unresolved phylogenetic mixture.” After exercising the “compare TyrA panels” option in the online version of Fig. 6 at the SEED (http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html), the color-coded cohesion groups containing intruders in panel 9 are specially labeled with a magnifiable cohesion group number. This allows navigation to the extended table (via the clickable links at the top) in order to view the entire membership of any cohesion groups of interest.
Substrate Specificities
The distributions of the various specificities for the cofactor substrate are shown in panel 10 of Fig. 6. Relatively few of these specificities are in doubt. In a few cases, it is uncertain whether the enzyme is specific for NADP+ or whether both pyridine nucleotides can be used (green highlighting). The overall variation in specificity is not particularly different when the two subhomology groupings are compared. Cofactor specificity sometimes varies within a single cohesion group, suggesting that some specificity changes have been quite recent.
The distributions of the three specificity patterns for the cyclohexadienyl substrate are shown in panel 11 of Fig. 6. At least within the TyrAα subhomology grouping, the broad-specificity cyclohexadienyl dehydrogenases appear to be most common. Relatively few specificities within the TyrAβ grouping are known. Both the cofactor substrate and cyclohexadienyl substrate specificities are listed in the right column of the extended table, to which panels 10 and 11 are linked online after choosing “compare TyrA panels.” Specificities that are considered to be certain are displayed in boldface type in the table; specificities thought to be probable but not certain are shown in lightface type.
Gene Fusions
The tyrA gene has been a popular fusion partner. Fusions of tyrA with various protein partners occur throughout the TyrAα and TyrAβ subhomology groupings, as displayed in Fig. 6, panel 13. The aroHI homolog of chorismate mutase has fused with tyrA in two cohesion groups within TyrAβ, and it is evident that these must have been independent gene fusions. In another case, the same two genes are fused, but here, aroHI is fused to the C terminus of tyrA (tyrA-aroHI). (Note that aroHI is well known in the literature as aroQ; consult a study by Okvist et al. [56] for an alternative classification of chorismate mutase subtypes). tyrA is fused to aroF in some of the upper Gammaproteobacteria and Betaproteobacteria (see below). The closest relatives of the latter that lack the fusion typically possess adjacent tyrA-aroF genes. To compare the gene organization at the level of cohesion group, one can use the “gene neighborhood” button in the extended table as described below. tyrA is fused to several known regulatory genes called ACT and REG, sometimes in combination with other structural genes such as aroHI and pheA. Other N-terminal or C-terminal extensions of tyrA exist (Fig. 6, panel 14), which could be regulatory domains. Some fusions are present in only a fraction of the cohesion group membership, indicating that the origin of these fusions is recent, i.e., a new fusion event or a recent LGT. Browsing the individual membership of a given cohesion group in the extended table allows one to view the existing fusion identities (in one of the right columns).
Gene Context of tyrA
tyrA is frequently adjacent to other aromatic pathway genes, often being within an operon or within a supraoperon. Panel 15 of Fig. 6 shows exceptions to the latter, namely, those cohesion groups or portions of cohesion groups where tyrA is encoded by an “isolated gene.” Even if other aromatic pathway genes are not far away, we refer to tyrA here as an isolated gene. Just because tyrA is unlinked to genes with obvious functional relationships does not necessarily mean that the surrounding gene organization is not conserved. Positioning of the cursor at the arrowhead at the top of each cohesion group in the extended table online (http://theseed.uchicago.edu/FIG/Html/TyrAExtended.html) activates a clickable pop-up that enables the comparative viewing of all tyrA gene neighborhoods in that cohesion group.
Gene organization is not highly conserved and can be quite erratic, even within short phylogenetic distances (33). Even operons are surprisingly vulnerable to disruption, as documented in detail with the trp operon (80). However, functionally related genes frequently retain linkage relationships over at least short phylogenetic distances, sometimes with distinct shuffling patterns. The comparative analysis of gene clusters can be extremely informative, yielding valuable functional and evolutionary clues. Examples of how this approach can elucidate functional roles for “missing genes” have been reported (30, 59, 61).
Each cohesion group section of the extended table has an arrowhead button after the cohesion group number, which allows navigation to a direct single-view comparison of the gene organizations surrounding tyrA within that cohesion group. These are extracted from all of the individual graphics that appear on the Protein Pages of each sequence at the SEED for which there is a current identification number. This accommodates a very convenient way to view the extent to which the gene organization is consistent within a cohesion group. Phylogenetic groupings at about the level of class often exhibit sufficient conservation of gene synteny that an ancestral gene organization can be deduced. Nevertheless, extensive gene shuffling occurs such that individual lineages will have highly scrambled (or even unrecognizable) versions of the consensus gene organization. The admixture in a given phylogeny of gene organizations conserved over relatively great phylogenetic distances (stability) in combination with dramatic gene shuffling over short phylogenetic distances (instability) is one of the intriguing mysteries of genomics. A detailed example of this was analyzed (67) in the upper Gammaproteobacteria and Betaproteobacteria, where a proposed ancestral supraoperon is gyrA>serC>aroQ-pheA>hisHb>tyrA>aroF>cmk>rpsA>himD. Only Ralstonia metallidurans in the Betaproteobacteria has a “perfect” ancestral supraoperon. Most of the other Betaproteobacteria exhibit very minor supraoperon alterations such as open reading frame insertions and single-gene deletions. Occasionally, more drastic gene shuffling (Chromobacterium violaceum) or partial supraoperon translocation (Nitrosomonas europaea) has occurred. At one extreme (species of Neisseria), the genes of the supraoperon have been completely dispersed. An entirely parallel situation is found in the upper Gammaproteobacteria, where most organisms house near-perfect ancestral supraoperons that differ only slightly in having gene insertions, gene deletions, or gene fusions. Pseudomonas aeruginosa, for example, possesses gyrA>serC>aroHI-pheA>hisHb>tyrA-aroF>cmk>rpsA>himD. Multiple fragmentation of the supraoperon has occurred elsewhere, e.g., in species of Xanthomonas and Xylella. It is quite striking that the supraoperon gene arrangement of R. metallidurans (Betaproteobacteria) is more similar to that of P. aeruginosa (upper Gammaproteobacteria) than to the supraoperon compositions of many other Betaproteobacteria. In reciprocal fashion, the P. aeruginosa supraoperon gene arrangement is more similar to that of R. metallidurans than to those of many other upper Gammaproteobacteria.
The data described above illustrate that within a manageable phylogeny (cohesion group), a particular order of dynamic events of gene ordering can be deduced, yielding a likely ancestral gene order. Parallel analyses at nearby phylogenetic nodes with a roughly equivalent hierarchical level can then lead to a systematic deduction of the ancestral synteny that predated those deduced for the sister nodes.
Data That Are Relevant to the Indel Hypothesis
Panels 12 to 18 of Fig. 6 are all relevant to the hypothesis that the existence of TyrAβ as a discrete subhomology group reflects functional dependence upon protein-protein contacts with either a fused domain or a complexed domain (see below). Essential functional regions may have become dispensable due to replacement by extra-TyrA contacts. This might be consistent with a shorter supradomain length (trimmed of any N-terminal or C-terminal extensions). Panel 12 of Fig. 6 shows that the lengths of TyrAα members is distinctly greater than those of TyrAβ members. The amino acid lengths shown at the ends of branches in panel 12 specify the length of the representative sequence for the cohesion group. These are quite consistent within the cohesion group. The individual cohesion group supradomain sequences can be downloaded for comparison from the pop-up menu provided with the online version of Fig. 2 (http://theseed.uchicago.edu/FIG/Html/tyrACGTree.html). Gene fusions shown in panel 13 are discussed above. In the context here, fusion events tend to have occurred most frequently within the TyrAβ subhomology group. Panels 16 to 18 are designed to examine motifs that are generally conserved within TyrAα but not within TyrAβ. The RxxxR motif (occupies positions 276 to 284 in Fig. 3, a numbering that takes into account an inserted three-residue gap for alignment purposes) is present in nearly all cohesion groups of TyrAα but is absent throughout most of TyrAβ. Panels 17 and 18 identify a number of motifs (having amino acid numbers given in Fig. 3) which again are generally conserved within TyrAα but not within TyrAβ. The color coding shows the cohesion groups that lack a given motif.
Examples of the application of the snapshot tool are pursued in some detail in later sections of this review.
ORGANISMS THAT CARRY MULTIPLE HOMOLOGS
PapC, a Functionally Specialized Paralog
Some dehydrogenases in the TyrA family utilize 4-amino-prephenate as a substrate in a reaction series that leads to 4-amino-phenylalanine and ultimately to the antibiotic chloramphenicol. The otherwise invariant residue at position 154 in Fig. 3 is histidine and is known to interact with the 4-hydroxy moiety of prephenate or l-arogenate. Since PapC utilizes a substrate that has an amino group in place of the hydroxyl moiety at the para position of the ring, the interaction at position 154 is necessarily different. PapC proteins have representation within both the TyrAα and TyrAβ subhomology groupings. Streptomyces coelicolor possesses two paralogs (TyrAa and PapC) that occupy the same cohesion group (TyrCG-17). The PapC paralog is encoded by a gene within the calcium-dependent antibiotic cluster (65) and possesses an alanine residue at position 154. The TyrA and PapC homologs in S. coelicolor are closely related intra-cohesion-group paralogs, of which PapC presumably arose recently by gene duplication, followed by a novel specialization of substrate specificity. It is interesting that calcium-dependent antibiotic contains a variety of nonprotein amino acids. (Note that this PapC paralog is not shown in the various figures and tables of this review in order to maintain focus upon the functional role of TyrA.)
Surprisingly, all remaining PapC paralogs (which have an asparagine residue at position 154) reside in a single cohesion group located in the TyrAβ assemblage (not shown in Fig. 2). These are present in Photorhabdus luminescens, Photorhabdus asymbiotica, Pseudomonas fluorescens, Streptomyces venezuela, Streptomyces pristinaespiralis, and Rhodococcus sp. The two Photorhabdus species also possess TyrA homologs within TyrCG-2, whereas Rhodococcus sp. appears to lack TyrA. TyrA homologs have not been identified in the remaining organisms, but these are unfinished genomes. The latter PapC proteins occupy a single cohesion group and thus probably share a fairly recent common ancestor. However, they are hosted by diverse taxa, so most or all of them might be xenologs. Another possible explanation for the unexpectedly close sequence similarity in diverse taxa is selective pressure for evolutionary convergence. If PapC proteins form a complex with one or more other proteins of the antibiotic synthesis pathway, similar but independently evolved constraints dictating crucial protein-protein interactions may have forced evolutionary convergence. This is similar to the convergence proposed to explain the TrpAa-TrpAb_PhzE clustering for proteins engaged in a step of phenazine pigment biosynthesis in species of Pseudomonas and Streptomyces (78). It is also similar to the indel hypothesis invoked in the following section of this review to explain the convergence of cohesion groups in the TyrAβ subhomology grouping.
Intra-Cohesion-Group TyrA Paralogs
Gene duplication is a frequent, ongoing process, with gene duplicates often being lost. Functionally redundant paralogs from a given organism that are present in the same cohesion group are of recent origin and likely exhibit little functional difference. Desulfuromonas acetoxidans provides one example of recent intra-cohesion-group paralogs (present in TyrCG-14). The only other example at present is the functionally differentiated PapC paralog of Streptomyces coelicolor, which occurs in TyrCG-17 with a TyrAa protein as discussed directly above.
Extra-Cohesion-Group TyrA Paralogs
Rhodospirillum rubrum and Silicibacter pomeroyi are finished genomes of the Alphaproteobacteria that each possess one TyrA species in cohesion group TyrCG-12 and one TyrA species in TyrCG-30. TyrCG-12 is a large group of sequences from Alphaproteobacteria that belong to the TyrAα subhomology group. TyrCG-30, on the other hand, belongs to the TyrAβ subhomology grouping and contains two TyrA sequences in addition to the paralogs from R. rubrum and S. pomeroyi. Maricaulis maris, a finished genome that also belongs to the Alphaproteobacteria, lacks a paralog member in TyrCG-12. Thus, M. maris is so far alone among the Alphaproteobacteria in its complete reliance upon a TyrAβ-specified dehydrogenase for tyrosine biosynthesis. The fourth member of TyrCG-30 is from Myxococcus xanthus (Deltaproteobacteria and an unfinished genome). The latter is provisionally labeled as an intruder sequence, although the alternative scenario, that the M. xanthus sequence is a native sequence from which the TyrAβ intruder sequences present in a few genera of Alphaproteobacteria originated, certainly cannot be ruled out. It is interesting that TyrA from M. xanthus is the only member of TyrCG-30 to have a fused chorismate mutase domain (tyrA-aroHI), distinctive from other chorismate mutase fusions because it is a C-terminal fusion. Regardless of whether M. xanthus was an LGT donor or recipient, the fusion must have occurred after the LGT event.
Ortholog/Xenolog Combinations
The above-described apparent extra-cohesion-group paralogs might be cases of ancient paralog divergence, but it is also possible that one apparent paralog is in fact a xenolog. However, we cannot be sure of the latter unless an LGT donor is identified. One clear example of an ortholog/xenolog combination is in two species of Nostoc where TyrA orthologs exist in TyrCG-16 (which contains TyrA orthologs from all cyanobacteria). In addition, the two species of Nostoc possess a xenolog intruder belonging to TyrCG-1. Hence, the LGT donor was a lower gammaproteobacterium. Interestingly, the Nostoc proteins have an N-terminal extension that appears to be a remnant of the fused chorismate mutase domain, which is present in all other members of TyrCG-1.
SIGNIFICANCE OF THE TyrAα/TyrAβ SCHISM
Panel 1 of Fig. 6 illustrates the distribution of all TyrA sequences from the Archaea and Eukaryota within the TyrAβ subhomology grouping. Whereas most bacterial TyrA sequences occupy TyrAα, many cohesion groups are also represented within TyrAβ. How is this to be explained? Firstly, the possibility must be considered that some or all bacterial sequences that belong to the TyrAβ subhomology grouping originated from an archaeal or eukaryal source by LGT. Secondly, the possibility is considered that members of TyrAα act as independent catalysts, whereas members of TyrAβ exhibit constraints that have driven convergence. These constraints reflect dependence upon contacts with a fused or complexed protein. These possibilities are discussed in turn.
Lateral Gene Transfer between Superkingdoms?
All of the TyrA sequences from the superkingdoms Archaea and Eukaryota are located in the TyrAβ subhomology group, and most of the TyrA sequences from the superkingdom Bacteria are located in the TyrAα subhomology group. However, a scattered number of bacterial sequences also belong to the TyrAβ grouping. Among the Proteobacteria, the latter include all of the lower Gammaproteobacteria (TyrCG-1), TyrCG-4 from the upper Gammaproteobacteria, a small group of TyrAα sequences from the Alphaproteobacteria (TyrCG-30) (also containing one intruder sequence carried by a deltaproteobacterium), and TyrCG-15, which is populated by two sequences from the Deltaproteobacteria. No Betaproteobacteria or Epsilonproteobacteria that host proteins belonging to the TyrAβ subhomology grouping are currently known. The phylum Bacteroidetes is represented by TyrCG-24 and TyrCG-23 in the TyrAα and TyrAβ subhomology groups, respectively.
The Alphaproteobacteria exhibit some novel variations. Most of them contribute to a 38-member cohesion group (TyrCG-12), which, along with an orphan sequence (Pelagibacter ubique), belong to the TyrAα subhomology group. Three Alphaproteobacteria have members that occupy the TyrAβ subhomology group (TyrCG-30). Two of the latter (Rhodospirillum rubrum and Silicibacter pomeroyi) also host paralogs among the above-mentioned group of 38, thus being the only organisms so far known to possess a TyrA member of each subhomology group. The third member of TyrCG-30 (Maricaulis maris) is the only alphaproteobacterium whose sole TyrA sequence belongs to TyrAβ.
Could all of the bacterial sequences that fall into the TyrAβ subhomology group be explained as acquisitions from archaeal or eukaryotic donors via LGT? If so, multiple LGT events would have had to occur independently in different bacterial lineages since those Bacteria whose sequences belong to the TyrAβ subhomology grouping do not cluster together in a common lineage. None of the seven cohesion groups within the TyrAβ subhomology grouping that have bacterial membership contain a sequence of the Archaea or Eukaryota that would implicate an LGT donor. This, of course, is also true for the two bacterial orphan sequences present in the TyrAβ subhomology grouping. Since genomic sampling is still quite minimal in the Archaea, it is possible that the LGT donors are simply unknown. However, the probability of this is lessened considering that a donor has not materialized on nine different occasions.
Does Membership within TyrAβ Reflect Protein-Protein Interactions?
We believe that it is likely that the TyrAβ subhomology group contains TyrA proteins that exhibit functionally critical protein contacts with either fused proteins or partnered members of a complex. In contrast, members of TyrAα are postulated to function independently of any protein partners. In a previous paper (67), it was noted that some TyrA sequences, such as that from E. coli, possessed distinctive indel structuring (insertions and deletions) in alignments with what are here called TyrAα subhomology group members. The above-described types of sequences (herein named TyrAβ) were originally named TyrAc_Δ (cyclohexadienyl dehydrogenases that have indel structuring). The previous TyrAc_Δ designation is herein abandoned in favor of the current TyrAβ designation (one which does not imply any substrate specificity). This indel hypothesis is stimulated largely by experimental work with E. coli and some close relatives. Thus, TyrA from E. coli (and all other lower Gammaproteobacteria) is fused at the N terminus with chorismate mutase (AroHI). Chen et al. (18) demonstrated that neither chorismate mutase nor cyclohexadienyl dehydrogenase reactions of E. coli are fully competent when isolated from one another. Sun et al. (71) cited a variety of other documentation to suggest that the two fused domains are functionally dependent. There is the suggestive correlation that lower Gammaproteobacteria have the fusion and belong to TyrAβ, whereas the closely related upper Gammaproteobacteria lack the fusion and belong to TyrAα. Xanthomonas and Xylella species (TyrCG-4) are exceptions among the upper Gammaproteobacteria in that they belong to the TyrAβ subhomology grouping. However, these TyrA species exhibit another fusion pattern: a C-terminal fusion with ACT, a broadly distributed regulatory domain. The intruder TyrA sequences present in species of Nostoc which are derived from the lower Gammaproteobacteria lineage possess an N-terminal extension that appears to be a remnant of the fused chorismate mutase, otherwise found in TyrCG-1. Key catalytic residues needed for chorismate mutase activity have not been conserved. It is interesting to consider that the extension nevertheless persists in order to maintain the domain-domain interactions proposed for TyrAβ enzyme species. This would be worthwhile to test experimentally since one can potentially evaluate what regions are needed to support TyrA activity without complications related to chorismate mutase activity. In addition to fusions with AroHI and the ACT domain, other members of TyrAβ exhibit fusions with a domain called REG (67) or have sequence extensions that may be unknown regulatory domains. Thus, cohesion groups that fall within the TyrAβ subhomology grouping consist of sequences that have experienced a wide variety of different and independent indel events postulated to be associated with functional domain-domain interactions. This variety plus normal phylogenetic divergence explain the separation of cohesion groups within the TyrAβ subhomology grouping. However, at the broadest level, the cohesion group members of TyrAβ have converged because they have the indel disruption of highly conserved motifs that are shared by members of TyrAα in common.
The indel hypothesis does not require that members of the TyrAα subhomology group lack TyrA fusions and that members of the TyrAβ subhomology group possess TyrA fusions, although this is certainly the trend (Fig. 6, panels 13 and 14). In some cases, TyrAα members do carry a fusion. Here, TyrA is presumably not dependent upon the fused domain for function. In support of this, Xie et al. (77) showed that TyrA from Pseudomonas stutzeri was not affected when separated from its AroF fusion partner. It is also suggestive in this context that closely related species of Burkholderia share membership in TyrCG-7 (TyrAα subhomology grouping), even though some of them possess a fusion of tyrA with aroF and some do not. In those cases where TyrAβ members have no fusion or sequence extensions, we suggest that these associate with another protein to form a complex and that such protein-protein contacts are functionally important. Panel 12 of Fig. 6 shows that the length of the core supradomain is typically shorter in members of TyrAβ than in members of TyrAα, an observation that is consistent with indel deletions that might be compensated for by an extradomain protein partner region.
Sequence convergence following the independent fusion of interacting domains in widely separated organisms was demonstrated (78) in a simpler case where only two interacting domains were involved. Xie et al. (78) showed that four different and large TrpAa (anthranilate synthase aminase) cohesion groups were populated by sequences from the Actinobacteridae, Cyanobacteria, upper Gammaproteobacteria/Betaproteobacteria, and Alphaproteobacteria, respectively. Four TrpAb (anthranilate synthase amidotransferase) cohesion groups were populated by sequences from exactly the same organisms. However, several organisms in each of the former taxa possessed TrpAa and TrpAb domains, which were fused to one another and which did not belong to the expected cohesion groups made up of free-standing TrpAa or TrpAb domains. In comparison with the four separated positions of free-standing TrpAa domains on a phylogenetic tree, the fused TrpAa- domains were all clustered together on a divergent branch of the tree. (The hyphen and its placement signify a fusion at the C terminus.) Similarly, in comparison with the positions of free-standing TrpAb domains on a phylogenetic tree, all of the fused -TrpAb domains were clustered together on one divergent branch of the tree. Evidence that TrpAa-TrpAb fusions have occurred independently as many as seven times and that the convergence observed for sequences from diverse taxa is the consequence of rigid constraints imposed for proper protein-protein interactions of these subunits was presented (76).
Utility of Cohesion Group Snapshots
In our system, any TyrA features deemed to be of interest are displayed by painting them on the cohesion group tree shown in Fig. 2. Thus, Fig. 6 contains 18 such mini-semblances. As new sequences become available and enter an existing cohesion group, a variety of character states already associated with the cohesion group become likely character states of the new sequence. Cases such as that of TyrCG-17, discussed above, where a distinct divergence separates sequences that are NAD+ specific from the broadly specific NAD(P)+ ones, show that alternative character states may partition within a defined section of a given cohesion group. Different proteins or groups of proteins will have individualistic features of interest that can be tracked in association with cohesion groups. The organization of TyrA character states within the assemblage of cohesion groups can serve as a springboard for experimental predictions as illustrated below.
Are Essential Extradomain Contacts Needed for TyrA Members of TyrAβ?
It is postulated that members of the TyrAβ subhomology group have a supradomain core region that is functionally dependent upon supradomain contacts with either a fused protein or a complexed protein. This was experimentally demonstrated for E. coli (18) and is reasonably extrapolated to all members of TyrCG-1. There is an excellent and well-organized information background to select key, well-spaced cohesion group members to test experimentally whether isolated catalytic core regions of TyrAα members are catalytically competent, in contrast to isolated supradomain regions of TyrAβ members, which are predicted to require contacts with extra-TyrA protein domains. In the case of E. coli, the fused chorismate mutase (AroHI) has a reciprocal dependence upon the fused TyrA for normal function. This raises the question of whether fused chorismate mutases and free-standing chorismate mutases of the AroHI homology class would also exhibit a bifurcated divergence similar to the TyrAα/TyrAβ dichotomy. This is certainly worthy of further examination.
Interesting Specificity Issues
Streptomyces coelicolor possesses two paralogs of recent divergence but functionally differentiated: tyrA and papC genes. As discussed above, S. coelicolor PapC is widely divergent from all other PapC proteins, the latter of which collect together within a single TyrAβ cohesion group (not shown in the figures and tables of this review). S. coelicolor PapC is assumed to play a role in the synthesis of calcium-dependent antibiotic because of the position of its encoding gene in the middle of the large CDA (calcium-dependent antibiotic) gene cluster (65). PapC proteins are generally assumed to utilize 4-amino-prephenate as a substrate, thereby producing 4-amino-phenylpyruvate as product. It is quite possible, however, that a given PapC could utilize 4-amino-arogenate instead. This specificity could apply if the order of dehydrogenase and transaminase steps was reversed (as can occur in tyrosine biosynthesis). Since the S. coelicolor PapC paralog is of recent origin and occupies the same cohesion group as its TyrA paralog, one might predict that the specificities of the two might be the same for the side chain of the substrate. Hence, we propose an experimentally testable idea, namely, that since TyrA from S. coelicolor is specific for l-arogenate (alanyl side chain), it is likely that the PapC paralog is specific for 4-amino-arogenate (alanyl side chain).
Expanding the Evolutionary Context across Subsystems
Cohesion groups can be formulated for single proteins, as exemplified by TyrA and the seven proteins of l-tryptophan biosynthesis, and the result can produce a picture of what features evolved in what lineages at what times. An evaluation of what character states evolved “purely” within a vertical genealogy and what character states were obtained by LGT can be deciphered. From the time of any new LGT acquisition that can be pinpointed, a new vertical genealogy can be tracked. Thus far, concatenates of the seven tryptophan pathway enzymes have been used to define supercohesion groups. The supercohesion groups, of course, have much more resolving power than do individual cohesion groups. The pathway of aromatic amino acid biosynthesis consists of a common trunk of seven reactions and three amino acid branches (http://www.aropath.lanl.gov/Visualizations/index.html). This can be thought of as four manageable metabolic subsystems that can eventually morph into a single subsystem. Since chorismate mutase and aromatic aminotransferase activities overlap the phenylalanine and tyrosine branches in a very intimate way, it would be logical to join TyrA, PheA, and the various homolog types responsible for chorismate mutase and aromatic aminotransferase in a single study, i.e., making up a single metabolic subsystem. It should be possible to assemble concatenates as a source of supercohesion groups that would represent the steps proceeding from chorismate to both phenylalanine and tyrosine. Finally, inclusion of the seven common pathway enzymes that feed all of the divergent branches of aromatic amino acid biosynthesis in the analysis will yield an integrated picture of what the milestone events were in each of the four individual subsystems and how these events may have impacted the gestalt of the overall pathway. An initial sense of how the larger picture can build may be gotten from a section below, which compares TyrA cohesion groups with tryptophan supercohesion groups. We anticipate that the eventual ability to “paint” the locations of cohesion groups corresponding to many metabolic subsystems on 16S rRNA trees would be valuable for a multitude of purposes.
CANDIDATE TyrA PROTEINS FOR X-RAY CRYSTAL STUDIES
Challenge of Broad-Specificity Reactions
Enzymes can range between those that are exquisitely demanding and precise in their catalytic requirements and those that accelerate reactions that can accommodate alternative substrates or cofactors. The former enzymes seem to be encoded by genes that are generally larger and more conserved than genes encoding broad-specificity reactions. Highly specific and highly conserved enzymes are exemplified by dehydroquinate synthase, the second enzyme of aromatic amino acid biosynthesis, or 5-enolpyruvylshikimate-3-phosphate synthase, the sixth enzyme of aromatic amino acid biosynthesis (12). The latter enzyme combines two phosphorylated substrates with a precise and intricate mechanism that cannot tolerate much deviation. The multistep mechanism of dehydroquinate synthase involves alcohol oxidation, phosphate β elimination, carbonyl reduction, ring opening, and intramolecular aldol condensation. In such cases, X-ray crystal studies with an enzyme from just a single organism can provide widely applicable information. On the other hand, enzymes catalyzing broad-specificity reactions may have the pliability to accept a range of related substrates, may readily mutate from a given specificity to a closely related one, or may readily mutate to a narrowed or broadened profile of substrate specificity. Even where specificity for a particular substrate is the same in different members of a broad-specificity enzyme family, the pliability to allow divergence to different active-site variations that still accomplish exactly the same reaction may exist. These aspects of enzymatic plasticity, albeit intriguing, mean that a relatively large number of coordinated crystal studies are required if one is to fully understand the complete array of important amino acid contacts that fall under the catalytic umbrella of pliant enzyme families such as TyrA. Thus, one challenge is that whereas amino acid motifs that correspond to important active-site residues can be conspicuously invariable for ultraspecific enzymes, motifs may be much more elusive in multiple alignments of broad-specificity enzyme sets. In the latter case, careful and comprehensive work might reveal a series of motifs, each conserved and typifying particular lineages that carry a set of TyrA proteins. The potential results of a comprehensive series of X-ray crystal studies with a small enzyme having substantial catalytic plasticity can reasonably be expected to contribute general insight into what is required to make accurate functional inferences for the very large number of such “difficult” enzymes.
Informative Selections from TyrAα Subhomology Group Members
The existing comprehensive sequence analysis, as exemplified in Fig. 3, should be helpful in guiding rational selections of subject TyrA proteins for X-ray crystal studies and other molecular characterizations that might be maximally instructive. Since we have not found any distinctive motifs associated with TyrA proteins that were sorted and aligned according to criteria of specificity for prephenate or specificity for l-arogenate or having broad cyclohexadienyl specificity, it seems likely that multiple active-site configurations that are able to confer a given specificity exist. With insight from a sufficient abundance of well-chosen X-ray crystal studies, it should eventually be possible to equate different substrate specificity patterns in a defined phylogenetic lineage with definitive sequence motifs.
Key variables of interest are TyrA crystals bound with any substrate for which it has catalytic competence. Given that enzymes specific for cyclohexadienyl substrate and pyridine nucleotide cofactor are known to occur in all combinations, this alone generates a qualitative total of nine comparative possibilities. An enzyme such as that from Ralstonia solanacearum (TyrCG-7) has roughly equal capabilities with NAD+ and NADP+ as well as roughly equal capabilities with l-arogenate and prephenate. Hence, there are four protein-substrate combinations that can be analyzed from this single TyrA species, each of which should be informative in comparison with TyrA proteins that can be selected for the various appropriate narrow specificities. Another dimension of complexity is that many broad-specificity TyrA species have order-of-magnitude preferences for one substrate or for one cofactor. These quantitative differences must have discernible parallels at the molecular level that distinguish them from the absolutely specific TyrA proteins or from broad-specificity TyrA proteins that accept alternative substrates about equally well.
Ideal TyrA candidates for initial crystal studies are those that have been well characterized, are produced from organisms with complete genomes, and have core supradomains that are uncomplicated by fused catalytic or regulatory domains. Examples of such organisms selected from the TyrAα subhomology grouping are Zymomonas mobilis (broad-specificity cyclohexadienyl dehydrogenase with a preference for l-arogenate) (NAD+ specific), Aquifex aeolicus (cyclohexadienyl dehydrogenase markedly favoring prephenate) (NAD+ specific), Rhodopseudomonas palustris (cyclohexadienyl dehydrogenase with a marked preference for prephenate) (NADP+ specific), Ralstonia eutropha (cyclohexadienyl dehydrogenase) {broad cofactor specificity [NAD(P)+]}, Neisseria gonorrhoeae (cyclohexadienyl dehydrogenase with marked preference for prephenate) (NAD+ specific), Nitrosomonas europaea (l-arogenate specific and NADP+ specific), Corynebacterium glutamicum (l-arogenate specific, with a marked preference for NADP+ over NAD+), Synechocystis sp. (l-arogenate-specific and NADP+ specific), Gluconobacter oxydans (prephenate specific and NADP+ specific), and Clostridium difficile (prephenate specific and NAD+ specific). Although many additional TyrA proteins from organisms whose genomes unfortunately are not yet sequenced have been well characterized, it seems likely that this will be largely ameliorated in the near future, considering the high and increasing rate of genome sequencing.
Although a well-spaced phylogenetic selection of TyrA proteins is generally desirable, in some cases, it might also be worthwhile to select TyrA proteins from a single cohesion group that have variant properties of substrate selectivity. This can be comparable to the approach of selecting specificity mutants for comparison with the wild-type parent in order to carry out structural analysis. For example, the entire cyanobacterial phylum possesses a TyrA member belonging to a single cohesion group (TyrCG-16). An extensive enzymological comparison indicated that most, if not all, cyanobacterial TyrA enzymes can utilize l-arogenate and NADP+ as substrates (29). Although some are absolutely specific for these two substrates, cyanobacteria frequently express broad-specificity enzymes that are capable of utilizing NAD+ (albeit always less well than NADP+). Less commonly, broad specificity for the cyclohexadienyl substrate exists, although l-arogenate is always utilized better than prephenate. (At one extreme, Synechocystis sp. strain PCC7509 uses prephenate 48% as well as l-arogenate at substrate saturation.)
A second example that offers interesting comparative possibilities is the collection of TyrA proteins from the Betaproteobacteria. All members of TyrCG-7, TyrCG-8, and TyrCG-10 and four orphans (Table 2) are broad-specificity cyclohexadienyl dehydrogenases that have the broad cofactor specificity motif pattern 36GxxRS40 (Fig. 4). Members of TyrCG-11 possess narrowed specificity for both substrates, being l-arogenate specific and NADP+ specific. On the other hand, members of TyrCG-9 possess the opposite pattern of narrowed specificities, being NAD+ specific and exhibiting a very marked preference for prephenate as the cyclohexadienyl substrate.
Informative Selections from TyrAβ Subhomology Group Members
The basis for the supposition that TyrA proteins that belong to the TyrAβ subhomology grouping are ones that exhibit functional interactions with attached catalytic or regulatory domains (or perhaps which do so via protein-protein complexes) is discussed above. Compared to the TyrAα subhomology group, relatively few TyrA enzymes from the TyrAβ subhomology group have been characterized. Of course, TyrAc from E. coli is an obvious selection choice because of the abundance of experimental work with it, including evidence upon which the indel hypothesis is based (see references 11 and 71 and references therein). TyrAc from E. coli and TyrAc from Aquifex aeolicus should be a good comparative match as selections taken from the TyrAβ and the TyrAα subhomology groups, respectively. Each of these is NAD+ specific, and each is a cyclohexadienyl dehydrogenase that has a marked preference for prephenate as a substrate. Each is sensitive to l-tyrosine inhibition.
Xanthomonas campestris and other members of TyrCG-4 are upper Gammaproteobacteria that possess a TyrA enzyme with a C-terminal ACT domain, with the latter perhaps being responsible for placement in the TyrAβ subhomology grouping. (Note that the presence of an attached ACT domain does not necessarily mean that a so-endowed TyrA species will be in the TyrAβ subhomology grouping since many gram-positive bacteria in the TyrAα subhomology grouping, e.g., all members of TyrCG-18, have an ACT domain.) In contrast to the members of TyrCG-4, all upper Gammaproteobacteria (TyrCG-2, TyrCG-3, TyrCG-5, TyrCG-6, and five orphans) lack an attached ACT domain and belong to the TyrAα subhomology grouping. X. campestris TyrA has been characterized as being NAD+ specific and broadly specific for cyclohexadienyl substrate. The best match for this substrate profile among the upper Gammaproteobacteria in the TyrAα subhomology grouping would be TyrAc produced by any of three orphans: Acidithiobacillus ferrooxidans, Methylococcus capsulatus, or Nitrosococcus oceani. The TyrA protein from Coxiella burnetii might also be worth considering for comparison. Like the X. campestris protein, it belongs to the TyrAβ subhomology grouping, but it lacks an ACT domain. This TyrA species is NAD specific, but its cyclohexadienyl specificity is uncertain. Also, we cannot be sure that this TyrA enzyme is a native upper Gammaproteobacteria protein since it resides in TyrCG-26, which is an unresolved phylogenetic mixture.
Finally, TyrA proteins from higher plants (TyrCG-95) are well characterized as being l-arogenate-specific and NADP+-specific enzymes. Since the Synechocystis sp. strain PCC6803 enzyme (TyrAα subhomology group) has the same specificity profile as TyrA from organisms such as Arabidopsis thaliana (TyrAβ subhomology group), X-ray crystal comparative studies should be illuminating.
Inhibition Properties: Insight into Binding of the 1-Carboxy Moiety?
For the simplest TyrA proteins where allosteric domains or interacting catalytic domains are not attached, it has been proposed (77) that the product inhibitors (either l-tyrosine or 4-hydroxyphenylpyruvate) act directly at the active site as classical competitive inhibitors. Thus, there are cases where an enzyme is specific for prephenate (having a pyruvyl side chain) and is inhibited by 4-hydroxyphenylpyruvate (also having a pyruvyl side chain) but is not inhibited by l-tyrosine (alanyl side chain). On the other hand, enzymes such as those from higher-plant plastids that are specific for l-arogenate (alanyl side chain) and are inhibited by l-tyrosine (alanyl side chain) but not by 4-hydroxyphenylpyuvate (pyruvyl side chain) are known. For simple TyrA enzymes that lack discrete allosteric domains or interacting fusions, it generality seems to hold that the specificity of this core supradomain for the side chain of any substrate accepted (i.e., pyruvyl and/or alanyl) will parallel the specificity of product inhibition. Neisseria gonorrhoeae possesses a cyclohexadienyl dehydrogenase that prefers prephenate markedly over l-arogenate as a substrate. Accordingly, inhibition by 4-hydroxyphenylpyruvate is potent, and inhibition by l-tyrosine is weak. Thus, whenever inhibition has been observed, the side chain specificities of inhibitor and substrate parallel one another. However, some TyrA proteins are completely insensitive to competitive inhibition by the product. Thus, Acidovorax facilis and Rubrivivax gelatinosus possess TyrAc enzymes that are not sensitive to inhibition by either l-tyrosine or 4-hydroxyphenylpyruvate, Zymonomas mobilis TyrAc is not sensitive to inhibition by either l-tyrosine or 4-hydroxyphenylpyruvate, and Nitrosomonas europaea TyrAa is not sensitive to inhibition by the l-tyrosine product. Presumably, the latter TyrA species require the ring carboxylate for binding, whereas TyrA species that are sensitive to product inhibition must not require the ring carboxylate for binding.
Comparison of reasonably close sets of TyrA proteins that differ in being resistant or sensitive to product inhibition could give insight into residue contacts that are important for binding of the ring carboxylate. For example, a reasonable choice for comparison might be two TyrA members of the Betaproteobacteria. TyrAc enzymes from Acidovorax facilis (TyrCG-10) and Burkholderia cepacia (TyrCG-7) are very similar in having broad specificities for the two cyclohexadienyl substrates and broad specificities for cofactor. The alternative substrates and alternative cofactors are accepted about equally well. However, the A. facilis enzyme is totally refractive to product inhibition, whereas the B. cepacia enzyme is sensitive to product inhibition. Sun et al. (71) pointed out that a glycine-rich region, 273-GGG-275, immediately preceding the 277-RxxxR-284 motif of Aquifex TyrA, seems to play a critical role in positioning 278-D′ into the active site within interacting distance of the ring carboxylate of prephenate (numbering as given in Fig. 3). 274-GG-275 of the glycine-rich region appears to be particularly conserved. It may be significant that TyrA enzymes from organisms (Pseudomonas aeruginosa, Burkholderia cepacia, Ralstonia solanacearum, and Ralstonia eutropha) that have been shown to be sensitive to product inhibition all possess 274-GG-275. In contrast, TyrA enzymes from Zymomonas mobilis (GS), Acidovorax sp. (PG), Nitrosomonas europeae (SS), and Rubrivivax gelatinosus (PG) are not inhibited by the reaction product and lack the GG signature.
TyrAc from Aquifex aeolicus, one of the two TyrA proteins for which X-ray crystal studies exist (71), has a marked preference for prephenate and is NAD+ specific. Since it is quite sensitive to tyrosine inhibition (11), one would expect even greater sensitivity to inhibition by 4-hydroxyphenylpyruvate, but this was not tested. This TyrA sequence is currently an orphan sequence, so comparisons with relatively close orthologs are not yet possible. The second subject of an X-ray crystal study is Synechocystis sp. (48). This l-arogenate-specific, NADP+-specific enzyme was reported to be insensitive to inhibition by l-tyrosine. Unfortunately, this is at odds with a report by Bonner et al. (10), who detailed good sensitivity of TyrAa from the same strain to competitive inhibition by l-tyrosine. Enzymes that become selectively desensitized to inhibition while maintaining catalytic competence are known, but these usually are enzymes that have a distinct allosteric domain (or subunit). Legrand et al. (48) suggested that the difference might be due to “mutations” in four amino acids very near the C terminus. However, this apparent difference in sequence was due to an inadvertent transposition of a glutamine residue in the preparation of Fig. 7 in the paper by Bonner et al. (10). A substantial amount of comparative enzymology (including determinations of sensitivity to inhibition by l-tyrosine) was done with TyrA species of various named Synechocystis species (29). Organismal differences in substrate specificity and sensitivity to inhibition by l-tyrosine that would fit the results of either research group were observed. Unfortunately, Synechocystis sp. strain PCC6803 was not included in the latter study. Hence, whether the TyrAa enzyme from Synechocystis sp. strain PCC6803 is sensitive or refractive to product inhibition by l-tyrosine must await further experimental clarification.
Selections Based upon Other TyrA Features
Thus far, in this review, a total of 15 organisms have been suggested as examples that could be selected for comparative studies from a perspective of (i) interest in the nature of variable specificities for cyclohexadienyl substrate or cofactor reactant, (ii) gaining insight into the distinct difference between the alpha and beta subhomology groupings, or (iii) elucidating what dictates whether the 1-carboxy moiety is required for binding and whether this determines sensitivity to product inhibition directly at the active site.
Still other features deemed to have significance could be used as criteria of significance with respect to organisms selected as a source of TyrA protein. These features would not necessarily be independent of some of the above-described considerations. For example, the motif RxxxR has been discussed above as a character state that has been suggested in the X-ray crystal study described Sun et al. (71) to be important in the mechanism employed by the TyrA protein of Aquifex aeolicus. The idea has been presented that in proteins belonging to the TyrAβ subhomology family (10, 71), this motif has been disrupted by extra-TyrA contacts extended from an attached or complexed domain. This is consistent with the near-total conservation of this motif throughout the TyrAα subhomology grouping and with its near-total absence in proteins belonging to the TyrAβ subhomology grouping. Thus, this motif seems intimately relevant to the second perspective described above. Scrolling through the extended table online shows that exceptions in the TyrAα subhomology grouping whereby the motif is disrupted include one member of TyrCG-5, some members of TyrCG-16, two members of TyrCG-11, the Flavobacteria component of TyrCG-13, half the members of TyrCG-16, most members of TyrCG-24, and one of the two members of TyrCG-31. Comparison of a motif-present member with a motif-absent member in the latter cohesion groups might be of particular value because the motif difference seen in each pair exists in a background of close phylogeny.
The X-ray crystal study of TyrA from Aquifex aeolicus (71) indicated that the RxxxR motif comprises part of an ionic network, which was proposed to support a gated mechanism for the access of substrate to the active site. However, the X-ray crystal study of TyrA from Synechocystis sp. (48) asserted that this patch of basic residues does not seem to play a critical role in the binding of substrate. Synechocystis sp. belongs to TyrCG-16, a cohesion group that contains a total of 16 cyanobacteria. Although the subject of the X-ray crystal study has the motif, it is absent in 10 members of TyrCG-16. This suggests the possibility that the presence of the motif in some cyanobacteria may be only coincidental, and it may not have the functional significance that generally applies in the TyrAα subhomology grouping. It was also noted (71) that the rightward R residue of the motif (R284 in Fig. 3) forms an ion pair with E160. In this context, it may be significant that the latter residue is completely conserved (sometimes substituted with a D) in the TyrAα subhomology grouping with only two exceptions, one of them being TyrA from Synechocystis sp.
The Snapshot Tool for Facilitating Selection Choices for Comparative Analysis
In this section, the further consideration of the RxxxR motif is pursued as an example of how the snapshot tool can be implemented to make rational choices for protein selection. Panel 16 of Fig. 6 displays the distribution of the motif in cohesion groups or parts of cohesion groups, and this can be viewed in parallel with up to two other panels with the online tool to clarify correlated patterns of distribution.
Example 1.
Suppose that one chooses to think about the Gammaproteobacteria (a taxon at the level of class) in terms of how it has diverged into cohesion groups, where these cohesion groups belong in terms of the two primary subhomology groupings, and what the distribution pattern is for the RxxxR motif. If panels 2 and 16 of Fig. 6 are displayed side by side using the tool at the SEED platform (http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html), 10 cohesion groups of Gammaproteobacteria are visualized in panel 2, and these can be compared to the presence or absence of the RxxxR motif in panel 16. The lower Gammaproteobacteria populate a single cohesion group (TyrCG-1) within the TyrAβ subhomology grouping. TyrA sequences from the upper Gammaproteobacteria separate into 10 cohesion groups, four of which are orphans. TyrA from Coxiella burnetii is a member of an unresolved phylogenetic mixture (TyrCG-26) and, together with members of TyrCG-4, is the only sequence from the upper Gammaproteobacteria that belongs to the TyrAβ subhomology grouping. The C. burnetii sequence as well as all members of TyrCG-1 and TyrCG-4 lack the RxxxR motif as is typical of the TyrAβ subhomology grouping. As expected of the TyrAα subhomology grouping, all of the remaining members of the upper Gammaproteobacteria possess the RxxxR motif, except for one member of TyrCG-5 (produced by Thermochromatium tepidum). Hence, it would be attractive to have definitive X-ray crystal results with the TyrA enzymes from (i) T. tepidum (exceptional in lacking the RxxxR motif), (ii) a member of the same cohesion group (where the motif is present), (iii) one of the other upper Gammaproteobacteria cohesion groups of the TyrAα subhomology grouping (motif is present), (iv) Xanthomonas campestris (a member of the upper Gammaproteobacteria belonging to the TyrAβ subhomology grouping and lacking the motif), and (v) E. coli (lower Gammaproteobacteria, TyrAβ subhomology grouping, and the motif is absent). These five choices offer potential for a wealth of comparative information that will reveal structural ties to functional properties. Evidence supporting refined evolutionary conclusions can also be anticipated. For example, one can speculate that the separation of the lower Gammaproteobacteria from the upper Gammaproteobacteria correlated with the attachment of a chorismate mutase domain to TyrA in the former group. Also, more recently, in the Xanthomonas/Xylella lineage, the attachment of an ACT domain to TyrA occurred after its diversion from the other upper Gammaproteobacteria. These two independent fusion events presumably account for membership of the latter TyrAs in the TyrAβ subhomology grouping. The uncoupling of motif presence with membership within the TyrAα subhomology group in a single organism (T. tepidum) whose close phylogenetic relatives in TyrCG-5 have maintained the normal coupling should be instructive.
Example 2.
Suppose panel 3 of Fig. 6 is viewed in parallel with panel 16. Panel 3 highlights cohesion groups and orphans that are represented by TyrA proteins from the Alphaproteobacteria, the Deltaproteobacteria, and the Epsilonproteobacteria. By using the snapshot tool and using the links to navigate to the extended table where necessary, one can develop a rationale for TyrA selections from these three classes of Proteobacteria that might be the most informative with respect to the significance of the RxxxR motif.
The Alphaproteobacteria mostly populate TyrCG-12 in the TyrAα subhomology grouping, where they consistently possess the RxxxR motif. One can see just by considering TyrCG-12 alone that the significance of this motif has some broader meaning than a relationship to substrate/cofactor specificity in view of the widely different specificities previously described for organisms such as Zymomonas mobilis, Rhodopseudomonas palustris, and Gluconobacter oxydans, all members of TyrCG-12. In spite of its overall sequence divergence from most other Alphaproteobacteria, the TyrAα orphan Pelagibacter ubique also possesses the RxxxR motif. The three members of TyrCG-30 are the only Alphaproteobacteria present in the TyrAβ subhomology group, and all of them lack the motif. Thus, at the taxon level of class, TyrA proteins from the Alphaproteobacteria have diverged to form one orphan, one small cohesion group, and one large cohesion group. Only the small cohesion group belongs to the TyrAβ subhomology grouping, and this correlates perfectly with the lack of the RxxxR motif. Both Rhodospirillum rubrum and Silicibacter pomeroyi possess paralog members of TyrCG-12 and TyrCG-30, so a comparison of one of these two paralog pairs should also be rewarding.
TyrA proteins from the Deltaproteobacteria populate three cohesion groups. Most of them are in TyrCG-14, which occupies the TyrAα subhomology grouping, and these have the RxxxR motif and are the only Deltaproteobacteria that are NAD+ specific. Members of TyrCG-15 and an orphan from Syntrophobacter fumaroxidans belong to the TyrAβ subhomology grouping and lack the motif, as expected. Selection of one TyrA from each of the two subhomology groupings yields a pair where the TyrAβ subhomology grouping member possesses a core supradomain length that is shortened (Fig. 6, panel 12) and yet where there are no extra core extensions (panel 14) or domain fusions (panel 13). Biochemical work may show that this TyrA protein partners with another protein that makes contacts in the complex that is needed for maximal activity. The TyrA enzymes of Epsilonproteobacteria all belong to a single cohesion group, TyrCG-13. This cohesion group also contains all members of the class Flavobacteria. It was concluded above that genes encoding the TyrA enzymes from Flavobacteria arrived from a donor in the Epsilonproteobacteria lineage via LGT. TyrCG-13 is in the TyrAα subhomology grouping. It is interesting that all TyrA members from the Epsilonproteobacteria possess the RxxxR motif, whereas all those from the Flavobacteria lack the motif. The latter members do not seem to have a shortened supracore domain, so it may be that in this case, the absence of the motif does not indicate a domain-domain interaction. If so, alternative amino acid contacts may substitute here for the otherwise highly conserved RxxxR motif.
Experimental Truncation of Fused Domains
We suggest that the catalytic function of TyrA for members of the TyrAα grouping is not dependent upon an attached domain even if such fusions are present. It is predicted that the removal of such attached domains will not directly affect the catalytic reaction. This has in fact been shown for the tyrAc-aroF fusion of Pseudomonas stutzeri, where removal of the C-terminal AroF catalytic domain had no effect upon the remaining TyrA domain (77). In addition, TyrCG-7 contains 11 members, only three of which possess a tyrA-aroF fusion. This recent fusion has not distanced the TyrA domain of the small clade of Burkholderia species that contain it from the unfused TyrA domains of the sister Burkholderia species and species of Ralstonia that occupy the cohesion group. If new indel contacts had developed in the newly evolved TyrA-AroF protein to create interdependent domains, one would expect these TyrA domains to have diverged away from the unfused TyrA domains in TyrCG-7.
TyrA proteins frequently possess a C-terminal ACT domain, as exemplified by the well-studied Bacillus subtilis enzyme (17), which belongs to the TyrAα subhomology grouping. It would be quite interesting to examine this enzyme following the removal of the ACT domain, which is an allosteric domain. This amino acid binding domain presumably accounts for the sensitivity of B. subtilis TyrAp to inhibition by l-tyrosine, l-phenylalanine, l-tryptophan, and d-tyrosine. Removal of the ACT domain should abolish these amino acid sensitivities, leaving only the sensitivity to inhibition by 4-hydroxyphenylpyruvate intact. This expectation is enhanced by the fact that exactly these properties were obtained with the selection of a d-tyrosine-resistant, tyrosine-excreting mutant in 1970 (17). Similar opportunities for examining the effects of removing a C-terminal ACT domain exist in other cohesion groups belonging to the TyrAα subhomology grouping, e.g., TyrCG-20, TyrCG-21, and TyrCG-22.
In contrast with the above-described expectations for TyrA proteins belonging to the TyrAα subhomology grouping, experimental truncations that remove attached catalytic or regulatory domains of TyrA proteins belonging to the TyrAβ subhomology grouping are expected to impact TyrA catalysis directly. This has already been demonstrated following removal of the N-terminal chorismate mutase domain from E. coli aroHI-tyrAc (18), and X-ray crystal results that demonstrate the projected domain-domain contacts projected by Bonvin et al. (11) would be most welcome. Xanthomonas campestris and other members of TyrCG-4 possess a C-terminal ACT domain, just like B. subtilis and other members of TyrCG-18. Since the former and latter represent the TyrAβ and TyrAα subhomology groupings, respectively, the differences in how this allosteric domain interacts should be fascinating. Another attached regulatory domain of potential interest is the C-terminal REG domain present in members of TyrCG-80 (Euryarchaea_1).
COMPARISON OF TYROSINE AND TRYPTOPHAN PATHWAY COHESION GROUPS
Background
Concatenated sequences of the seven tryptophan pathway enzymes that specifically participate in primary biosynthesis were previously assembled and used to construct trees. This produced seven supercohesion groups and 11 unnumbered orphans (78). The compositions of these multimembered and orphan tryptophan supercohesion groups obtained from 47 organisms are compared with the TyrA cohesion groups present in the same organisms (see below).
Tyrosine pathway cohesion groups and tryptophan pathway supercohesion groups cannot be expected to correspond with one another perfectly for the following reasons. First, intruder sequences that become established in a given organism for one pathway will not generally be present for another pathway. Second, the sequence length and degree of conservation of the protein(s) upon which cohesion groups are based will dictate different relative resolving powers. Because the Trp enzyme concatenate trees are more robust than the single-enzyme TyrA trees, it is expected that some Trp supercohesion groups would correspond to multiple TyrA cohesion groups. Finally, aside from the differential resolving powers of the particular proteins used to make trees, dynamic evolutionary changes that sometimes occur in a short time frame (evolutionary jumps) drive accelerated divergence that leads to separated cohesion groups or supercohesion groups. Thus, for example, TrpSCG-6 contains concatenates from Bacillus subtilis, B. stearothermophilus, and B. halodurans that are clearly separated from concatenates from other Bacillus species and from certain sister firmicute species (Lactococcus/Listeria/Staphylococcus/Streptococcus) that populate TrpSCG-7 (80). Dynamic and recent evolutionary events in the smaller clade that have driven rapid divergence are the insertion of the trp operon into a six-gene aro operon; the loss of a gene encoding a histidine pathway aminotransferase from the histidine operon, forcing an aromatic aminotransferase in the aro operon to take on a dual function; and the loss of trpAb from the trp operon, forcing pabAb to assume a dual function. In contrast, TyrCG-18 is a large cohesion group that contains TyrA members from all of the organisms corresponding to TrpSCG-6 and TrpSCG-7. Thus, on the one hand, the B. subtilis/B. halodurans/B. stearothermophilus trio has experienced an evolutionary jump that led to a dramatic divergence with respect to the tryptophan pathway (see “Intra-Cohesion-Group Intruders” above for a proposed scenario for this evolutionary jump). On the other hand, only a shallow, graded divergence occurred for TyrA throughout this large clade of firmicutes, with the result that TyrA from the B. subtilis/B. halodurans/B. stearothermophilus trio occupies a common cohesion group with TyrA proteins from Bacillus, Listeria, Staphylococcus, Streptococcus, and Lactococcus.
In previous studies of the tryptophan pathway (78, 80), a substantial fraction of the genomes and the corresponding taxonomic representation were absent compared to the much greater abundance of genomes available for the TyrA cohesion group study. Thus, in the following sections, discussion is limited to those TyrA cohesion groups existing in organisms where Trp supercohesion groups were also studied.
Lower Gammaproteobacteria
Lower Gammaproteobacteria (67) refers to a lineage within the Gammaproteobacteria that we consider to be the equivalent of a superorder. Its membership is drawn from the orders Enterobacterales, Vibrionales, and Pasteurellales and most of the Alteromonadales. Except for intruder sequences, TrpSCG-1 and TyrCG-1 possess sequences from exactly the same phylogenetic grouping, namely, the lower Gammaproteobacteria. Both the pathway of l-tryptophan biosynthesis and the TyrA subsystem can be considered to have experienced evolutionary jumps that, together with other features of aromatic amino acid biosynthesis, have separated the lower Gammaproteobacteria from the upper Gammaproteobacteria. The suite of evolutionary events relevant to l-tryptophan biosynthesis is discussed above. The evolutionary jump for TyrA is presumably tied to the gene fusion event with the gene encoding chorismate mutase.
TrpSCG-1 contains whole-operon intruders that reside in contemporary Helicobacter pylori and in coryneform bacteria. TyrCG-1 contains intruder sequences that reside in species of Nostoc (a lineage within cyanobacteria). The trp operon LGT events resulted in a total displacement of the native trp genes, but the functional role of performing l-tryptophan biosynthesis remained exactly the same. In contrast, the tyrA intruders in Nostoc did not displace the native orthologs and are thought to exercise another functional role in secondary metabolism (68). Each of the three LGT events was relatively recent, since the intruder sequences in H. pylori are absent from other Epsilonproteobacteria, those present in coryneform bacteria are absent from other actinomycete bacteria, and those present in Nostoc are absent from other cyanobacteria.
Upper Gammaproteobacteria and Betaproteobacteria
We consider the upper Gammaproteobacteria to be the equivalent of a second superorder within the class Gammaproteobacteria. Their TyrA membership is drawn sparsely from the order Alteromonadales as well as from the remaining orders not listed above for the lower Gammaproteobacteria. TrpSCG-2 contained concatenate sequences from not only the upper Gammaproteobacteria but also the Betaproteobacteria. In contrast, TyrA sequences from the upper Gammaproteobacteria have been placed into 10 cohesion groups (visualized in green on Fig. 6, panel 2), and sequences from the Betaproteobacteria have been placed into 10 additional cohesion groups (Fig. 6, panel 4). It seems likely that many of the latter cohesion groups will merge into a single group in view of the precedent of TrpSCG-2, the finding that PheA sequences from upper Gammaproteobacteria and Betaproteobacteria appear to join together on a phylogenetic tree (44), and the finding that common aromatic pathway proteins of upper Gammaproteobacteria and Betaproteobacteria also form single, cohesive groupings (data not shown). Indeed, seven upper Gammaproteobacteria TyrA cohesion groups and orphans (Microbulbifer degradans, TyrCG-6, TyrCG-3, TyrCG-5, TyrCG-2, Methylococcus capsulatus, and Nitrosococcus oceani) join one another at a common node to the exclusion of TyrA proteins from any other phylogenetic grouping, as is also the case for eight Betaproteobacteria cohesion groups (TyrCG-9, TyrCG-8, TyrCG-7, Thiobacillus denitrificans, Methylobacillus flagellatus, Dechloromonas aromatica, Azoarcus sp., and Chromobacterium violaceum). The latter two nodes are indicated by arrowheads in Fig. 2, and these nodes perhaps could already be collapsed had a less rigorous bootstrap cutoff been implemented. Among the Betaproteobacteria groups, only TyrCG-9 and TyrCG-10 diverge before the major Betaproteobacteria node. One upper Gammaproteobacteria TyrA orphan (Acidithiobacillus ferrooxidans) branches near the major upper Gammaproteobacteria node. Cohesion group TyrCG-4 is clearly divergent from cohesion groups present in all the other upper Gammaproteobacteria, of course, because it is located in the TyrAβ subhomology region of the TyrA protein tree (Fig. 2). TyrA from Coxiella burnetii in TyrCG-26 (TyrAβ subhomology grouping) might exemplify another divergence, but this TyrA protein might be a xenolog intruder since TyrCG-26 is presently an unresolved phylogenetic mixture. The TyrA protein from the Xanthomonas/Xylella lineage (TyrCG-4) is considered to have made an evolutionary jump by virtue of its acquisition of a fused ACT domain. The Xanthomonas/Xylella lineage is thus one case where evolution has been more dynamic for TyrA than for the l-tryptophan pathway.
Whereas TrpSCG-2 contains two cases of partial-pathway operon LGT, no intruders have so far been found to be present in any of the 20 TyrA cohesion groups that populate the upper Gammaproteobacteria and Betaproteobacteria (although, as mentioned above, the TyrA protein from C. burnetii in TyrCG-26 could possibly be a xenolog intruder).
Alphaproteobacteria
TrpSCG-3 contained Trp concatenates from the three Alphaproteobacteria genomes available at the time. TyrCG-12 presently contains TyrA sequences from the same organisms plus from an additional 34 organisms. Only an orphan (Pelagibacter ubique), a member of TyrCG-26 (unresolved phylogenetic mixture), and the small membership of TyrCG30 possess TyrA sequences that do not belong to TyrCG-12 (Fig. 6, panel 3). The TyrCG30 cohesion group differs due to its placement in the TyrAβ subhomology section of the tree (Fig. 2). It contains TyrA sequences from Rhodospirillum rubrum and from Maricaulis maris as well as a xenolog intruder located in Myxococcus xanthus (Deltaproteobacteria). (The single TyrA orphan from Pelagibacter ubique, provisionally considered to be divergent, could prove to be a xenolog intruder within the enlarged future population of the cohesion group that is projected.) It will be interesting to reexamine Trp concatenates in the genomes of Alphaproteobacteria having TyrA proteins not in TyrCG-12 to see the extent to which l-tryptophan biosynthesis might have diverged in the same organisms having divergent TyrA proteins.
Epsilonproteobacteria
A Trp concatenate was previously available from only a single epsilonproteobacterium, this being the above-mentioned whole-operon intruder present in Helicobacter pylori. However, differences in gene organization and gene fusion noted for three other Epsilonproteobacteria (even though they lack status as complete genomes) indicated that this group has experienced dynamic evolutionary changes with respect to the Trp pathway. In contrast, the TyrA sequences from the 10 currently available genomes of Epsilonproteobacteria coexist as a cohesive grouping in TyrCG-13. The Epsilonproteobacteria exemplify a second case where dynamic evolutionary events in tryptophan biosynthesis have driven dramatic cohesion group divergence, in contrast to the modest tyrosine pathway divergence that accounts for a single TyrA cohesion group. Interestingly, TyrCG-13 also contains nine TyrA sequences that reside in the class Flavobacteria of the phylum Bacteroidetes. Given the occupation of TyrCG-13 by a mixture of nearly equal numbers of sequences from the class Epsilonproteobacteria and from the class Flavobacteria, a common ancestor of either group could a priori have been the recipient of a xenolog intruder originating from the other. It is concluded that the intruder sequences are the ones hosted by Flavobacteria based upon the rationale developed in the last section of this article and summarized by Fig. 9.
Deltaproteobacteria
Trp concatenates from the only two Deltaproteobacteria previously available for study were divergent orphans. TyrA sequences from these same bacteria are also clearly divergent. One of them, from Geobacter sulfurreducens, occupies TyrCG-14 along with seven other sequences (TyrAα subhomology grouping). The other, from Desulfovibrio desulfuricans, occupies TyrCG-15 along with one other sequence (TyrAβ subhomology grouping). Three additional Deltaproteobacteria contain TyrA sequences that do not belong to the former two cohesion groups. The TyrA sequence from Synthrophobacter fumaroxidans is an orphan (TyrAβ subhomology grouping); TyrA from Anaeromyxobacter dehalogenans belongs to TyrCG-27 (TyrAα subhomology grouping), which is an unresolved phylogenetic mixture; and TyrA from Myxococcus xanthus is a xenolog intruder of TyrCG-30 (TyrAβ subhomology grouping).
Firmicutes
Tryptophan pathway concatenates found in the firmicute bacteria partitioned into two multisequence cohesion groups and two orphan sequences. TyrA sequences were distributed as four multisequence cohesion groups and two orphans. As discussed above, the membership of TrpSCG-6 and TrpSCG-7 was contributed by organisms whose TyrA proteins fell into a single TyrA cohesion group, TyrCG-18 (referred to as Firmicutes_1 in Table 2). Both the Trp pathway concatenate and TyrA from Desulfitobacterium hafniense were orphans. The orphan Trp pathway concatenate from Clostridium acetobutylicum corresponds to the three-member TyrCG-19 (referred to as Firmicutes_2 in Table 2). Interestingly, one of the TyrA sequences in TyrCG-19 is from C. difficile, an organism which lacks the tryptophan pathway altogether. TyrCG-20 (Firmicutes_3), TyrCG-21 (Firmicutes_4), and the orphan from Syntrophomonas wolfei contain TyrA sequences from organisms that were not available for the Trp pathway concatenate analysis.
Cyanobacteria
All of the Trp pathway concatenates fell into a single cohesion group, TrpSCG-4, and all TyrA sequences that function for primary tyrosine biosynthesis also fell into a single cohesion group, TyrCG-16. As discussed above, Nostoc species contain additional TyrA sequences that are intruder sequences belonging to TyrCG-1 and that have a specialized function in the synthesis of an indole alkaloid sunscreen agent (68).
Actinomycetes
Three tryptophan pathway concatenates from actinomycetes belonged to a single cohesion group, TrpSCG-5. Two additional actinomycete concatenates belonged to TrpSCG-1, but this was not due to divergence but was due to xenolog intrusion. TyrA sequences from 33 organisms all belong to TyrCG-17. Only TyrA from Symbiobacterium thermophilum is a divergent orphan.
Emerging Perspective
Supercohesion groups and cohesion groups have now been formulated for tryptophan pathway enzyme concatenates and for the TyrA assemblage of proteins, respectively. Once events of LGT have been sorted out, substantial parallelism is seen in the organisms that share members of a Trp cohesion group or a Tyr cohesion group. Dynamic evolutionary jumps that drive rapid divergence have been discussed for both the Trp pathway and the TyrA subsystem. Such evolutionary jumps have the effect of compressing the cohesion group to a smaller membership, as was seen with the multiple events which attended the insertion of a trp operon into an aro operon in Geobacillus species and a small clade of Bacillus species (80). It is noteworthy that the distinct separation of Gammaproteobacteria into lower Gammaproteobacteria and upper Gammaproteobacteria on the criterion of Trp cohesion groups is exactly paralleled on the criterion of Tyr cohesion groups. Other character states of enzymes performing early aromatic pathway reactions have been noted to exhibit qualitative differences that define the same taxonomic split. Kleeb et al. (44) observed two clusters on a phylogenetic tree of PheA sequences from Gammaproteobacteria (called by them Gammaproteobacteria I and Gammaproteobacteria II). The latter clusters correspond to lower Gammaproteobacteria and upper Gammaproteobacteria. It is noteworthy that Trp concatenates from upper Gammaproteobacteria and from Betaproteobacteria defined a single supercohesion group. The above-mentioned phylogenetic tree of PheA sequences, having less resolving power than Trp concatenates, nevertheless exhibited neighboring clusters, albeit with weak bootstrap support (44). A phylogenetic tree of TyrA sequences (Fig. 2) with even less resolving power also exhibited weakly supported neighboring clusters for TyrA proteins from upper Gammaproteobacteria and Betaproteobacteria. Finally, an imperfectly conserved gene organization featuring a large supraoperon containing many genes relevant to aromatic biosynthesis is clearly visible in both the upper Gammaproteobacteria and the Betaproteobacteria but not in the lower Gammaproteobacteria, where a different gene organization is imperfectly conserved (this is covered in more detail in the next section). All of this suggests that with additional sequences from new genomes or after utilizing advanced concatenation strategies, the TyrA cohesion groups currently derived from upper Gammaproteobacteria and Betaproteobacteria may merge.
Identification in painstaking detail of qualitatively different character states of genes and their encoded products, their evolutionary progression in the vertical genealogy, and evolutionary acquisitions made via LGT can feasibly be accomplished for relatively small metabolic segments, such as the individual terminal branches of aromatic biosynthesis. Once coverage is completed for the entire pathway, including the minor vitamin-like branches, it should be apparent that evolutionary conclusions arrived at separately via steps that are essentially atomistic can be combined to describe evolutionary progressions at the whole-pathway level that reveal a larger gestalt of interlocking relationships. The next section illustrates examples of this approach.
TRACKING MILESTONE EVOLUTIONARY EVENTS ACROSS SUBSYSTEMS
Gene Fusion
The tyrA gene exhibits a very similar environment of neighboring genes in the upper Gammaproteobacteria and Betaproteobacteria (67). (In contrast, the lower Gammaproteobacteria have a much different gene synteny.) The proposed ancestral synteny is one in which tyrA is closely followed by aroF, and this gene order has been largely conserved in the upper Gammaproteobacteria and the Betaproteobacteria that reside in the TyrAa subhomology grouping. Given the tenacity of these gene proximities, it is not surprising that tyrA-aroF fusions exist in both the upper Gammaproteobacteria and the Betaproteobacteria (Fig. 6, compare panels 2, 4, and 13). Among the upper Gammaproteobacteria, this fusion includes the orphan Microbulbifer degradans as well as all members of TyrCG-2, TyrCG-3, and TyrCG-6; but it is absent in members of TyrCG-5 and the orphans Methylococcus capsulatus, Nitrosococcus oceani, and Acidithiobacillus ferrooxidans. In the Betaproteobacteria, the tyrA-aroF fusion is limited to a small clade of Burkholderia within TyrCG-7. The Ralstonia and remaining Burkholderia species that are represented within TyrCG-7 lack the fusion. It seems clear that the fusion in the Betaproteobacteria occurred recently and was independent of the same fusion event which occurred in the upper Gammaproteobacteria. Acquisition of the tyrA-aroF fusion via LGT from an upper Gammaproteobacteria source can be ruled out because (i) these TyrA supradomains from the small clade of Betaproteobacteria should then belong to one of the upper Gammaproteobacteria cohesion groups and (ii) one would not expect the fused TyrA domains from the small clade of Betaproteobacteria to be located in TyrCG-7 with unfused TyrA proteins from other Betaproteobacteria.
Did the tyrA-aroF fusion occur on a single occasion in the upper Gammaproteobacteria? An inspection of Fig. 2 indicates that the position of TyrCG-5 is inconsistent with a single common ancestor that acquired the tyrA-aroF fusion. If the order of branching shown was correct, the fusion either occurred twice (once in the common ancestor of Microbulbifer degradans, TyrCG-6, and TyrCG-3 and once in the common ancestor of the members of TyrCG-2) or occurred once in the common ancestor of all the organisms hosting the fusion but was subsequently lost in the common ancestor of TyrCG-5.
Since the cohesion groups are defined such that there is little confidence in the order of branching, a tree that was based upon an alignment of all the TyrA-AroF fusion sequences with concatenated TyrA and AroF sequences from upper Gammaproteobacteria and Betaproteobacteria that lack the fusion was assembled (Fig. 7). This creates a much more reliable protein tree since AroF is a much longer and much more conserved protein than TyrA. The results shown in Fig. 7 are indeed consistent with an order of branching in upper Gammaproteobacteria such that a single tyrA-aroF fusion occurred in the common ancestor of Microbulbifer degradans and in the organisms hosting the members of TyrCG-2, TyrCG-3, and TyrCG-6. Hence, it seems clear that a recent tyrA-aroF fusion occurred in the ancestor of a clade of the upper Gammaproteobacteria and that an even more recent, second fusion occurred in the ancestor of a very small clade of the Betaproteobacteria. As suggested previously (35), this use of gene fusions has great potential for ordering phylogenetic progressions of related organisms. This section illustrates how analysis of the relationships between AroF (the sixth enzyme of the erythrose-4-phosphate to chorismate portion of aromatic biosynthesis) and TyrA provides a small glimpse of the potential of different subsystems to merge, thereby expanding to an ever-wider and more insightful view.
Aromatic Biosynthesis in the Subclass Actinobacteridae
Figure 8 illustrates how information from different studies can be assembled to catalog milestone evolutionary events that are associated with different subsystems. Here, the analysis is limited to dynamic events that are associated with the node that defines the subclass Actinobacteridae. All TyrA proteins of the Actinobacteridae populate a single cohesion group, TyrCG-17. Character states A to C (orange) existed in the common ancestor of the Actinobacteridae. The utilization of a single broad-specificity isomerase (Pri) to function in both tryptophan biosynthesis and histidine biosynthesis (6) (character state A) has not been observed elsewhere and may have originated uniquely around this time. TyrA must have been specific for l-arogenate (character state B) but broadly specific for NAD(P)+, the pyridine nucleotide cofactor (character state C). After divergence of the orders Actinomycetales and Bifidobacteriales, the cofactor specificity of TyrA narrowed to become NAD+ specific (character state D) in all families of the Actinomycetales but one. In the family Corynebacterinceae, we have speculated that while the ancestral character state of broad cofactor specificity was technically preserved, a marked quantitative preference for NADP+ emerged. The genera Corynebacterium, Mycobacterium, and Nocardia have been shown to comprise a monophyletic taxon within the Actinobacteria (22). In the mycobacterial arm of the Corynebacterineae, a periplasmic chorismate mutase was acquired (character state E). This chorismate mutase is an AroHI homolog that possesses an N-terminal extension encoding a signal peptide and a substantial C-terminal extension that has no known free-standing homologs. Whether this secreted chorismate mutase evolved from a duplicate of AroHI in a common ancestor of the genus Mycobacteria or was obtained via LGT is difficult to say at present. In the coryneform arm of the Corynebacterineae, the entire tryptophan operon was acquired from the lower Gammaproteobacteria by LGT (character state F) (80). In Corynebacterium diptheriae, the LGT-derived trp operon has been disrupted by the insertion of several native genes into the alien operon (character state G). In the Pseudocardineae/Streptosporangineae, the genes encoding TrpAa and TrpAb have become fused (character state H). The taxonomic distribution of this fusion would predict that the latter two families are the most proximal of those shown in Fig. 8. In the Bifidobacteriaceae, aroE (encoding shikimate kinase) and aroB (encoding dehydroquinate synthase) have become fused (aroE-aroB). This fusion event is indicated as character state I in Fig. 8. Adjacently positioned aroE and aroB have been noted to be one of the most persistent gene linkages in the superkingdom Bacteria (79). This ubiquitous positioning can be expected to enhance the probability of occasional gene fusion. Indeed, in addition to the Bifidobacteriaceae, independent fusions exist in (i) a clade of the Thermotogaceae, which so far includes the genera Thermotoga and Petrotoga; (ii) a clade of the Alphaproteobacteria, which includes Gluconobacter oxidans, Granulibacter bethesdensis, and Acidiphilium cryptum of the order Rhodospiralles as well as Xanthobacter autotrophicus and Bradyrhizobium sp. of the order Rhizobiales; (iii) a clade of the Betaproteobacteria that includes Herminiimonas arsenicoxydans and Janthinobacterium sp., both belonging to the family Oxalobacteraceae; (iv) Carboxydothermus hydrogenoformans (Clostridia); and (v) Herpetosiphon aurantiacus (Chloroflexi). These six fusions are of relatively recent origin since they are narrowly distributed among their phylogenetic neighbors. None of the six AroE-AroB fusion groups resemble each other enough to suggest an LGT donor within the group. It is further unlikely that any of the fusions were acquired by LGT from perhaps an unsampled genome because the shikimate kinase and dehydroquinate synthase domains of each fusion group are most similar to unfused, monofunctional domains present in the closest phylogenetic neighbors that lack the fusion.
FIG. 8.
Tracking milestone evolutionary events in the Actinobacteridae. The dendrogram for the subclass Actinobacteridae of the Bacteria (not drawn to scale) includes the family Bifidobacteriaceae of the order Bifidobacteriales (top) and the various families belonging to the order Actinomycetales. Character states asserted to exist in the common ancestor are indicated by orange encircled letters. More recent evolutionary events are shown as yellow encircled letters.
Aromatic Biosynthesis in the Superphylum Bacteroidetes/Chlorobi
The Bacteroidetes/Chlorobi group, as a superphylum taxon, represents an ancient phylogenetic section. This taxon is chosen to illustrate how the evolutionary history of TyrA can be tracked in concert with other milestone events of aromatic biosynthesis (Fig. 9). Currently, there is better genome representation in the phylum Bacteroidetes (three classes represented) than within the phylum Chlorobi (only one class represented). TyrA cohesion groups separate cleanly at the level of class. Thus, within the phylum Bacteroidetes, cohesion groups TyrCG-13, TyrCG-23, and TyrGC-26 coincide with the classes Flavobacteria, Bacteroidetes, and Sphingobacteria, respectively. All 10 of the currently available genomes in the phylum Chlorobi belong to the class Chlorobium, and TyrA sequences from each of these belong to TyrCG-24.
In this superphylum, all TyrA proteins are NAD+ specific and of unknown specificity for the cyclohexadienyl substrate (NADTyrAx). Both the TyrAα and TyrAβ subhomology groups are represented in this superphylum, and it is suggested (Fig. 9) that a single transition from TyrAα to TyrAβ occurred in a common ancestor of the classes Bacteroidetes and Sphingobacteria. After divergence of the two phyla, aroAIβ (encodes DAHP synthase) became fused with aroHI (encodes chorismate mutase) in the phylum Bacteroidetes (aroAIβ-aroHI). Members of the class Bacilli also have this fusion, albeit in the opposite orientation (aroHI-aroAIβ). The latter fusion has long been known to be the basis for a pattern of allosteric regulation (sequential feedback inhibition), whereby the substrate (chorismate) and product (prephenate) of chorismate mutase double up as feedback inhibitors of DAHP synthase (37). The putative ancestral operon (pheA>aspC>tyrA>aroAIβ>aroHI) joins all of the enzymes that divert chorismate to l-phenylalanine and l-tyrosine. AroHI can supply prephenate to both amino acid branches, and AspC undoubtedly is an aromatic aminotransferase that is capable of catalyzing very similar transaminase reactions in both amino acid branches. The fusion shown in Fig. 9 (aroAIβ-aroHI) can reasonably be considered to be a regulatory innovation. Based upon the rationale asserted by Xie et al. (80), an ancestral classical trp operon similar to that which still persists in the class Sphingobacteria is presumed to exist. Members of the class Flavobacteria have retained the original aro operon as well as the original trp operon (albeit with an open reading frame insertion between trpAa and trpAb). In the class Bacteroidetes, both the trp and aro operons have been slightly scrambled, with trpEb being translocated to the beginning of the operon and with an exchanged positioning of tyrA and aroAIβ-aroHI. The trp operon remains intact in the Sphingobacteria, but the aro operon has been disrupted. In the phylum Chlorobi, both the trp and aro operons have been dispersed.
The TyrCG-13 cohesion group is populated by TyrA sequences from not only the class Flavobacteria but also the class Epsilonproteobacteria. Hence, a fairly ancient event of LGT is implicated. As an isolated observation, it is difficult to know which of these two classes is likely to be the host of the intruder sequences and which is likely to be the donor. If these classes diverged within their phyla at different times, the most recently emerged class could not have been the LGT donor to a common ancestor of the more ancient class. The phylogenetic tree of organisms reported by Olsen et al. (57) shows the class Epsilonproteobacteria diverging from its sister classes of Proteobacteria at an earlier time than the class Flavobacteria diverged from sister classes of the phylum Bacteroidetes. It thus appears that the class Flavobacteria did not yet exist at the time of the common ancestor of Epsilonproteobacteria, and hence, no Flavobacteria could have been the donor. On the other hand, a member of Epsilonproteobacteria could have been an LGT donor of tyrA to a common ancestor of the Flavobacteria. If so, it appears that the resident tyrA gene was replaced by homologous recombination without disrupting the aro operon, of which tyrA is a member; i.e., the context of gene organization surrounding tyrA in the Flavobacteria fits into the larger context of the superphylum (Fig. 9). Thus, the aro operons of the class Flavobacteria (TyrCG-13) and the class Bacteroidetes (TyrCG-23) share the distinctive fusion gene and a nearly identical gene order.
OVERVIEW PERSPECTIVE
Small proteins that are not highly conserved represent a contemporary challenge for functional annotation. A difficult hurdle is a determination of what evolutionary distance is valid for making annotation transfers with respect to phylogenomic inference. The degree to which various functional alternatives will persist over evolutionary distance will vary for different protein families.
The extent to which current annotations are correct depends upon generations of previous experimental work and is hugely assisted by a fraction of genes that are highly conserved and evolve in the face of many limitations and constraints due to their elegant and complex mechanisms. Within the aromatic pathway, an example would be 5-enolpyruvylshikimate-3-phosphate synthase, a highly specific enzyme that utilizes a complex catalytic mechanism. Such complexity facilitates reliable annotations. On the other hand, enzymes having the plasticity to catalyze broad-specificity reactions can be represented by entirely different homology groups or by distinctly different subhomology groups that can make functional predictions elusive. A multitude of proteins (exemplified by such enzymes as kinases, phosphatases, and dehydrogenases) that illustrate the many and varied challenges for correct calls of functional role exist. The TyrA protein family of dehydrogenases benefits from a treasure trove background of wide-ranging comparative enzymology. The current analysis, together with previous work, has been a labor-intensive effort. Comparable efforts are not easily fitted to goals of high-throughput annotations for thousands of sequences in many hundreds of organisms, hence the dilemma of rapid results achieved with a lesser quality of annotation accuracy than one would like. “Difficult” gene products require a labor-intensive effort as a useful step in order to generate and preserve the information needed to allow the rich array of bioinformatic tools available to succeed in increasing the quality of high-throughput annotation efforts.
Acknowledgments
We acknowledge partial support from contract HHSN266200400042C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, and from grant G13 LM008297 from the National Library of Medicine.
APPENDIX
Determination of Cohesion Groups
TyrA sequences were collected from the SEED and from other public databases. A file of trimmed core supradomain TyrA sequences was created by trimming away obvious fused domains or extensions. At the N termini, all sequences were uniformly trimmed to begin five residues ahead of the GxGxxG motif, i.e., to match the beginning of the Wierenga fingerprint (73). C termini were trimmed of unconserved residues using the endpoints of some of the shortest TyrA proteins that have been fully characterized for guidance. ClustalX was used to create a preliminary alignment. This alignment was imported into the BioEdit sequence alignment editor. Manual adjustments were made to obtain a high-quality alignment. This alignment was used as input into a phylogenetic tree program (Phylip software, unweighted-pair group method using average linkages) (27). Trees were visualized with the TREEVIEW application (62).
In the initial tree of 347 trimmed sequences, nodes were collapsed at bootstrap values of 68%. An arbitrarily chosen member of the collapsed groups was selected as a representative sequence of that node position. The resulting 64 sequences were used to obtain a second Phylip tree, which yielded 60 sequences with the collapse of a few more nodes when a bootstrap value of 68% was applied as a cutoff. An additional repetition of this process resulted in a final tally of 58 cohesion groups. The ultimate collapsed tree (Fig. 2) exhibited nodes with bootstrap values below 58%.
Web Resources at the SEED
TyrA subsystem home page.
Resources relevant to the TyrA subsystem, individually described below, are linked to the home page at http://theseed.uchicago.edu/FIG/Html/tyrASubsystem.html. This includes the online interactive version of Fig. 2, which is linked to the online version of Table 2, the “extended table,” where sources of TyrA sequences and many properties of the corresponding TyrA proteins are tabulated, and a list of trimmed sequences corresponding to the core supradomain of TyrA proteins. The latter three pages can also be accessed directly from the TyrA subsystem home page. The online version (http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html) of the snapshot panels shown in Fig. 6 is integrated with a tool that can be used to compare up to three panels and which is linked to the extended table.
Navigating to and within the Protein Pages.
The version of Fig. 2 installed at http://theseed.uchicago.edu/FIG/Html/tyrACGTree.html is the portal to hyperlinked cohesion group tables (the short Table 2 version or a comprehensive “extended table”) that in turn are linked to the Protein Pages at the SEED. Each of the latter prominently display a clickable graphic showing the location of a given tyrA gene within an array of flanking genes, and many links are provided to allow navigation to a variety of detailed bioinformatic information. Tools are available. For example, one can ask for a comparison of the displayed gene organization with similar gene organizations present in other organisms. Mousing over any given cohesion group of Fig. 2 also delivers a drop-down menu that gives access to the relevant group of trimmed TyrA sequences.
One innovation in the extended table is a “gene neighborhood” button within each cohesion group section, which delivers a comparison of gene organization flanking tyrA within the cohesion group.
Sortable character state snapshots.
The individual panels of Fig. 6 can be viewed at http://theseed.uchicago.edu/FIG/Html/TyrAPanels.html. Choosing “compare TyrA panels” activates an option to compare up to three side-by-side panels. For example, one might want to choose and display the Proteobacteria (Fig. 6, panel 2) side by side with the view of cofactor specificities (panel 7) or with instances of gene fusion (panel 9). These individual, sortable panels identify the cohesion group numbers for all of the cohesion groups that are color coded. One can then view the complete membership of any cohesion groups of interest by linking to the extended table via links provided at the top of the screen. A JavaScript magnifying tool is provided when mousing over a given panel with the cursor. Depression of the up or down key on the keyboard increases or decreases the zoom ratio, respectively. Depression of the right or left key increases or decreases the window size, respectively.
Semiautomation of cohesion groups.
An important accomplishment would be to lock in and build upon the manual effort represented by this project with continuing semiautomatic follow-up. The technology to support the creation, curation, and advanced development of subsystems at the SEED was described previously (60). Tools to preserve the trimmed sequence alignment, accurately add newly available sequences, and update the tree and cohesion group assemblages are being implemented.
Web Resources at AroPath
The nomenclature of genes and gene products follows the rules posted under “Nomenclature: Genes/Enzymes” on the AroPath home page (http://aropath.lanl.gov). Aromatic pathway diagrams with complete biochemical structures, a list of attenuator structures associated with tyrA operons, and a tool (phyloTreeBuilder) to build 16S rRNA trees from selected organisms can be accessed from the home page.
A universal four-letter system for coding organisms to the species level with unambiguous acronyms has been developed (the first letter of the genus in capital letters followed by the first three letters of the species in lowercase type). When necessary to disambiguate a four-letter acronym, a number is attached. For example, Escherichia coli is designated Ecol, whereas Enterococcus columbae is designated Ecol-1. If the species has not been determined, the first four letters of the genus are used (all in caps). To find a given four-letter acronym associated with an organism, a list of organisms currently in the system can be browsed by clicking the link under “organisms” entitled “browse organism acronyms” at the AroPath home page. Each organism is hyperlinked to the NCBI taxonomy browser. Each species entry can be expanded to show all of the component strains and their corresponding absolute acronyms (see below).
In addition, a tool to generate an acronym that is unique at the level of a specific strain, designated an absolute acronym, is provided. A given strain or list of strains can be uploaded to AroPath by clicking the link under “organisms” entitled “get absolute acronym.” This will enable the return of an absolute acronym that is a unique identifier at the strain level. Any strain for which an absolute acronym has not been previously requested will automatically be assigned a unique designation, which will be held permanently in the database.
Finally, a useful tool is provided to amend personal sequence files to be used for obtaining multiple sequence alignments and phylogenetic trees such that key acronym information for both organism and protein are displayed in the sequence names. FASTA sequence files can be uploaded to AroPath by clicking the link under “organisms” entitled “convert sequence files,” and a converted output will be returned. For example, a sequence name returned that begins “>Ecol_J_F_AroA_b,” when used as input in a tree-building program, will appear in that form as an informative label. It will indicate that the sequence is from Escherichia coli (Ecol) strain CFT073 (_J), that the sequence is from a finished genome (_F) rather than an unfinished genome (_U), and that the sequence is one of multiple AroA paralogs (AroA_b). If a hypothetical organism possessed a single gene product, two paralogs, or three paralogs, the corresponding designations would be AroA; AroA_a and AroA_b; and AroA_a, AroA_b, and AroA_c.
REFERENCES
- 1.Abou-Zeid, A., G. Euverink, G. I. Hessels, R. A. Jensen, and L. Dijkhuizen. 1995. Biosynthesis of l-phenylalanine and l-tyrosine in the actinomycete Amycolatopsis methanolica. Appl. Environ. Microbiol. 611298-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afriat, L., C. Roodveldt, G. Manco, and D. S. Tawfik. 2006. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 4513677-13686. [DOI] [PubMed] [Google Scholar]
- 3.Aharoni, A., L. Gaidukov, O. Khersonsky, S. M. Gould, C. Roodveldt, and D. S. Tawfik. 2005. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 3773-76. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad, S., and R. A. Jensen. 1988. The phylogenetic origin of the bifunctional tyrosine-pathway protein in the enteric lineage of bacteria. Mol. Biol. Evol. 5282-297. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad, S., and R. A. Jensen. 1987. The prephenate dehydrogenase component of the bifunctional T-protein in enteric bacteria can utilize L-arogenate. FEBS Lett. 216133-139. [DOI] [PubMed] [Google Scholar]
- 6.Barona-Gómez, F., and D. A. Hodgson. 2003. Occurence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis. EMBO Rep. 4296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanc, V., P. Gil, N. Bamas-Jacques, S. Lorenzon, M. Zagorec, J. Schleuniger, D. Bisch, F. Blanche, L. Debussche, J. Crouzet, and D. Thibaut. 1997. Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-L-phenylalanine precursor of pristinamycin I. Mol. Microbiol. 23191-202. [DOI] [PubMed] [Google Scholar]
- 8.Bonner, C. A., R. S. Fischer, S. Ahmad, and R. A. Jensen. 1990. Remnants of an ancient pathway to l-phenylalanine and l-tyrosine in enteric bacteria: evolutionary implications and biotechnological impact. Appl. Environ. Microbiol. 563741-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner, C. A., R. S. Fischer, R. R. Schmidt, P. W. Miller, and R. A. Jensen. 1995. Distinctive enzymes of aromatic amino acid biosynthesis that are highly conserved in land plants are also present in the chlorophyte alga Chlorella sorokiniana. Plant Cell Physiol. 361013-1022. [Google Scholar]
- 10.Bonner, C. A., R. A. Jensen, J. E. Gander, and N. O. Keyhani. 2004. A core catalytic domain of the TyrA protein family: arogenate dehydrogenase from Synechocystis. Biochem. J. 382279-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonvin, J., R. A. Aponte, M. Marcantonio, S. Singh, D. Christendat, and J. L. Turnbull. 2006. Biochemical characterization of prephenate dehydrogenase from the hyperthermophilic bacterium Aquifex aeolicus. Protein Sci. 151417-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, K. A., E. P. Carpenter, K. A. Watson, J. R. Coggins, A. R. Hawkins, M. H. Koch, and D. I. Svergun. 2003. Twists and turns: a tale of two shikimate-pathway enzymes. Biochem. Soc. Trans. 31543-547. [DOI] [PubMed] [Google Scholar]
- 13.Byng, G. S., R. J. Whitaker, R. L. Gherna, and R. A. Jensen. 1980. Variable enzymological patterning in tyrosine biosynthesis as a means of determining natural relatedness among the Pseudomonadaceae. J. Bacteriol. 144247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byng, G. S., R. J. Whitaker, C. L. Shapiro, and R. A. Jensen. 1981. The aromatic amino acid pathway branches at l-arogenate in Euglena gracilis. Mol. Cell. Biol. 1426-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calhoun, D. H., D. L. Pierson, and R. A. Jensen. 1973. Channel-shuttle mechanism for the regulation of phenylalanine and tyrosine synthesis at a metabolic branch point in Pseudomonas aeruginosa. J. Bacteriol. 113241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catrina, I., P. J. O'Brien, J. Purcell, I. Nikolic-Hughes, J. G. Zalatan, A. C. Hengge, and D. Herschlag. 2007. Probing the origin of the compromised catalysis of E. coli alkaline phosphatase in its promiscuous sulfatase reaction. J. Am. Chem. Soc. 1295760-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Champney, W. S., and R. A. Jensen. 1970. The enzymology of prephenate dehydrogenase in Bacillus subtilis. J. Biol. Chem. 2453763-3770. [PubMed] [Google Scholar]
- 18.Chen, S., S. Vincent, D. B. Wilson, and B. Ganem. 2003. Mapping of chorismate mutase and prephenate dehydrogenase domains in the Escherichia coli T-protein. Eur. J. Biochem. 270757-763. [DOI] [PubMed] [Google Scholar]
- 19.de Kraker, J. W., K. Luck, S. Textor, J. G. Tokuhisa, and J. Gershenzon. 2006. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 143970-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domenech, J., and J. Ferrer. 2006. A new D-2-hydroxyacid dehydrogenase with dual coenzyme-specificity from Haloferax mediterranei, sequence analysis and heterologous overexpression. Biochim. Biophys. Acta 17601667-1674. [DOI] [PubMed] [Google Scholar]
- 21.Doong, R.-L., R. Ganson, and R. A. Jensen. 1993. Plastid-localized 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (DS-Mn): the early-pathway target of sequential feedback inhibition in higher plants. Plant Cell Environ. 16393-402. [Google Scholar]
- 22.Embley, T. M., and E. Stackebrandt. 1994. The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 48257-289. [DOI] [PubMed] [Google Scholar]
- 23.Fani, R., M. Brilli, and P. Lio. 2005. The origin and evolution of operons: the piecewise building of the proteobacterial histidine operon. J. Mol. Evol. 60378-390. [DOI] [PubMed] [Google Scholar]
- 24.Fazel, A. M., J. R. Bowen, and R. A. Jensen. 1980. Arogenate (pretyrosine) is an obligatory intermediate of L-tyrosine biosynthesis: confirmation in a microbial mutant. Proc. Natl. Acad. Sci. USA 771270-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fazel, A. M., and R. A. Jensen. 1979. Obligatory biosynthesis of L-tyrosine via the pretyrosine branchlet in coryneform bacteria. J. Bacteriol. 138805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazel, A. M., and R. A. Jensen. 1980. Regulation of prephenate dehydratase in coryneform species of bacteria by L-phenylalanine and by remote effectors. Arch. Biochem. Biophys. 200165-176. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 28.Gutierrez-Preciado, A., R. A. Jensen, C. Yanofsky, and E. Merino. 2005. New insights into regulation of the tryptophan biosynthetic operon in gram-positive bacteria. Trends Genet. 21432-436. [DOI] [PubMed] [Google Scholar]
- 29.Hall, G. C., M. B. Flick, R. L. Gherna, and R. A. Jensen. 1982. Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J. Bacteriol. 14965-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heath, R. J., and C. O. Rock. 2000. A triclosan-resistant bacterial enzyme. Nature 406145-146. [DOI] [PubMed] [Google Scholar]
- 31.Hirano, S. I., M. Morikawa, K. Takano, T. Imanaka, and S. Kanaya. 2007. Gentisate 1,2-dioxygenase from Xanthobacter polyaromaticivorans 127W. Biosci. Biotechnol. Biochem. 71192-199. [DOI] [PubMed] [Google Scholar]
- 32.Ingram-Smith, C., and K. S. Smith. 2006. AMP-forming acetyl-CoA synthetases in Archaea show unexpected diversity in substrate utilization. Archaea 295-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh, T., K. Takemoto, H. Mori, and T. Gojobori. 1999. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16332-346. [DOI] [PubMed] [Google Scholar]
- 34.Jensen, R. A. 1976. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30409-425. [DOI] [PubMed] [Google Scholar]
- 35.Jensen, R. A., and S. Ahmad. 1990. Nested gene fusions as markers of phylogenetic branchpoints in prokaryotes. Trends Ecol. Evol. 5219-224. [DOI] [PubMed] [Google Scholar]
- 36.Jensen, R. A., and W. Gu. 1996. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 1782161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen, R. A., and E. W. Nester. 1965. The regulatory significance of intermediary metabolites: control of aromatic acid biosynthesis by feedback inhibition in Bacillus subtilis. J. Mol. Biol. 12468-481. [DOI] [PubMed] [Google Scholar]
- 38.Jensen, R. A., G. Xie, D. H. Calhoun, and C. A. Bonner. 2002. The correct phylogenetic relationship of KdsA (3-deoxy-d-manno-octulosonate 8-phosphate synthase) with one of two independently evolved classes of AroA (3-deoxy-d-arabino-heptulosonate 7-phosphate synthase). J. Mol. Evol. 54416-423. [DOI] [PubMed] [Google Scholar]
- 39.Keller, B., E. Keller, H. Gorisch, and F. Lingens. 1983. Biosynthesis of phenylalanine and tyrosine in streptomycetes. Hoppe Seyler's Z. Physiol. Chem. 364455-459. (In German.) [PubMed] [Google Scholar]
- 40.Keller, B., E. Keller, and F. Lingens. 1985. Arogenate dehydrogenase from Streptomyces phaeochromogenes. Purification and properties. Biol. Chem. Hoppe-Seyler 3661063-1066. [DOI] [PubMed] [Google Scholar]
- 41.Khersonsky, O., C. Roodveldt, and D. S. Tawfik. 2006. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10498-508. [DOI] [PubMed] [Google Scholar]
- 42.Kino, K., S. Kuratsu, A. Noguchi, M. Kokubo, Y. Nakazawa, T. Arai, M. Yagasaki, and K. Kirimura. 2007. Novel substrate specificity of glutathione synthesis enzymes from Streptococcus agalactiae and Clostridium acetobutylicum. Biochem. Biophys. Res. Commun. 352351-359. [DOI] [PubMed] [Google Scholar]
- 43.Kino, K., M. Sato, M. Yoneyama, and K. Kirimura. 2007. Synthesis of DL-tryptophan by modified broad specificity amino acid racemase from Pseudomonas putida IFO 12996. Appl. Microbiol. Biotechnol. 731299-1305. [DOI] [PubMed] [Google Scholar]
- 44.Kleeb, A. C., P. Kast, and D. Hilvert. 2006. A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii. Biochemistry 4514101-14110. [DOI] [PubMed] [Google Scholar]
- 45.Kunzler, D. E., S. Sasso, M. Gamper, D. Hilvert, and P. Kast. 2005. Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from combinatorial mutagenesis and selection. J. Biol. Chem. 28032827-32834. [DOI] [PubMed] [Google Scholar]
- 46.Kurakin, A. 2007. Self-organization versus Watchmaker: ambiguity of molecular recognition and design charts of cellular circuitry. J. Mol. Recog. 20205-214. [DOI] [PubMed] [Google Scholar]
- 47.Lawrence, J. G. 1999. Gene transfer, speciation, and the evolution of genomes. Curr. Opin. Microbiol. 2519-523. [DOI] [PubMed] [Google Scholar]
- 48.Legrand, P., R. Dumas, M. Seux, P. Rippert, R. Ravelli, J. L. Ferrer, and M. Matringe. 2006. Biochemical characterization and crystal structure of Synechocystis arogenate dehydrogenase provide insights into catalytic reaction. Structure 14767-776. [DOI] [PubMed] [Google Scholar]
- 49.Liberles, J. S., M. Thorolfsson, and A. Martinez. 2005. Allosteric mechanisms in ACT domain containing enzymes involved in amino acid metabolism. Amino Acids 281-12. [DOI] [PubMed] [Google Scholar]
- 50.Macchiarulo, A., I. Nobeli, and J. M. Thornton. 2004. Ligand selectivity and competition between enzymes in silico. Nat. Biotechnol. 221039-1045. [DOI] [PubMed] [Google Scholar]
- 51.Mayer, E., S. Waldner-Sander, B. Keller, E. Keller, and F. Lingens. 1985. Purification of arogenate dehydrogenase from Phenylobacterium immobile. FEBS Lett. 179208-212. [DOI] [PubMed] [Google Scholar]
- 52.Miller, B. G., and R. T. Raines. 2004. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 436387-6392. [DOI] [PubMed] [Google Scholar]
- 53.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51433-446. [DOI] [PubMed] [Google Scholar]
- 54.Nishimasu, H., S. Fushinobu, H. Shoun, and T. Wakagi. 2007. Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii. J. Biol. Chem. 2829923-9931. [DOI] [PubMed] [Google Scholar]
- 55.O'Brien, P. J., and D. Herschlag. 1999. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6R91-R105. [DOI] [PubMed] [Google Scholar]
- 56.Okvist, M., R. Dey, S. Sasso, E. Grahn, P. Kast, and U. Krengel. 2006. 1.6 A crystal structure of the secreted chorismate mutase from Mycobacterium tuberculosis: novel fold topology revealed. J. Mol. Biol. 3571483-1499. [DOI] [PubMed] [Google Scholar]
- 57.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 1761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osterman, A. 2006. A hidden metabolic pathway exposed. Proc. Natl. Acad. Sci. USA 1035637-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osterman, A., and R. Overbeek. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7238-251. [DOI] [PubMed] [Google Scholar]
- 60.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 962896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 63.Porat, I., B. W. Waters, Q. Teng, and W. B. Whitman. 2004. Two biosynthetic pathways for aromatic amino acids in the archaeon Methanococcus maripaludis. J. Bacteriol. 1864940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rippert, P., and M. Matringe. 2002. Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly similar and active protein domains. Plant Mol. Biol. 48361-368. [DOI] [PubMed] [Google Scholar]
- 65.Ryding, N. J., T. B. Anderson, and W. C. Champness. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwab, W. 2003. Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62837-849. [DOI] [PubMed] [Google Scholar]
- 67.Song, J., C. A. Bonner, M. Wolinsky, and R. A. Jensen. 2005. The TyrA family of aromatic-pathway dehydrogenases in phylogenetic context. BMC Biol. 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soule, T., V. Stout, W. D. Swingley, J. C. Meeks, and F. Garcia-Pichel. 2007. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 1894465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stenmark, S. L., D. L. Pierson, F. I. Glover, and R. A. Jensen. 1974. Blue-green bacteria synthesize L-tyrosine by the pretyrosine pathway. Nature 247290-292. [DOI] [PubMed] [Google Scholar]
- 70.Subramaniam, P., R. Bhatnagar, A. Hooper, and R. A. Jensen. 1994. The dynamic progression of evolved character states for aromatic amino acid biosynthesis in gram-negative bacteria. Microbiology 1403431-3440. [DOI] [PubMed] [Google Scholar]
- 71.Sun, W., S. Singh, R. Zhang, J. L. Turnbull, and D. Christendat. 2006. Crystal structure of prephenate dehydrogenase from Aquifex aeolicus. Insights into the catalytic mechanism. J. Biol. Chem. 28112919-12928. [DOI] [PubMed] [Google Scholar]
- 72.Vogel, C., M. Bashton, N. D. Kerrison, C. Chothia, and S. A. Teichmann. 2004. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 14208-216. [DOI] [PubMed] [Google Scholar]
- 73.Wierenga, R. K., P. Terpstra, and W. G. Hol. 1986. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187101-107. [DOI] [PubMed] [Google Scholar]
- 74.Wolterink-van Loo, S., A. van Eerde, M. A. Siemerink, J. Akerboom, B. W. Dijkstra, and J. van der Oost. 2006. Biochemical and structural exploration of the catalytic capacity of Sulfolobus KDG aldolases. Biochem. J. 403421-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia, T., G. Zhao, R. S. Fischer, and R. A. Jensen. 1992. A monofunctional prephenate dehydrogenase created by cleavage of the 5′ 109 bp of the tyrA gene from Erwinia herbicola. J. Gen. Microbiol. 1381309-1316. [DOI] [PubMed] [Google Scholar]
- 76.Xie, G., C. A. Bonner, T. Brettin, R. Gottardo, N. O. Keyhani, and R. A. Jensen. 2003. Lateral gene transfer and ancient paralogy of operons containing redundant copies of tryptophan-pathway genes in Xylella species and in heterocystous cyanobacteria. Genome Biol. 4R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie, G., C. A. Bonner, and R. A. Jensen. 2000. Cyclohexadienyl dehydrogenase from Pseudomonas stutzeri exemplifies a widespread type of tyrosine-pathway dehydrogenase in the TyrA protein family. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 12565-83. [DOI] [PubMed] [Google Scholar]
- 78.Xie, G., C. A. Bonner, J. Song, N. O. Keyhani, and R. A. Jensen. 2004. Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy. BMC Biol. 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie, G., T. S. Brettin, C. A. Bonner, and R. A. Jensen. 1999. Mixed-function supraoperons that exhibit overall conservation, albeit shuffled gene organization, across wide intergenomic distances within eubacteria. Microb. Comp. Genomics 45-28. [DOI] [PubMed] [Google Scholar]
- 80.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yanai, K., N. Sumida, K. Okakura, T. Moriya, M. Watanabe, and T. Murakami. 2004. para-Position derivatives of fungal anthelmintic cyclodepsipeptides engineered with Streptomyces venezuelae antibiotic biosynthetic genes. Nat. Biotechnol. 22848-855. [DOI] [PubMed] [Google Scholar]
- 82.Zamir, L. O., R. A. Jensen, B. Arison, A. Douglas, G. Albers-Schonberg, and J. R. Bowen. 1980. Structure of arogenate (pretyrosine), an amino acid intermediate of aromatic biosynthesis. J. Am. Chem. Soc. 1024499-4504. [Google Scholar]
- 83.Zamir, L. O., R. Tiberio, K. A. Devor, F. Sauriol, S. Ahmad, and R. A. Jensen. 1988. Structure of D-prephenyllactate. A carboxycyclohexadienyl metabolite from Neurospora crassa. J. Biol. Chem. 26317284-17290. [PubMed] [Google Scholar]
- 84.Zeigler, D. R. 2005. Application of a recN sequence similarity analysis to the identification of species within the bacterial genus Geobacillus. Int. J. Syst. Evol. Microbiol. 551171-1179. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, L., B. Ahvazi, R. Szittner, A. Vrielink, and E. Meighen. 1999. Change of nucleotide specificity and enhancement of catalytic efficiency in single point mutants of Vibrio harveyi aldehyde dehydrogenase. Biochemistry 3811440-11447. [DOI] [PubMed] [Google Scholar]
- 86.Zhao, G., T. Xia, L. O. Ingram, and R. A. Jensen. 1993. An allosterically insensitive class of cyclohexadienyl dehydrogenase from Zymomonas mobilis. Eur. J. Biochem. 212157-165. [DOI] [PubMed] [Google Scholar]