Abstract
Although it is recognized that acidification of freshwater systems results in decreased overall species richness of plants and animals, little is known about the response of aquatic microbial communities to acidification. In this study we examined bacterioplankton community diversity and structure in 18 lakes located in the Adirondack Park (in the state of New York in the United States) that were affected to various degrees by acidic deposition and assessed correlations with 31 physical and chemical parameters. The pH of these lakes ranged from 4.9 to 7.8. These studies were conducted as a component of the Adirondack Effects Assessment Program supported by the U.S. Environmental Protection Agency. Thirty-one independent 16S rRNA gene libraries consisting of 2,135 clones were constructed from epilimnion and hypolimnion water samples. Bacterioplankton community composition was determined by sequencing and amplified ribosomal DNA restriction analysis of the clone libraries. Nineteen bacterial classes representing 95 subclasses were observed, but clone libraries were dominated by representatives of the Actinobacteria and Betaproteobacteria classes. Although the diversity and richness of bacterioplankton communities were positively correlated with pH, the overall community composition assessed by principal component analysis was not. The strongest correlations were observed between bacterioplankton communities and lake depth, hydraulic retention time, dissolved inorganic carbon, and nonlabile monomeric aluminum concentrations. While there was not an overall correlation between bacterioplankton community structure and pH, several bacterial classes, including the Alphaproteobacteria, were directly correlated with acidity. These results indicate that unlike more identifiable correlations between acidity and species richness for higher trophic levels, controls on bacterioplankton community structure are likely more complex, involving both direct and indirect processes.
Freshwater, although accounting for less than 1% of the Earth's liquid water, is arguably the most important natural resource on the planet. Microbial communities associated with freshwater environments form the foundation of freshwater food webs and are the primary biogeochemical agents involved in nutrient cycling; yet they remain relatively understudied. During the past several decades, our appreciation of the diversity and complexity of microbial systems has dramatically increased, largely due to the use of new molecular genetic tools in environmental microbiology. Our knowledge of marine microbial communities has been the focus of considerable study and has been growing at an exponential rate. Freshwater microbial populations have also attracted attention, but to date, there has been considerably less research on these populations (64).
Before culture-independent molecular genetics-based techniques were available, freshwater microbial communities were believed to be more similar to marine and soil communities than different from marine and soil communities (41). However, more recent examinations of bacterial communities in freshwater environments have suggested that they are distinguishable from marine communities, largely by the dominance of members of the Betaproteobacteria class and representatives of the Actinobacteria class (16, 33, 58, 59, 64). Representation of these groups appears to be intermediate in estuarine environments (6, 7, 40). Investigations of bacterial communities in Adirondack lakes in the state of New York were among the first to recognize unique freshwater bacterial lineages of freshwater Betaproteobacteria and Actinobacteria (22, 33). Additionally, recent studies are revealing diversity of freshwater picoeukaryote communities that rival the diversity seen in marine environments (42). Less is known about freshwater Archaea communities, although several studies have investigated these communities (8, 26, 39, 53).
The importance and numerical abundance of these “typical” freshwater bacterial groups have been confirmed in studies employing fluorescence in situ hybridization approaches (17). Interestingly, the abundance and distribution of Actinobacteria and Betaproteobacteria suggest that these two groups are influenced differently by lake hydrologic type, nutrient conditions, seasonality, and grazing pressures (9, 29, 61). Recently, Hahn and others have succeeded in isolating representatives of some of the freshwater Actinobacteria and Betaproteobacteria species and have begun to demonstrate the ecological significance of some phylogenetically distinct groups of these freshwater bacteria (19, 20, 38, 47, 59).
One of the longest and most comprehensive investigations of freshwater systems has focused on lakes and ponds in the Adirondack Park located in northeastern section of New York State in the United States. The Adirondack Park is a 6-million-acre forested upland and mountainous region, which contains approximately 2,770 lakes and ponds in six different watersheds (11, 25). The Adirondack region receives an average of 100 to 150 cm of precipitation annually, and due to the low acid-neutralizing capacity of most Adirondack waters, the region has been severely impacted by acid precipitation (25). In recent years, Adirondack lakes have been on a trajectory to recovery from acidification in response to decreasing levels of air pollution (11, 25). Lakes in the Adirondacks are characterized hydrologically and physiochemically into distinct types defined by their geomorphology, drainage, and chloride, calcium, and dissolved organic carbon (DOC) concentrations (25, 37). Most Adirondack lakes are small in terms of surface area and volume, and they have moderate drainage areas, resulting in short hydraulic retention times. Detailed hydrographic and chemical characteristics of many Adirondack lakes can be obtained from the Adirondack Lake Survey Corporation at the website www.adirondacklakessurvey.org. From 1994 to 2007, under the auspices of the Adirondack Effects Assessment Program (AEAP) supported by the U.S. Environmental Protection Agency, the potential recovery of Adirondack lakes from acidification has been investigated. The AEAP has focused on the long-term monitoring of water chemistry and biota, including microorganisms, in 30 representative lakes. In this study we report on bacterial communities in 17 of these lakes, as well as an 18th Adirondack lake, Lake George, that has not been significantly impacted by acid deposition.
Lake George is a 62.5-km-long glacial lake located in the southeastern corner of the Adirondack Park. It is relatively large with respect to other lakes in the region, with a surface area of 114 km2. On the basis of nutrient concentrations, Lake George is classified as an oligo- to mesotrophic lake with moderate alkalinity (35). Continuous long-term monitoring of water chemistry in Lake George has been ongoing since 1979 (35) (http://www.rpi.edu/dept/DFWI/).
The deleterious impact of atmospheric acid deposition on the biota of freshwater environments, particularly macroorganisms (plants and animals), has been shown to be dramatic, particularly with respect to large invertebrate and vertebrate predators (21, 44). Most acutely in the northeastern portion of North America, acid deposition in freshwater environments has resulted in the loss of aquatic species, including algae, fish, submerged vegetation, amphibians, and aquatic invertebrates (44). In recent years, primarily due to reduced atmospheric sulfur deposition, lakes, rivers, and streams across Europe and North America have experienced increasing pH with concomitant decreases in SO4 and other chemical indicators of acidic precipitation (49, 51, 57). In some, but not all, cases, the biotic communities (submerged vegetation, invertebrates, and fish) are also recovering, although generally there has been a substantial lag period between chemical and biological recovery (11, 24, 48). Because relatively little is known about the response, potential recovery, or changes of bacterial communities in association with acid deposition processes, we investigated the composition and diversity of bacterial communities in Adirondack lakes recovering from acidification (along with one lake that has not been impacted). We also examined the relationship between bacterioplankton community structure and lake physiochemical and hydrological properties to elucidate some of the factors that may influence microbial diversity in an effort to develop specific testable hypotheses concerning environmental controls on the composition of freshwater bacterial communities.
MATERIALS AND METHODS
Study sites.
The study lakes and their locations and physical characteristics are shown in Table 1. Seventeen of the 30 lakes and ponds in the AEAP suite of lakes were selected for studying bacterioplankton community structure. These lakes, located in the southwestern portion of the Adirondack Park in New York State, included different hydrogeomorphic types as characterized on the basis of the classification scheme developed by Newton and Driscoll (37). Additional information on the physical and chemical properties of these lakes has been provided by Jenkins et al. (25).
TABLE 1.
Locations and physical characteristics of the 18 study lakes
| Lake name | Lake code | Location (latitude, longitude) | Size (Ha)a | Maximum depth (m) | Hydrological typeb | Hydraulic retention time (days)a | Carlson's TSIc |
|---|---|---|---|---|---|---|---|
| Big Moose Lake | BM | 43°49.02′N, 74°51.23′W | 512 | 21.2 | TDL | 174 | 33.9 |
| Brooktrout Lake | BT | 43°36.00′N, 73°39.45′W | 29 | 20.0 | TDL | 521 | 48.3 |
| Cascade Lake | CS | 43°49.46′N, 74°26.12′W | 40 | 5.8 | MDL | 159 | 38.0 |
| Dart Lake | DT | 43°47.36′N, 74°52.16′W | 52 | 16.0 | TDL | 17 | 30.4 |
| Grass Lake | GR | 43°52.24′N, 74°34.33′W | 5 | 5.0 | MDH | 26 | 36.6 |
| Hell Diver Pondd | HD | 43°40.10′N, 74°42.00′W | 6.5 | 2.4 | TDH | 63 | |
| Ice House Lake | IH | 43°39.58′N, 74°42.13′W | 3 | 13.0 | TSH | 365 | 44.6 |
| Lake George | LG | 43°33.49′N, 73°39.05′W | 11,400 | 60.0 | TDL | 3,042 | 39.4 |
| Long Lake | LN | 43°50.15′N, 74°28.50′W | 2 | 4.0 | TDH | 61 | 39.4 |
| Middle Settlement Lake | MS | 43°41.02′N, 75°06.00′W | 16 | 9.0 | TDL | 261 | 31.9 |
| Moss Lake | MO | 43°46.52′N, 74°51.11′W | 46 | 13.9 | MDL | 228 | 42.8 |
| Raquette Reservoir | RQ | 43°47.42′N, 74°39.05′W | 1.5 | 2.8 | MDH | 6 | 48.2 |
| Lake Rondaxe | RX | 43°45.23′N, 74°54.59′W | 90.5 | 9.7 | TDL | 9 | 38.6 |
| Sagamore Lake | SG | 43°45.57′N, 74°37.43′W | 68 | 20.0 | MDH | 72 | 35.5 |
| South Lake | SO | 43°30.34′N, 74°52.38′W | 202 | 19.0 | TDL | NA | 34.3 |
| Squaw Lake | SW | 43°38.10′N, 74°42.20′W | 36 | 6.6 | TDL | 281 | 34.6 |
| Willis Pond | WS | 43°22.17′N, 74°14.47′W | 15 | 2.3 | MDL | 79 | 42.5 |
| Willy's Lake | WY | 43°58.20′N, 74°57.20′W | 24 | 11.9 | MDL | 365 | 32.0 |
Data from the Adirondack Lakes Survey Corporation website (www.adirondacklakessurvey.org), except for Lake George (data for Lake George from reference 45). NA, data not available.
The lakes were classified by the classification scheme of Newton and Driscoll (37). Abbreviations: T, thin tilled; M, mounded; D, drainage; S, seepage; H, high DOC; L, low DOC.
Carlson's trophic state index (TSI) score was calculated on the basis of epilimnion chlorophyll a (CHL) concentrations by the following equation: TSI (CHL) = 9.81 ln(CHL) + 30.6. The trophic categories of the lakes were assigned as follows: TSI scores of <39, oligotrophic; TSI scores of 40 to 50 mesotrophic; TSI scores of 50 to 60, eutrophic.
Hell Diver Pond is a brown water dystrophic pond and thus not appropriate for Carlson's TSI classification.
In addition to the AEAP lakes, Lake George was included in this study (Table 1) as a representative Adirondack lake that has not been negatively impacted by regional acid deposition. Although the Lake George samples were collected 2 years earlier than the AEAP lakes, our intuition is that the variability of bacterial community composition between lakes is much higher than any temporal differences between Lake George and the AEAP lakes. Therefore, it is appropriate to make comparisons between clones collected from Lake George in 2000 with clones collected from other Adirondack lakes in 2002. A detailed description of Lake George has been provided by Frischer et al. (14) and Shuster et al. (45).
Sample collection.
With the exception of Lake George, water samples from lakes that exhibited thermal stratification were collected from the epilimnion (well-mixed surface layer) and the hypolimnion (1 meter above the lake bottom). Not all sample lakes contained a hypolimnion layer during sampling (see Table 3). Of the 17 lakes that were sampled, 8 had both layers, 9 did not have a hypolimnetic layer developed, and 1 did not have an epilimnion. Depth-integrated epilimnetic water samples were collected by lowering a 2.54-cm (1-in.)-diameter hose to 1 m above the thermocline, plugging the top with a rubber cork to create a vacuum, and retrieving the water into a well-rinsed bucket. Water samples from the hypolimnion were collected using a Wildco Alpha VanDorn style water bottle. Two liters of water from each sample was transferred to sterile darkened containers, stored on ice, and transported to the lab. All samples were processed within 12 h of collection. In Lake George, three epilimnion samples were collected from the microsurface layer (top 60 μm) and from the epilimnion (top 10 m). The microsurface layer was sampled as previously described (36) using a custom-modified towed skimming sled. Integrated water samples from the top 10 m (epilimnion) were collected using a modified zooplankton pump by the method of Frischer et al. (14). Samples were kept on ice, transported to the lab, and processed within 3 h of collection.
TABLE 3.
Distribution of clones in each lake sample
| Lake name | Sample typea | No. of clones | % of bacteria in the following bacterial class in each lake sample
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Betaproteobacteria | Actinobacteria | Sphingobacteria | Alphaproteobacteria | Gammaproteobacteria | Bacteroidetes | Cyanobacteria | Deltaproteobacteria | Unclassified Bacteria | Verrucomicrobia | Planctomycetacia | Flavobacteria | Genera incertae sedis OP10 | Unclassified proteobacteria | Fibrobacteres | Chloroflexi | Fusobacteria | Chlorobia | Spirochaetes | |||
| Big Moose Lake | e | 25 | 80.00 | 4.00 | 4.00 | 4.00 | 4.00 | 0.00 | 4.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Brooktrout Lake | e | 78 | 25.64 | 0.00 | 17.95 | 33.33 | 6.41 | 0.00 | 1.28 | 11.54 | 2.56 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Brooktrout Lake | h | 85 | 23.53 | 2.35 | 9.41 | 27.06 | 16.47 | 2.35 | 2.35 | 5.88 | 7.06 | 2.35 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Cascade Lake | e | 65 | 18.46 | 4.62 | 1.54 | 0.00 | 73.85 | 0.00 | 1.54 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Dart Lake | e | 90 | 6.67 | 73.33 | 17.78 | 2.22 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Grass Lake | e | 73 | 24.66 | 24.66 | 19.18 | 10.96 | 15.07 | 0.00 | 0.00 | 2.74 | 0.00 | 0.00 | 0.00 | 2.74 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Hell Diver Pond | e | 94 | 56.38 | 10.64 | 1.06 | 25.53 | 3.19 | 0.00 | 2.13 | 0.00 | 1.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Hell Diver Pond | h | 97 | 56.52 | 15.22 | 2.17 | 19.57 | 0.00 | 1.09 | 1.09 | 0.00 | 1.09 | 1.09 | 0.00 | 0.00 | 0.00 | 2.17 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ice House Lake | e | 41 | 31.71 | 26.83 | 14.63 | 12.20 | 12.20 | 0.00 | 0.00 | 2.44 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ice House Lake | h | 60 | 18.33 | 6.67 | 20.00 | 6.67 | 28.33 | 13.33 | 0.00 | 1.67 | 3.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.67 | 0.00 |
| Lake George | e | 228 | 17.50 | 37.30 | 12.70 | 4.40 | 6.10 | 0.40 | 11.00 | 0.00 | 3.10 | 0.00 | 3.10 | 3.10 | 0.90 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Long Lake | e | 46 | 50.00 | 30.43 | 0.00 | 4.35 | 6.52 | 2.17 | 6.52 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Middle Settlement Lake | e | 70 | 51.43 | 28.57 | 14.29 | 0.00 | 2.86 | 0.00 | 0.00 | 2.86 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Middle Settlement Lake | h | 95 | 5.26 | 41.05 | 35.79 | 17.89 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Moss Lake | e | 87 | 34.48 | 8.05 | 31.03 | 6.90 | 10.34 | 0.00 | 4.60 | 0.00 | 1.15 | 0.00 | 0.00 | 1.15 | 0.00 | 0.00 | 0.00 | 2.30 | 0.00 | 0.00 | 0.00 |
| Moss Lake | h | 91 | 37.36 | 14.29 | 2.20 | 6.59 | 18.68 | 4.40 | 0.00 | 6.59 | 2.20 | 1.10 | 1.10 | 0.00 | 1.10 | 0.00 | 2.20 | 0.00 | 1.10 | 0.00 | 1.10 |
| Raquette Reservoir | h | 71 | 56.34 | 22.54 | 5.63 | 5.63 | 1.41 | 1.41 | 2.82 | 4.23 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Raquette Reservoir | e | 48 | 56.25 | 4.17 | 16.67 | 16.67 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 4.17 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Lake Rondaxe | e | 94 | 63.83 | 14.89 | 8.51 | 1.06 | 9.57 | 0.00 | 1.06 | 0.00 | 0.00 | 1.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Sagamore Lake | e | 82 | 47.56 | 34.15 | 4.88 | 8.54 | 1.22 | 0.00 | 0.00 | 1.22 | 1.22 | 1.22 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Sagamore Lake | h | 93 | 56.99 | 9.68 | 7.53 | 13.98 | 3.23 | 0.00 | 1.08 | 2.15 | 2.15 | 0.00 | 3.23 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| South Lake | e | 87 | 49.43 | 13.79 | 20.69 | 2.30 | 5.75 | 3.45 | 0.00 | 3.45 | 0.00 | 1.15 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| South Lake | h | 94 | 85.11 | 4.26 | 4.26 | 3.19 | 2.13 | 1.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Squaw Lake | e | 89 | 23.60 | 55.06 | 10.11 | 2.25 | 3.37 | 0.00 | 0.00 | 4.49 | 0.00 | 1.12 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Willis Pond | e | 79 | 22.78 | 48.10 | 12.66 | 7.59 | 1.27 | 0.00 | 1.27 | 1.27 | 2.53 | 2.53 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Willy's Lake | h | 77 | 20.21 | 3.19 | 4.26 | 22.34 | 4.26 | 0.00 | 37.23 | 0.00 | 7.45 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
e, epilimnion; h, hypolimnion.
Auxiliary analytical measurements.
In addition to examination of the composition of bacterial communities, 31 physical and chemical properties were determined on replicate water samples. Dissolved oxygen and temperature were measured in situ using an YSI dissolved oxygen meter (model 57; YSI Life Sciences). The meter was calibrated at each lake immediately prior to sample collection. Secchi depth was also estimated in each AEAP lake at the time of sampling. Water samples were processed, preserved (where appropriate), and analyzed for chemical analytes and nutrients listed in Table S1 in the supplemental material. All analyses were conducted at the Darrin Fresh Water Institute's New York State Environmental Laboratory Approval Program-certified laboratories following appropriate analytical and quality assurance/quality control procedures approved by the U.S. Environmental Protection Agency. Environmental Laboratory Approval Program certification is required for all laboratories performing environmental analyses on samples originating from New York State to ensure the accuracy and reliability of these analyses. Detailed methodological protocols can be found at www.cee.rpi.edu/res_ee_keck.cfm.
DNA purification, PCR amplification, cloning, and sequencing.
AEAP lakes were sampled for bacterioplankton from 10 to 24 September 2002. Lake George was sampled in November 2000. For AEAP lakes, epilimnetic and hypolimnetic subsamples (900 to 1,000 ml each) were collected and prefiltered (5 μm), and bacterioplankton communities were then collected on 0.22-μm mixed cellulose ester filters (Millipore) by gentle vacuum using Nalgene MityvacII hand pumps and stored at −80°C until further processed. For all lakes, total genomic DNA was purified using the soil DNA extraction kit (Q-Biogene) with a bead-beating step using filters cut into small pieces as the “soil.” Purified DNA was quantified by spectrophotometry and analyzed for quality by visual assessment after agarose gel electrophoresis on 1% gels. Total genomic DNA was used for PCR amplification of the 16S rRNA genes using universal forward 8F (5′ AGA GTT TGA TCM TGG CTT CAG) and reverse 1492R (5′ GGT TAC CTT GTT ACG ACT T) primers (1). Amplification products were resolved by gel electrophoresis and subcloned into the pCR 2.1 vector using the TA Cloning kit (Invitrogen). After the amplification product was cloned into the plasmid vector, the 16S rRNA gene insert was reamplified with primers M13 Forward (−20) (5′ GTA AAA CGA CGG CCA GTG) and M13 Reverse (−27) (5′ GGA AAC AGC TAT GAC CAT G) targeting the M13 cloning site. The amplified insert was verified visually by agarose gel electrophoresis (1% gels) and purified using the Qiagen MinElute purification kit following the manufacturer's instruction (Qiagen). Purified PCR amplicon (typically 5 to 125 ng μl−1) was eluted in 10 μl EB buffer (10 mM Tris-Cl, pH 8.5). Sequencing was accomplished using the Big Dye Terminator v3.0 cycle sequencing kit (Applied Biosystems) in-house starting with 1 to 2 μl of purified PCR amplicon using the M13 primers. Automated sequencing was performed using an ABI 3700 sequencer at the Center for Functional Genomics at the State University of New York at Albany. The identity of each clone sequence was determined by identifying its nearest neighbor using the Sequence Match and Classifier tools at the Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/) on 18 May 2006 (RDP update 39).
Clone identification.
A total of 1,907 clones from 25 libraries produced from 17 AEAP study lakes and 228 clones from Lake George in six libraries were isolated and analyzed. Because all of the clones from the Lake George libraries were derived from epilimnetic samples, they were pooled and analyzed as a single library. Initially, 96 clones were picked from each library but depending on downstream processing, fewer clones were usually analyzed. For the AEAP lakes, the number of clones successfully analyzed from each library ranged from 25 (Big Moose Lake, epilimnion) to 97 (Hell Diver Pond, hypolimnion; 97 because there was low recovery from the first library, a second library was also processed). The average number of clones analyzed per lake was 76 ± 19, and the specific number of clones analyzed in each lake is presented below (see Table 3). Initially, amplified rRNA gene inserts from clones were verified visually by agarose gel electrophoresis (1% gels) to confirm the presence of full-length rRNA gene inserts. Clone inserts were preliminarily identified on the basis of restriction digestion profiling of amplified 16S rRNA gene fragments (amplified rRNA gene restriction analysis [ARDRA]) essentially as previously described by Vergin et al. (56) except that the isoschizomer of BsuRI, HaeIII was utilized. ARDRA patterns were assessed after visualization by agarose gel electrophoresis on 3% agarose gels. Restriction profiles were visually sorted into patterns differentiated by the number and size of fragments. Pattern recognition was further facilitated by digital image analysis using the Kodak 1D Image Analysis Software Package v3.6 (Eastman Kodak, NY). ARDRA samples were first pooled by lake layer and then between epilimnion and hypolimnion layers of the same lake. Each unique ARDRA profile was assigned an identification code, and on average, 62% of the representative clones of each ARDRA pattern were sequenced. In the case of the same ARDRA pattern being discovered in different lakes, representative clones from each lake were always sequenced, which provided identical taxonomic classifications at the class and subclass levels as determined using the RDP Classifier tool. However, in some cases when the same ARDRA pattern was observed in a single lake, but in different layers, representatives from both layers were not always sequenced. Of the 1,907 clones obtained from the AEAP lakes and classified into 86 unique ARDRA profiles, 431 were sequenced. There was evidence of microdiversity among clones that had been classified by ARDRA (i.e., multiple ARDRA patterns were identified as belonging to the same class or subclass and very minor sequence differences between some clones were observed). From the Lake George libraries, 228 clones were obtained, all of which were sequenced. In 10 instances, when the ARDRA patterns were very distinct, less than 20% of the clones with identical ARDRA patterns were sequenced.
Each clone was phylogenetically classified to the taxonomic level of class and subclass according to the hierarchical taxonomy proposed by Garrity et al. (15) using the RDP Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp) release 9.0. The taxonomic classification of subclass is an intermediate taxonomic rank between class and order used by Garrity et al. (15) and adopted by the RDP. Although the majority of sequences were reliably classified to much lower taxonomic ranks, it was not possible to consistently assign all 2,135 clone identities to less than a class designation. Therefore, the generic term subclass was used to define classifications below the rank of class but higher than order. A comprehensive phylogenetic analysis of the Adirondack lake bacterial clones obtained in this study was beyond the scope of the current study and will be presented elsewhere. Likewise, since all clones could not be assigned to taxonomic ranks directly comparable to the recently proposed ecologically significant classifications of the freshwater betaproteobacteria (19) and Actinobacteria (58), comparison on the basis of these rankings was also considered outside the scope of this study.
Database organization.
All clone information is maintained in an Excel spreadsheet and identified by a unique clone identification number consisting of an ADK (Adirondack) designation code, lake identifier, date (year) identifier, and clone number. Epilimnetic and hypolimnetic samples are designated by an e or h, respectively, following the lake identifier. The lake identifier codes are shown in Table 1. For example, the clone identifier ADK-DTe02-58 uniquely identifies clone 58 obtained from the epilimnion of Dart Lake in 2002. Complete clone information, sequences, and classification are maintained on the Darrin Fresh Water Institute website (www.rpi.edu/dept/DFWI/index.html) and are available for research purposes with permission.
Statistical analyses.
Richness at the taxonomic levels of class and subclass in all clone libraries was estimated using the software package EstimateS v7.5.1 (R. K. Colwell, EstimateS [http://purl.oclc.org/estimates]), which gives a statistical estimation of species richness and shared species from samples. The richness estimator Chao2 without bias correction is reported, since this metric is recommended as among the most robust for sample sets with many rare species (5) although the Chao1, ICE, and ACE indices were also calculated and resulted in similar richness estimates. Richness of the complete Adirondack data set was estimated on the basis of the total number (2,135 clones) of phylogenetically classified clones (Table 1). Sampling sufficiency of each library (lake sample) was determined as described by Kemp and Aller (27) using the “Large Enough” estimator available online at http://www.aslo.org/lomethods/free/2004/0114a.html. The Large Enough software provides a convenient calculator for the Chao1 and ACE richness estimates based on the number of uniquely classified clones in each clone library (see Table 3). The actual clone numbers for each lake sample can be calculated by multiplying the number of clones per sample (n) by the percent occurrence of each bacterial class. Diversity indexes including the Shannon diversity index, the Brillouin index, Simpson's index, McIntosh's index, and the Berger-Parker index were all calculated using the BIO-DAP software package (G. M. Thomas [http://nhsbig.inhs.uiuc.edu]), a biodiversity analysis package, although only the Shannon diversity index is reported. Diversity indices were calculated on the basis of the number of uniquely classified clones in each clone library. The overall bacterioplankton community composition was explored by principal component analysis (PCA) using the Unscrambler software package v9.2 (CAMO, Norway). Principal component models were built with full leverage correction and based on percent normalized (by lake sample) abundance of clones in each library (see Table 3). The statistical significance of PCAs were assessed by analysis of variance of PCA scores between lake samples. The significance of correlations between bacterial classes and the 31 physical and chemical properties was assessed by correlation analysis between each chemical and physical parameter and frequency of occurrence of each phylogenetic bacterial class in each lake sample. In cases where normality and equality of variance of least square residuals assumptions were satisfied, the parametric Pearson correlation procedure was used. In cases where normality and equality of variance assumptions were not met, the nonparametric Spearman rank procedure was utilized. All analysis of variance and correlation analyses were facilitated using the software package SigmaStat v3.0 (SPSS Inc.).
Nucleotide sequence accession numbers.
The sequences of all unique clones obtained in this study have been deposited in GenBank (accession numbers EF520350 to EF520639).
RESULTS
Characterization of study lakes.
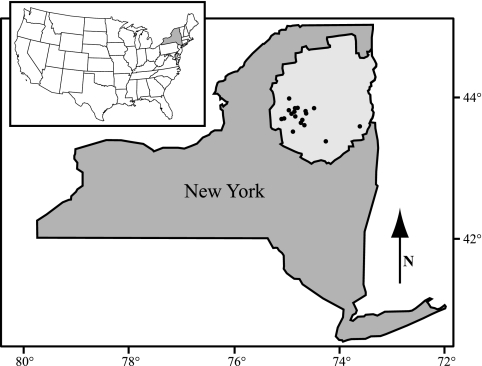
Eighteen lakes located in the southern region of the Adirondack Park in New York State were examined in this study (Fig. 1). All samples were collected in September 2002, except for Lake George, which was sampled in November 2000. With the exception of Lake George, which has not become acidified during recent decades, all the lakes are exhibiting various degrees of recovery from the effects of regional acid deposition (11, 25). The location, size, depth, hydrological classification, and estimated hydraulic retention time of each of the 18 lakes are shown in Table 1. The size of lakes ranged from as small as 1.5 ha to as large as 11,400 ha. Excluding Lake George, which is anomalously large for the region, the average size of the lakes in this study was 67.5 ha. The estimated hydraulic retention times of this set of lakes vary from 6.2 to >3,000 days. The average hydraulic retention time for all lakes (excluding Lake George) was 167 days. Lake depths ranged from 2.3 to 60 m with the majority of the lakes exhibiting thermal stratification at the time of sampling (14 of 18). Five hydrological types of lakes as classified by Newton and Driscoll (37) were studied and included: eight thin-tilled drainage lakes with low DOC concentrations (TDL), two thin-tilled drainage lakes with high DOC concentrations (TDH), one thin-tilled seepage lake with high DOC concentration (TSH), four mounded drainage lakes with low DOC concentrations (MDL), and three mounded drainage lakes with high DOC concentrations (MDH). On the basis of chlorophyll a concentrations (see Table S1B in the supplemental material), the lakes ranged from oligotrophic to mesotrophic as classified according to Carlson's trophic state index (3). One lake, Hell Diver Pond, is a brown water lake with anomalously high DOC concentrations and therefore not appropriately classified using this index. The pH of the lakes ranged from 4.86 (Long Lake, epilimnion) to 7.76 (hypolimnion of Ice House Lake). During the 1980s, Ice House Lake was limed (J. Harrison, personal communication). All of the physical and chemical parameters measured in each lake are presented in Tables S1A to C in the supplemental material.
FIG. 1.
Map of New York State with the Adirondack Park showing the 18 lakes sampled during this study. The locations of the 18 lakes sampled during the study are indicated (•). The inset indicates the location of New York State within the United States.
Clone classification.
A total of 2,135 16S rRNA gene fragment clones derived from 31 independent libraries from the 18 lakes were identified and classified. These clone libraries have been considered collectively (pooled) to provide an overall estimate of bacterial diversity of Adirondack lake bacteria. Of these clones, approximately 30% (659 clones) were sequenced, and the identity of the other clones was inferred on the basis of ARDRA using the restriction endonuclease HaeIII and compared to sequenced clones with identical restriction patterns. The classification of unique clones from all samples is shown in Table 2. Of the 2,135 clones that were identified, 19 phylogenetic classes represented by 95 unique subclasses occurred in this Adirondack lake data set. As reported elsewhere, freshwater representatives of the Actinobacteria and Betaproteobacteria classes accounted for the majority of 16S rRNA gene Adirondack clones recovered. Over half of the clones (59.1%) belonged to these groups. Bacteria belonging to the Sphingobacteria (12.1%), Alphaproteobacteria (10.3%) and Gammaproteobacteria (8.2%) classes also comprised a significant component of the bacterial community inferred from the abundance of recovered 16S rRNA genes. The distribution of clones in each lake is presented in Table 3.
TABLE 2.
Classified rRNA gene clones from Adirondack lakes
| Phylogenetic class | No. of ARDRA patterns per classa | No. of subclasses | No. of clones | GenBank accession no.b |
|---|---|---|---|---|
| Betaproteobacteria | 9 | 16 | 785 | EF520438-EF520508 |
| Actinobacteria | 4 | 8 | 476 | EF520350-EF520391 |
| Sphingobacteria | 11 | 11 | 258 | EF520589-EF520614 |
| Alphaproteobacteria | 12 | 20 | 219 | EF520392-EF520434 |
| Gammaproteobacteria | 4 | 10 | 175 | EF520558-EF520582c |
| Bacteroidetes | 1 | 1 | 21 | EF520435-EF520437 |
| Cyanobacteria | 5 | 4 | 79 | EF520512-EF520528 |
| Deltaproteobacteria | 9 | 9 | 40 | EF520529-EF520547 |
| Unclassified Bacteria | 18 | 1 | 36 | EF520616-EF520634c |
| Verrucomicrobiae | 1 | 3 | 12 | EF520636-EF520639 |
| Planctomycetacia | 1 | 2 | 11 | EF520584-EF520588 |
| Flavobacteria | 1 | 3 | 10 | EF520551-EF520556 |
| Genera incertae sedis OP10 | 1 | 1 | 3 | EF520583 |
| Unclassified proteobacteria | 3 | 1 | 2 | EF520635 |
| Fibrobacteres | 1 | 1 | 2 | EF520548-EF520550 |
| Chloroflexi | 1 | 1 | 2 | EF520511 |
| Fusobacteria | 1 | 1 | 1 | EF520557 |
| Chlorobia | 1 | 1 | 2 | EF520509-EF520510 |
| Spirochaetes | 1 | 1 | 1 | EF520615 |
| Total | 85 | 95 | 2,135 |
The ARDRA patterns for water samples from Lake George were not determined because all clones were sequenced. Lake George contained nine unique subclasses that were not present in the AEAP lake sample set.
Representative sequences of all unique 16S rRNA gene clones recovered were deposited in GenBank.
The sequences associated with accession numbers EF520579 (gammaproteobacteria) and EF520617 (unclassified bacteria) were withdrawn from GenBank as nonunique sequences.
Of the 785 clones classified in the Betaproteobacteria class, six sequences comprised over half (53.4%) of the clones. Ninety-nine clones were most similar (96.1% nucleotide similarity) to an uncultured betaproteobacterium FukuN33 clone (GenBank accession number AJ289997) classified in the Polynucleobacter necessarius subcluster PnecC (19). An additional 53 clones were most similar (93.9% nucleotide similarity) to another Polynucleobacter species betaproteobacterium clone PRD01a011B (GenBank accession number AF289159) also classified in the PnecC P. necessarius subcluster. Ninety-seven clones were most similar (92.6% nucleotide similarity) to betaproteobacterium F1021 clone, a representative of the Acidovorax subclass (GenBank accession number AF236005). Eighty-seven clones were most similar (98.4% nucleotide similarity) to a cultivated Polynucleobacter species HTCC543 (GenBank accession number AY584579) isolated from Crater Lake, Oregon, and classified in the P. necessarius subcluster PnecD. Forty-two clones were most similar (91.6% nucleotide similarity) to an unidentified Betaproteobacteria clone ACK-L6 originally identified by our group from Limekiln Lake in the Adirondacks (GenBank accession number U85123). ACK-L6 is a member of the P. necessarius PnecA subcluster. Forty-one clones were most similar (98.6% nucleotide similarity) to another uncultured betaproteobacterium identified from a water sample from Toolik Lake in Alaska and identified by Crump et al. (9) as a member of the Rhodoferax subclass (GenBank accession number AF534429). The remaining 366 clones were distributed among 13 different subclasses of the Betaproteobacteria, none of which were represented by more than 4% of the Betaproteobacteria clones.
Of the 476 Actinobacteria clones, 207 were most similar (98.3% nucleotide similarity) to an uncultured Actinobacterium clone (GenBank accession number AJ575501) belonging to the acI-A clade as first described by Warnecke et al. (58) and originally detected in the Řimov Reservoir, a eutrophic dimictic reservoir located in southern Bohemia. The second most abundant group (116 clones) was most similar (92.5% nucleotide similarity) to an uncultured Actinobacterium clone FukuS81 (GenBank accession number AJ290054). This clone (FukuS81) was described by Glöckner et al. (17) and originally identified from a water sample collected from Lake Fuchskuhle, a meso- to acidotrophic forest lake in the Brandeburg Mecklenburg lake district (Germany). According to the phylogenetic analysis presented by Warnecke et al. (58), this organism belongs to the acI-B cluster of the freshwater Actinobacteria. At the subclass level, both of these clones were classified as unclassified Actinomycetales using the RDP classifier tool release 9.0 (http://rdp.cme.msu.edu/classifier/classifier.jsp). The remaining 153 Actinobacteria clones were distributed among seven additional subclasses with nearest neighbors to 41 different uncultured clone sequences. However, none of these subclasses were represented by more than 16 clones, and in most cases, these subclasses were represented by three or fewer clones. GenBank accession numbers of all unique clones recovered and sequenced in this study are provided in Table 2.
Richness, diversity, and similarity of Adirondack lake bacterial communities.
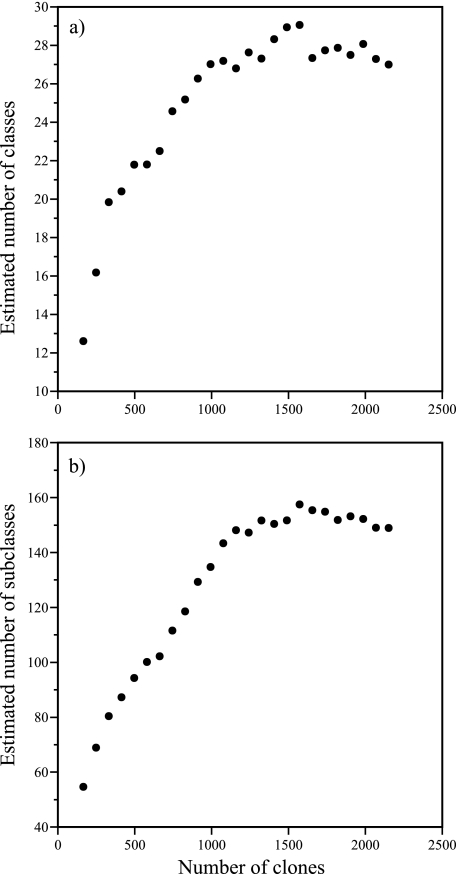
The overall richness of bacterial communities in the study lakes was estimated using the Chao2 estimator of richness (4) (Fig. 2). Total richness at the phylogenetic class and subclass levels was estimated to be 27 ± 6.5 and 149 ± 12, respectively. In both analyses, saturation of the Chao2 estimator was reached. Several other richness estimators, including the ACE, ICE, Chao1, and Jack estimators were also calculated with similar results. Richness estimates at the class level ranged from 23 to 31 and from 120 to 173 at the subclass level in these analyses.
FIG. 2.
Richness estimates of 16S rRNA gene fragment clone collections from AEAP lakes and Lake George at the phylogenetic level of class (a) and subclass (b). A total of 2,135 individual clones were classified using the RDP Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp) from 26 samples derived from 18 different Adirondack lakes. The richness estimator Chao2 was calculated as described by Chao (4) and implemented in the EstimateS software package v7.5.1 (http://purl.oclc.org/estimates). The Chao2 richness estimator was utilized without bias correction.
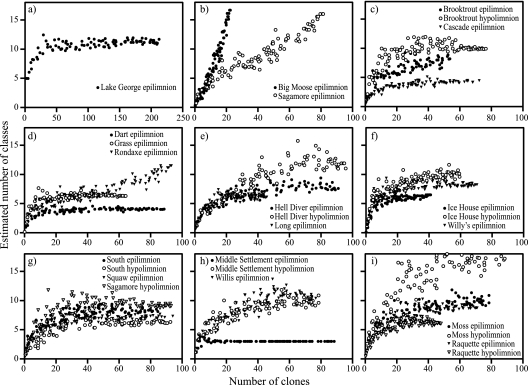
Sampling sufficiency was estimated independently in each library to determine whether libraries were statistically large enough as described by Kemp and Aller (27). At the class level, with the exception of two libraries (Big Moose Lake epilimnion and Sagamore Lake epilimnion [Fig. 3b]), each library met sampling requirements (Fig. 3a and c to i). Therefore, between-lake comparisons of bacterial community structure were possible at the class level. At the subclass phylogenetic level, most of the libraries were not sufficiently sampled, so comparisons of bacterial community structure between lakes at this level should be interpreted with caution.
FIG. 3.
Sampling sufficiency estimates of clone libraries from AEAP study lakes and Lake George. The “Large Enough” calculator (27) was used to determine whether individual clone libraries were sampled sufficiently at the class level. If the estimated phylotype richness reached an asymptote, we inferred that the library was large enough to yield a stable estimate of phylotype richness. With the exception of the libraries derived from the Big Moose Lake and Sagamore Lake epilimnion samples (b), all other samples appeared to have been sufficiently sampled.
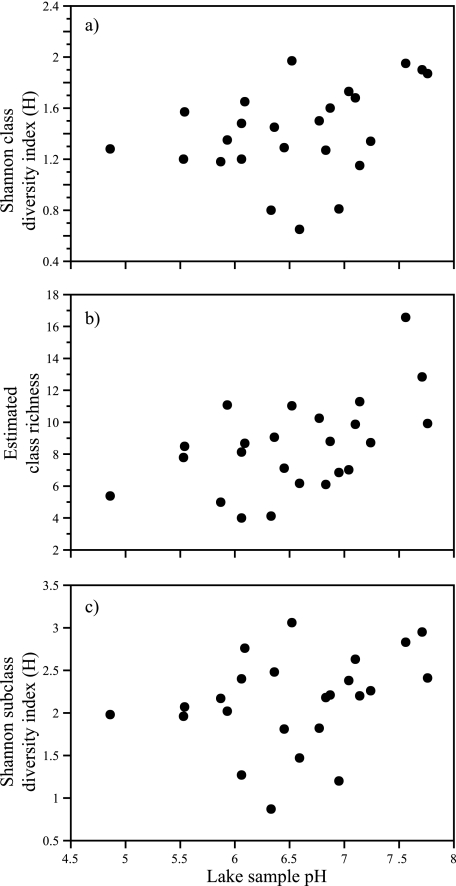
The diversity of the bacterial communities in each lake sample was estimated using the Shannon diversity index. At the class level (Fig. 4a), the Shannon diversity index ranged from 0.65 to 1.97 (average, 1.41). The highest diversity was observed in two hypolimnetic samples, Brooktrout Lake and Moss Lake. The lowest diversity was observed in the hypolimnetic sample from South Lake. At the subclass level (Fig. 4c), the Shannon diversity index ranged from 0.87 to 3.06 (average, 2.14). The highest diversity at this phylogenetic level was observed in the hypolimnion of Brooktrout Lake, and the lowest diversity was observed in the epilimnion of Dart Lake. The estimated richness (number of bacterial classes per lake sample), using the Chao2 statistic as implemented in the EstimateS package (EstimateS version 7.5 [http://purl.oclc.org/estimates]) ranged from 4 to 16.6 (Fig. 4b).
FIG. 4.
pH versus estimated diversity (a and c) and richness (b) of bacterial community composition based on 16S rRNA gene fragment clone collections from AEAP lakes and Lake George at the phylogenetic level of class (a and b) and subclass (c). Richness was not estimated at the subclass level because sampling at this level was not sufficient. Significant correlation between diversity and pH (r = 0.387, P = 0.05) (a) and between richness and pH (r = 0.5, P = 0.01) (b) were observed. Trends at the subclass level were similar, but statistical correlations were nonsignificant (r = 0.3, P = 0.13). Diversity was estimated using the Shannon index as implemented in the BIO-DAP software (http://nhsbig.inhs.uiuc.edu). Richness for each lake sample was estimated using the Chao1 statistic as implemented using the “Large Enough” estimator (27). Richness estimates for libraries derived from the epilimnion of Big Moose Lake and Sagamore Lake were omitted because analysis suggested that these libraries were insufficiently sampled. A total of 2,135 individual clones were classified using the RDP Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp) from 26 samples derived from 18 Adirondack lakes. RDP classifications were assigned on 18 May 2006 (RDP update 39).
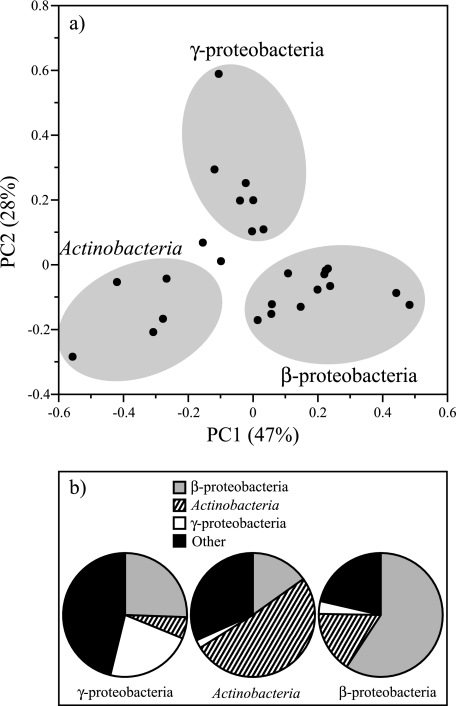
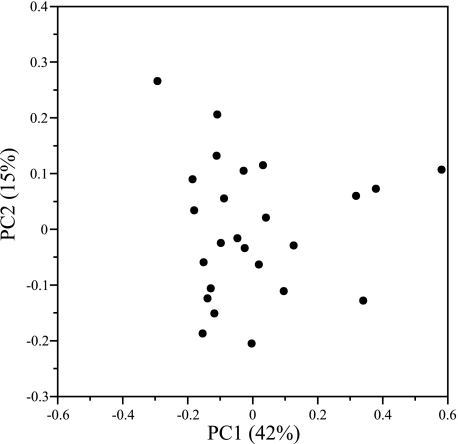
Bacterial community composition in each lake sample (with epilimnion and hypolimnion samples treated independently) was compared using PCA techniques. Comparisons based on the occurrence frequency of different classes of bacteria in each lake resolved three groups of bacterial assemblages (Fig. 5). Seventy-five percent of the total variability in the composition of the bacterial communities was explained by three classes of bacteria, the Actinobacteria, Betaproteobacteria, and Gammaproteobacteria. Twelve lake libraries were dominated by the Betaproteobacteria, including the epilimnion of Big Moose Lake (80%), Lake Rondaxe (64%), Hell Diver Pond (56%), Raquette Reservoir (56%), Middle Settlement Lake (51%), Long Lake (50%), South Lake (49%), and Sagamore Lake (48%), and hypolimnetic samples from South Lake (85%), Sagamore Lake (57%), Hell Diver Pond (57%), and Raquette Reservoir (56%). Five lake libraries were dominated by Actinobacteria, including Dart Lake epilimnion (73%), Squaw Lake epilimnion (55%), Willis Pond epilimnion (48%), Middle Settlement Lake hypolimnion (41%), and Lake George (37%). Lake samples that were distinguished by the occurrence of Gammaproteobacteria included the epilimnion of Cascade Pond (74%), Brooktrout Lake (7%), and Big Moose Lake (4%) and hypolimnetic samples from Ice House Lake (28%), Moss Lake (19%), Brooktrout Lake (7%), and Willy's Lake (4%). The bacterial assemblages found in the epilimnion of Grass and Ice House Lakes were intermediate with respect to the occurrence of these three classes of bacteria. The average distribution of the Actinobacteria, Betaproteobacteria, and Gammaproteobacteria are shown in each group of lakes discriminated by PCA is shown in Fig. 5b. Similar discrimination of bacterial communities based on classification at the subclass phylogenetic level was not apparent (Fig. 6), although 57% of the total variability could be explained by differences in the occurrence frequency of unclassified Betaproteobacteria and unclassified Actinobacteria species. Surprisingly, bacterial community assemblages observed in epilimnetic and hypolimnetic samples were not uniquely distinguishable (data not shown).
FIG. 5.
(a) Principal component analysis of bacterial community composition based on 16S rRNA gene fragment clone collections from AEAP lakes and Lake George at the phylogenetic level of class. The Actinobacteria, Betaproteobacteria, and Gammaproteobacteria accounted for significant differences (P = 0.008) between samples. A total of 75% of the variation was explained by the first two principal components (the first principal component [PC1] and the second principal component [PC2]). (b) Average distribution of the Actinobacteria, betaproteobacteria (β-proteobacteria), and gammaproteobacteria (γ-proteobacteria), and all other bacteria in each PCA-discriminated group. A total of 2,135 individual clones were classified using the RDP Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp) from 26 samples derived from 18 Adirondack lakes. RDP classifications were assigned on 18 May 2006 (RDP update 39).
FIG. 6.
Principal component analysis of bacterial community composition based on 16S rRNA gene fragment clone collections from AEAP lakes and Lake George at the phylogenetic level of subclass. At the phylogenetic level of subclass, most differences could be attributed to the unclassified betaproteobacteria and an unclassified Actinobacteria subclass, but differences were not significant (P > 0.05). The first two principal components (the first principal component [PC1]) and the second principal component [PC2]) explained 57% of the overall variation. The clone library from the epilimnion of Middle Settlement Lake was omitted from the analysis as an outlier. A total of 2,135 individual clones were classified using the RDP Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp) from 26 samples derived from 18 Adirondack lakes. RDP classifications were assigned on 18 May 2006 (RDP update 39).
Relationship between bacterial communities and acidity.
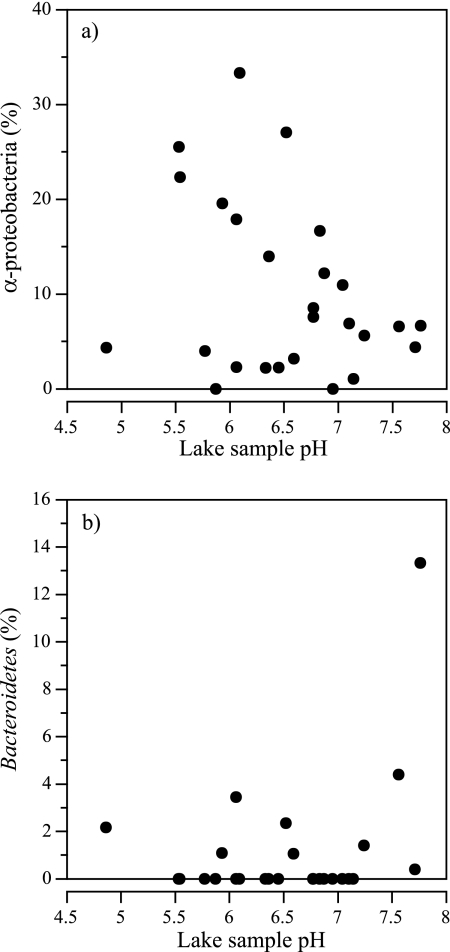
The relationship between the composition of bacterial communities and lake acidity was specifically investigated. Bacterial community richness and diversity were both positively correlated with pH, suggesting a relationship between bacterial community composition and lake acidity (Fig. 4). Significant correlation between diversity and pH (r = 0.4, P = 0.05) and richness and pH (r = 0.5, P = 0.01) were observed. Four phylogenetic classes of bacteria, including the Alphaproteobacteria (r = −0.45, P = 0.03), Bacteroidetes (r = 0.5, P = 0.018), Flavobacteria (r = 0.4, P = 0.04), and the genera incertae sedis OP10 (r = 0.45, P = 0.02) class were also significantly correlated with pH (Fig. 7) (see Table S2 in the supplemental material). Of these groups, only the Alphaproteobacteria were numerically important in the clone libraries (ca. 10%), but this class was not responsible for explaining the majority of variation of bacterial communities among the study lakes. The negative correlation between pH and the abundance of Alphaproteobacteria suggests that neutral pH selectively inhibits members of this bacterial class. The two most abundant subclasses of Alphaproteobacteria were the Caulobacter (22%) and an unclassified group of Methylocystaceae (19%). Significant correlations between pH and a specific subclass of Alphaproteobacteria were not observed (data not shown). However, because of the small sample size (219 clones in 26 samples) and the large number of Alphaproteobacteria subclasses (19), the lack of correlation is likely due to insufficient sampling. Negative findings should therefore be interpreted cautiously. Each of the other classes (Bacteroidetes, Flavobacteria, and genera incertae sedis OP10) that exhibited a positive correlation with pH was generally observed only in lake samples with pH above 7, although five Bacteroidetes clones were recovered from lake samples with pH < 7, including one clone from Long Lake with a pH of 4.86. However, since so few of these classes were recovered, the significance of these correlations are uncertain.
FIG. 7.
Relationship between the percentage of Alphaproteobacteria and Bacteroidetes in each lake clone library and pH. The percentage of 16S rRNA gene clones in each lake sample clone library is shown. (a) Relationship between the percentage of alphaproteobacteria (α-proteobacteria) and pH (r = −0.78, P = 0.0002); (b) relationship between the percentage of Bacteroidetes and pH (r = 0.48, P = 0.02).
Relationship between bacterial communities and other physiochemical characteristics.
Other investigators have reported significant relationships between the structure of bacterial communities and a variety of limnological properties, including hydraulic retention time, nutrient concentration, and pH (28, 30, 63). Table S2 in the supplemental material shows the significant correlations (P ≤ 0.05) of bacterial classes with the physiochemical properties of the Adirondack lakes in this study. Of the most abundant bacterial classes, the Alphaproteobacteria, Actinobacteria, and Betaproteobacteria classes were significantly correlated with lake physiochemical properties. In addition to these properties, the abundance (proportion) of Alphaproteobacteria was also significantly correlated with total phosphorus (r = 0.5, P = 0.01), molybdate-reactive phosphorus (MRP) (r = 0.6, P = 0.03), chlorophyll a (r = 0.6, P = 0.0007), total monomeric aluminum (r = 0.4, P = 0.05), and nonlabile monomeric aluminum (r = 0.4, P = 0.04). The abundance of Actinobacteria was positively correlated to MRP (r = 0.4, P = 0.01). The abundance of Betaproteobacteria was negatively correlated (r = −0.5, P = 0.05) with lake hydraulic retention times. However, since most of the lakes had relatively short retention times, it is difficult to interpret these results. Second to properties directly related to acidity (pH and acid-neutralizing capacity), the concentrations of reactive phosphorus appeared to most significantly explain the abundance and distribution of the Actinobacteria and Alphaproteobacteria within the Adirondack lakes. In the complete data set, there were 81 significant correlations between lake physiochemical properties and the abundance of specific groups of bacteria (see Table S2 in the supplemental material), but the full exploration of these correlations are beyond the scope of this report.
DISCUSSION
In this study we investigated the relationship between the composition, richness, diversity, and community structure of bacterioplankton communities with 31 physical and chemical parameters in 18 lakes, ponds, and reservoirs in the Adirondack mountain region in New York State in the United States. During the past several decades, many of the lakes and ponds in the Adirondacks, particularly in the southwestern region, have been impacted by anthropogenic acidic deposition. While impacts of lake acidification on a variety of aquatic biota have been well documented in the Adirondacks (21, 44, 52), relatively little is known concerning the response of prokaryotic communities. Comparison of bacterioplankton communities between a relatively large number of ponds and lakes with pHs spanning 4.86 to 7.76 provided a unique opportunity to specifically investigate the influence of acid deposition on the freshwater prokaryotic communities.
Bacterial communities in the Adirondack lake set were, in many ways, similar to bacterioplankton communities described in other freshwater systems. Communities were dominated by “typical” freshwater assemblages consisting of Betaproteobacteria and Actinobacteria lineages (54, 55, 60). The dominant groups of these bacteria observed in the Adirondack lakes have been previously described and phylogenetically classified, and some have even been isolated (19, 33, 38, 58). In 15 of the 26 lake samples examined, the Betaproteobacteria were the most abundant group with the largest number of clones (291 clones) belonging to the P. necessarius cluster. Of these clones, 152 (52%) were classified within the PnecC subcluster. The most abundant group within the Actinobacteria was the Actinomycetales subclass belonging to the acI-A clade (391 of 476 clones) as originally described by Warnecke et al. (58). Interestingly, seven lake samples were distinguished by the abundance of Gammaproteobacteria and in two samples (Cascade Pond epilimnion and Ice House Lake hypolimnion), Gammaproteobacteria were the most dominant group. To the best of our knowledge, this is the first report of the dominance of Gammaproteobacteria in any freshwater lake, although Gammaproteobacteria are common in marine systems (16). Two clone libraries (Brooktrout Lake epilimnion and hypolimnion) were dominated by representatives of the Gammaproteobacteria. Dominance of the Alphaproteobacteria is also more typical in marine systems (18, 40), but unlike marine systems that are dominated by the SAR and Roseobacter clades of the Alphaproteobacteria, the Alphaproteobacteria most commonly observed in the Adirondack lakes belonged to representatives of the Caulobacter (22%), Methylocystaceae (19%), and Rhizobiales (14%) taxa.
Several recent studies have investigated the relationship between environmental factors and the composition of bacterioplankton in freshwater lakes and rivers based on direct analysis of amplified rRNA gene and rRNA gene clone libraries (64). These studies have examined a variety of lake types from the tropics to polar regions and have included lakes of various sizes, hydrological types, and trophic status, but only a few have compared communities between large suites of lakes (30, 31, 62). Although these studies included acidity (pH) as a variable, none of them were specifically designed to investigate the influence of pH on bacterioplankton communities. On the basis of these investigations, it has been reported that lake size and hydraulic retention time (30, 31) and regional/landscape factors, including location and hydrological type (32, 62), appear to be primary variables responsible for shaping the composition of freshwater prokaryotic communities.
The association of pH with distribution of freshwater bacterial groups has also been reported (31, 50, 55, 62). Lindström et al. (31) examined 15 diverse lakes in northern Europe and identified that the pH, temperature, and hydraulic retention time were most strongly related to variations in the distribution of bacterial taxa. Similarly, in a study of four shallow eutrophic lakes in Belgium, a significant relationship between pH and the composition of lake bacterioplankton communities was reported (54).
In the Adirondack lakes, the diversity and richness of bacterial communities were significantly correlated with pH, mirroring similar relationships observed between higher organisms and pH in lakes (12, 23, 43, 46). However, pH was not significantly correlated with the overall structure of bacterioplankton communities as investigated by PCA, since the most dominant groups of bacteria (Betaproteobacteria and Actinobacteria) were not correlated with pH. Although these most abundant bacterial classes were not correlated with pH, the Alphaproteobacteria and Bacteroidetes taxa were. In the study of Lindström et al. (31), a relationship between these groups and pH was not reported, but they observed that two actinobacterial groups were correlated with pH. The ACK-M1 group was associated with high pH; Sta2-30 was associated with low pH. A third actinobacterial group (Urk-014) was not correlated with pH. These groups correspond to the acI-A, acI-B, and acIV actinobacterial lineages described by Warnecke et al. (58), respectively. In the present study, in which we recovered 476 Actinobacteria clones, the acI-A and acI-B groups exhibited essentially the same distribution among the lakes and did not correlate with pH. Lindström et al. (31) also reported an association between low pH and betaproteobacteria belonging to the P. necessarius group, which was not observed in the present study.
Possible relationships between other physiochemical parameters and bacterial communities were also examined. Significant correlations between the abundance of different bacterial classes and 26 of the 31 parameters examined were observed (see Table S2 in the supplemental material). Of the more abundant taxa, the Alphaproteobacteria, Bacteroidetes, and Verrucomicrobiae were correlated with a number of lake parameters other than acidity. The Bacteroidetes (largely composed of the unclassified Rikenellaceae subclass) was significantly correlated with 12 parameters (pH, dissolved oxygen, temperature, acid-neutralizing capacity, NH4, dissolved inorganic carbon, SO4, total P, MRP, total filterable P, total N, and Fe). The Adirondack Bacteroidetes clones were most similar to bacteria recovered from other acidic environments, including Lake Fuchskule, a mesotrophic acidotrophic forest lake (17). This group of organisms may be of particular interest for investigating bacteria associated with aquatic systems with low pH. The Verrucomicrobiae taxon was also significantly correlated with four parameters (water clarity, DOC, TFP, and total aluminum). This group of organisms has been reported from a wide variety of environments (10), including lakes and rivers, acidic forest soils, Antarctic sediments, and manure leakage, but little is known about the physiology of the Verrucomicrobiae.
The roles of regional and landscape factors have been suggested to be the most important factors influencing the composition of freshwater bacterial communities (31, 32, 62). Specifically, the influence of geographic region and the landscape level factors of lake type (seepage versus drainage) and hydraulic retention time. In this study, the lakes examined were within close proximity to each other and were predominantly drainage lakes; therefore, it was not possible to investigate regional differences or lake hydrological type. However, the most dominant group of bacteria observed in the Adirondack lakes, the Betaproteobacteria, was correlated with hydraulic retention time, supporting previous reports.
The correlation of some bacterial groups, particularly the Alphaproteobacteria, may be useful as biological indicators of recovery from acidification in Adirondack lakes. Initially, it was hypothesized that because of their short generation times and small size, bacterioplankton might be excellent as rapid biological indicators of recovery from acidification. However, relatively weak and inconsistent correlations between bacterial community and lake physiochemical properties suggest that bacterioplankton communities are indirectly rather than directly influenced by acid deposition. For example, bacterial growth rates may be influenced by alterations in populations of primary producer communities and chemical effects on nutrient cycling that affect the availability of dissolved organic and inorganic nutrients (13). Alternatively, the composition of bacterial communities may be selectively controlled by the composition of bactivore communities (2). If this were the case, then the restoration of Adirondack bacterioplankton communities to conditions that resemble similar but nonimpacted freshwater lakes might be interpreted as an end point metric in lake recovery rather than an early indicator of ongoing biological recovery.
Preliminary comparisons between a small data set collected in 1994 (22, 33, 34) and the 2002 studies described here (comparison of five lakes) are potentially useful for this purpose. Over this 8-year period, the AEAP study lakes experienced an average pH increase of 0.6 ± 0.3, while the pH of Lake George remained circumneutral (S. A. Nierzwicki-Bauer, unpublished). Although a robust analysis between these data sets is not possible because of methodological differences and the small sample size of the 1994 data set, several trends were observed. Bacterioplankton communities in the AEAP study lakes changed significantly from 1994 to 2002, while those in Lake George did not. Further, in the AEAP lake set, the abundance of Alphaproteobacteria changed, although this change was not significantly correlated with acidity. Interestingly, a significant negative correlation (r = −0.4, P = 0.02) between the abundance of Gammaproteobacteria and pH was observed in the lakes studied in 1994 and 2002 (data not shown), which was not suggested by examination of the 2002 data alone (18 study lakes). The significance of these preliminary observations with respect to recovery from acidification is not yet clear, but ongoing larger-scale studies are under way to explore this process.
Although it is difficult to reach conclusions regarding the functional significance or mechanisms responsible for relationships between acidity and the composition of bacterioplankton communities based only on ribosomal markers, the results of this study indicate that at least some groups of freshwater bacterioplankton are influenced by parameters associated with acid deposition. We observed that it was not the most abundant groups of bacterioplankton that were affected by pH, but several less dominant groups. In the future, it will be important to investigate the physiological and functional roles of these organisms to make predictions concerning potential biogeochemical and ecological significance. Perhaps in freshwater environments, the Betaproteobacteria and Actinobacteria can be considered ecological generalists, while other groups are more specialized. If this is the case, loss or gain of specialized communities may be of importance to particular biogeochemical or other processes.
Supplementary Material
Acknowledgments
This study is part of the Adirondack Effects Assessment Program (AEAP) that is supported by a grant from the U.S. Environmental Protection Agency (contract number 68D20171) to S. Nierzwicki-Bauer and C. Boylen at the Rensselaer Polytechnic Institute. Additional support was provided by the Helen V. Froehlich Foundation.
Although this work has been funded by the U.S. Environmental Protection Agency, it has not been subjected to the agency's review and, therefore, does not necessarily reflect the views of the agency, and no official endorsement should be inferred.
We are greatly indebted to our colleagues in the AEAP that have made significant contributions to the collection, analysis and/or interpretation of samples and results. In particular, Robert Bombard, Larry Eichler, Laurie Ahrens-Franklin, James Harrison, Jim Sutherland, Alexey Vepritskiy, and David Winkler made significant contributions. We also gratefully acknowledge the outstanding contributions of numerous undergraduate students in carrying out the molecular studies. Katie Opper, Mike Schaaff, Christy Stagnar, and Claudia Weigl particularly made outstanding contributions. Anna Boyette (Skidaway Institute of Oceanography) prepared the figures. We thank the useful and constructive comments of two anonymous reviewers.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Balkwill, D. L., R. H. Reeves, G. R. Drake, J. Y. Reeves, F. H. Crocker, M. B. King, and D. R. Boone. 1997. Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol. Rev. 20:201-216. [DOI] [PubMed] [Google Scholar]
- 2.Boenigk, J., P. Stadler, A. Wiedlroither, and M. W. Hahn. 2004. Strain-specific differences in the grazing sensitivities of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl. Environ. Microbiol. 70:5787-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson, R. E. 1977. A trophic state index for lakes. Limnol. Oceanogr. 22:361-369. [Google Scholar]
- 4.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 5.Chao, A., R. L. Chazdon, R. K. Colwell, and T. J. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159. [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 7.Cottrell, M. T., L. A. Waidner, L. Y. Yu, and D. L. Kirchman. 2005. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ. Microbiol. 7:1883-1895. [DOI] [PubMed] [Google Scholar]
- 8.Crump, B. C., and J. A. Baross. 2000. Archaeaplankton in the Columbia River, its estuary and the adjacent coastal ocean, USA. FEMS Microbiol. Ecol. 31:231-239. [DOI] [PubMed] [Google Scholar]
- 9.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driscoll, C. T., K. M. Driscoll, K. M. Roy, and M. J. Mitchell. 2003. Chemical response of lakes in the Adirondack region of New York to declines in acidic deposition. Environ. Sci. Technol. 37:2036-2042. [DOI] [PubMed] [Google Scholar]
- 12.Findlay, D. L. 2003. Response of phytoplankton communities to acidification and recovery in Killarney Park and the Experimental Lakes Area, Ontario. Ambio 32:190-195. [DOI] [PubMed] [Google Scholar]
- 13.Findlay, S. E. G., R. L. Sinsabaugh, W. V. Sobczak, and M. Hoostal. 2003. Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnol. Oceanogr. 48:1608-1617. [Google Scholar]
- 14.Frischer, M. E., B. R. McGrath, A. S. Hansen, P. A. Vescio, J. A. Wyllie, J. Wimbush, and S. A. Nierzwicki-Bauer. 2005. Introduction pathways, differential survival of adult and larval zebra mussels (Dreissena polymorpha), and possible management strategies, in an Adirondack lake, Lake George, NY. Lake Reservoir Manag. 21:391-402. [Google Scholar]
- 15.Garrity, G., J. A. Bell, and T. G. Lilburn. 2004. Taxonomic outline of the prokaryotes, p. 1-399. In G. M. Garrity et al. (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer-Verlag, New York, NY.
- 16.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haines, T. A. 1981. Acidic precipitation and its consequences for aquatic ecosystems—a review. Trans. Am. Fish. Soc. 110:669-707. [Google Scholar]
- 22.Hiorns, W. D., B. A. Methé, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornstrom, E. 2002. Phytoplankton in 63 limed lakes in comparison with the distribution in 500 untreated lakes with varying pH. Hydrobiologia 470:115-126. [Google Scholar]
- 24.Jeffries, D. S., T. A. Clair, S. Couture, P. J. Dillon, J. Dupont, W. Keller, D. K. McNicol, M. A. Turner, R. Vet, and R. Weeber. 2003. Assessing the recovery of lakes in southeastern Canada from the effects of acidic deposition. Ambio 32:176-182. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins, J., K. Roy, C. T. Driscoll, and C. Buerkett. 2005. Acid rain and the Adirondacks: a research summary. Adirondack Lakes Survey Corporation, Ray Brook, NY. http://www.adirondacklakessurvey.org/sosindex.htm.
- 26.Jurgens, G., F. O. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 27.Kemp, P. F., and J. Y. Aller. 2004. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol. Oceanogr. Methods 2:114-125. [Google Scholar]
- 28.Lindstrom, E. S., and A. K. Bergstrom. 2005. Community composition of bacterioplankton and cell transport in lakes in two different drainage areas. Aquat. Sci. 67:210-219. [Google Scholar]
- 29.Lindstrom, E. S., and A. K. Bergstrom. 2004. Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol. Oceanogr. 49:125-136. [Google Scholar]
- 30.Lindstrom, E. S., M. Forslund, G. Algesten, and A. K. Bergstrom. 2006. External control of bacterial community structure in lakes. Limnol. Oceanogr. 51:339-342. [Google Scholar]
- 31.Lindström, E. S., M. P. Kamst-Van Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 71:8201-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindstrom, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Methe, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 34.Methe, B. A., and J. P. Zehr. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77-96. [Google Scholar]
- 35.Momen, B., L. W. Eichler, C. W. Boylen, and J. P. Zehr. 1996. Application of multivariate statistics in detecting temporal and spatial patterns of water chemistry in Lake George, New York. Ecol. Model. 91:183-192. [Google Scholar]
- 36.Nelson, R. K., N. M. Frew, N. Wizell, T. Thwaites, and C. G. Johnson. 2002. SCIMS—a semi-autonomous system for sampling and extraction of surfactants in the sea-surface microlayer, abstr. OS12A-120. Eos Trans. AGU, Oceans Sci. Meet. Suppl., vol. 83. American Geophysical Union, Washington, DC.
- 37.Newton, R. M., and C. T. Driscoll. 1990. Classification of ALSC lakes, p. 2-70 to 2-91. Adirondack Lake Survey: an interpretive analysis of fish communities and water chemistry 1984-1987. Adirondack Lakes Survey Corporation, Ray Brook, NY.
- 38.Page, K. A., S. A. Connon, and S. J. Giovannoni. 2004. Representative freshwater bacterioplankton isolated from Crater Lake, Oregon. Appl. Environ. Microbiol. 70:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernthaler, J., F. O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rappe, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 41.Reheinheimer, G. 1992. Aquatic microbiology, 4th ed. John Wiley and Sons, New York, NY.
- 42.Richards, T. A., A. A. Vepritskiy, D. E. Gouliamova, and S. A. Nierzwicki-Bauer. 2005. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ. Microbiol. 7:1413-1425. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, D. A., R. Singer, and C. W. Boylen. 1985. The submersed macrophyte communities of Adirondack lakes (New-York, USA) of varying degrees of acidity. Aquat. Bot. 21:219-235. [Google Scholar]
- 44.Schindler, D. W. 1988. Effects of acid-rain on fresh-water ecosystems. Science 239:149-157. [DOI] [PubMed] [Google Scholar]
- 45.Shuster, E. L., R. G. LaFleur, and C. W. Boylen. 1994. The hydrologic budget of Lake George, Southeastern Adirondack mountains of New York. Northeast. Geol. 16:94-108. [Google Scholar]
- 46.Siegfried, C. A., J. A. Bloomfield, and J. W. Sutherland. 1989. Acidity status and phytoplankton species richness, standing crop, and community composition in Adirondack, New-York, USA lakes. Hydrobiologia 175:13-32. [Google Scholar]
- 47.Simek, K., K. Hornak, J. Jezbera, J. Nedoma, J. Vrba, V. Straskrabova, M. Macek, J. R. Dolan, and M. W. Hahn. 2006. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ. Microbiol. 8:1613-1624. [DOI] [PubMed] [Google Scholar]
- 48.Skjelkvale, B. L., C. Evans, T. Larssen, A. Hindar, and G. G. Raddum. 2003. Recovery from acidification in European surface waters: a view to the future. Ambio 32:170-175. [DOI] [PubMed] [Google Scholar]
- 49.Skjelkvale, B. L., J. L. Stoddard, D. S. Jeffries, K. Torseth, T. Hogasen, J. Bowman, J. Mannio, D. T. Monteith, R. Mosello, M. Rogora, D. Rzychon, J. Vesely, J. Wieting, A. Wilander, and A. Worsztynowicz. 2005. Regional scale evidence for improvements in surface water chemistry 1990-2001. Environ. Pollut. 137:165-176. [DOI] [PubMed] [Google Scholar]
- 50.Stepanauskas, R., M. A. Moran, B. A. Bergamaschi, and J. T. Hollibaugh. 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat. Microb. Ecol. 31:85-98. [Google Scholar]
- 51.Stoddard, J. L., D. S. Jeffries, A. Lukewille, T. A. Clair, P. J. Dillon, C. T. Driscoll, M. Forsius, M. Johannessen, J. S. Kahl, J. H. Kellogg, A. Kemp, J. Mannio, D. T. Monteith, P. S. Murdoch, S. Patrick, A. Rebsdorf, B. L. Skjelkvale, M. P. Stainton, T. Traaen, H. van Dam, K. E. Webster, J. Wieting, and A. Wilander. 1999. Regional trends in aquatic recovery from acidification in North America and Europe. Nature 401:575-578. [Google Scholar]
- 52.Sullivan, T. J. 2000. Aquatic effects of acid deposition. CRC Press, Boca Raton, FL.
- 53.Urbach, E., K. L. Vergin, G. L. Larson, and S. J. Giovannoni. 2007. Bacterioplankton communities of Crater Lake, OR: dynamic changes with euphotic zone food web structure and stable deep water populations. Hydrobiologia 574:161-177. [Google Scholar]
- 54.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 55.Van der Gucht, K., T. Vandekerckhove, N. Vloemans, S. Cousin, K. Muylaert, K. Sabbe, M. Gillis, S. Declerk, L. De Meester, and W. Vyverman. 2005. Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microbiol. Ecol. 53:205-220. [DOI] [PubMed] [Google Scholar]
- 56.Vergin, K. L., M. S. Rappe, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-944. [DOI] [PubMed] [Google Scholar]
- 57.Warby, R. A. F., C. E. Johnson, and C. T. Driscoll. 2005. Chemical recovery of surface waters across the northeastern United States from reduced inputs of acidic deposition: 1984-2001. Environ. Sci. Technol. 39:6548-6554. [DOI] [PubMed] [Google Scholar]
- 58.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Q. L., and M. W. Hahn. 2006. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol. Ecol. 57:67-79. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Q. L., and M. W. Hahn. 2006. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol. 8:1660-1666. [DOI] [PubMed] [Google Scholar]
- 61.Yannarell, A. C., A. D. Kent, G. H. Lauster, T. K. Kratz, and E. W. Triplett. 2003. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391-405. [DOI] [PubMed] [Google Scholar]
- 62.Yannarell, A. C., and E. W. Triplett. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.