Abstract
Since heterotrophic prokaryotes play an important biogeochemical role in aquatic ecosystems and have a high capacity to survive in extreme environments, easy-to-perform protocols that probe their physiological states and the effects of environmental variables on those states are highly desired. Some methodologies combine a general nucleic acid stain with a membrane integrity probe. We calibrated one of these, the nucleic acid double-staining (NADS) protocol (G. Grégori, S. Citterio, A. Ghiani, M. Labra, S. Sgorbati, S. Brown, and M. Denis, Appl. Environ. Microbiol. 67:4662-4670, 2001), determining the optimal stain concentrations in seawater and the response to conditions that generate prokaryote death (such as heat) and to conditions that are known to produce death in plankton, such as nutrient limitation or flagellate grazing. The protocol was validated by comparison to two methods used to detect viability: active respiration by 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) and incorporation of tritiated leucine. We show that concentrations in the range of 5 to 20 μg ml−1 of propidium iodide, simultaneous to a 10× concentration of Sybr green I, are best for detecting two separated populations of “live” (green cells) and “dead” (red cells) organisms. During exposure to heat and UVC, we observed that the number of live cells declined concurrently with that of actively respiring cells (CTC positive) and with total leucine incorporation. In seawater mesocosms, the NADS protocol allowed detection of bacterioplankton starvation-related death and flagellate predation. The protocol was also tested in deep profiles in the northwest Atlantic, demonstrating its potential for routine characterization of this fraction of the physiological diversity of marine heterotrophic prokaryotic plankton.
Heterotrophic prokaryotes (Bacteria and Archaea) play a relevant role in planktonic marine microbial food webs (e.g., reference 5). As a relevant difference from other planktonic organisms, prokaryotes are known to sometimes be inactive or dead, but in the way that they are commonly enumerated (e.g., with DAPI [4′,6′-diamidino-2-phenylindole]), these cells are accounted for in budgets for prokaryote biomass since they are not distinguished from live and active cells.
From an ecological point of view, at least three cell categories within microbial communities can be distinguished: the viable and active cells, which play a functional role and participate in biomass production at the time of sampling; the live but inactive cells (often called dormant cells), which do not participate in production at the time of sampling but have the potential for doing so; and the dead and therefore inactive cells, which do not have a role in the cycling of chemical elements, even though they might be retaining nutrients. The discrimination of these cellular categories is not without dispute: for example, the “active” term is often confounded with the “live” one and the “inactive” with the “dead” one (26). Moreover, different intrinsic levels of metabolic activity and vital states must be considered (21, 22, 62) and the question is not whether a prokaryote is active or not but how active it is. Recent studies have reported that marine bacterial cells are not uniformly active, and subsets of the assemblage appear to be more or less active at any one time (e.g., references 16, 18, 21, 22, 36, 43, and 62).
All this results in a major set of questions in microbial ecology: which members of the bacterial assemblages are responsible for the overall activity and/or productivity, and which members are inactive? There is interest in determining the affiliations of active and live cells (by, e.g., microautoradiography combined with fluorescence in situ hybridization [FISH]) (17) and interest in determining the numbers of cells in each physiological state because, for example, bulk ecosystem properties scaled to different numbers of highly participating organisms produce different values of cell-specific activity (22). In fact, by themselves, cell-specific production values (or specific growth rates) might be uninformative unless some knowledge of the number of participating cells exists.
The discrimination of active organisms among the live ones is done with consideration of some aspect of their physiological statuses, such as reproductive potential (55), respiratory activity (60), enzymatic activities, etc. Other aspects of the cell structure and physiology are used to discriminate dead cells from cells with at least potential for growth. Membrane integrity is one of the better-accepted criteria for distinguishing live cells from dead cells (22, 29, 32, 35) because of the assumption that cells with damaged membranes cannot sustain any electrochemical gradient and are not able to resume growth. The paradox in which a method classifies a given cell as dead because of a lack of membrane potential but the same cell still has enough ribosomes to be detected by FISH or has enough internal enzymes to be detected as active with other methods might exist (e.g., reference 67).
Different protocols and methodologies have been proposed to detect membrane damage in bacterioplankton cells. Apart from direct inspection by transmission electron microscopy (32), membrane polarity and integrity have been measured with probes such as ethidium bromide, calcofluor white, oxonols, carbocyanines, TO-PRO, Sytox Green, etc. (21, 39, 45, 48, 59). The most popular probe is probably propidium iodide (PI) (e.g., reference 67). Most of these stains are nucleic acid-labeling molecules that are either sized so that they are not incorporated through a healthy membrane but cross the membrane when it is damaged or charged so that they do not cross a negatively charged membrane. Lack of membrane permeability to large molecules is one of the characteristics of membrane integrity.
Often, the only evidence that a method might work when applied to plankton prokaryotes is given by laboratory tests with laboratory cultures, in combination with typical laboratory sources of mortality. The abiotic methods used to generate “death” in the laboratory (heat, irradiation, antibiotics, and osmotic and chemical stress) (7) may damage or kill the bacterial cells in a way that is fundamentally different from the way in which cells die in nature (i.e., viral lysis, protozoan grazing, cell aging, nutrient starvation, etc.) (10, 40, 47), generating disagreement between researchers on how to translate typical mortality laboratory experiments to field situations. In addition, there is a lack of a standard, undisputed way of determining the performance of newly developed methodologies as applied to natural prokaryotic communities. Total leucine incorporation is one universal tracer of heterotrophic activity that could play this role, but this method is seldom used as such.
Recently, several double-staining methodologies that combine PI with a generic nucleic acid stain have been developed. Among these, the Live/Dead BacLight viability kit, developed by Molecular Probes, and the nucleic acid double-staining (NADS) protocol have been applied in various areas of bacteriological research (12, 23, 26, 33). The latter methodology has an advantage in that it uses a stain (Sybr green [SG]) that is also used to enumerate total bacterial abundance (46) and can be bought separately, thus making this methodology cheaper and more flexible than the use of closed commercial kits. This protocol has been used to evaluate cultures (6, 57), freshwater and marine bacterioplankton (1, 29), and activated sludge samples (24, 70). All these double-staining protocols are based on the simultaneous use of two stains targeting the nucleic acid, the first (Syto9, SG, etc.) being a membrane-permeable green dye (31) and the second (PI) being a membrane-impermeant dye (34). The principle of these protocols is based on energy transfer from an excited donor (the green stain) to an acceptor molecule (the red stain) according to the fluorescence resonance energy transfer (FRET) phenomenon: the green stain is quenched in the presence of the red stain, and energy is transferred to the latter one if present.
This methodology has been used in field studies, but with different, nonstandardized protocol details (Table 1). Furthermore, it has not been tested in field-realistic death situations and has not been formally compared to other undisputed activity measurements, such as leucine incorporation. Here, we establish a working protocol for application of the NADS method and compare its performance in cases where bacterioplankton is faced with typical laboratory death-causing factors and with naturally occurring sources of mortality. We also test the performance of the methodology as applied to vertical profiles down to the mesopelagic zone in the central Atlantic Ocean. Our results show that when using a standardized protocol, the NADS method can be used to trace natural heterotrophic prokaryote mortality and can inform us of its temporal and spatial variations.
TABLE 1.
Characteristics of the different versions of the NADS protocol found in the literaturea
| Type of sample | Stain 1 | Concn (×) | Dilution (vol/vol) | Stain 2 | Concn (μg ml−1) | Note | Reference |
|---|---|---|---|---|---|---|---|
| Freshwater | SG1 | 0.1 | 1:100,000 | PI | 1 | For fewer than 0.5 × 106 bacteria ml−1 | 29 |
| Freshwater | SG1 | 1 | 1:10,000 | PI | 10 | For up to 20 × 106 bacteria ml−1 | 29 |
| Seawater | SG2 | 10 | 1:1,000 | PI | 10 | 29 | |
| Seawater | SG2 | 10 | 1:1,000 | PI | 10 | 30 | |
| Seawater | SG1 | 1 | 1:10,000 | PI | 1-40 | Range of PI studied | 44 |
| Drinking water | SG2 | 5 | 1:2,000 | PI | 10 | 56 | |
| Seawater | SG1 | 25 | 1:400 | PI | 0.6 | 58 | |
| Seawater | SG1 | 1 | 1:10,000 | PI | 10 | 1 | |
| Freshwater | SG | 10 | 1:1,000 | PI | 2 | Does not specify which SG | 9 |
SG2, Sybr green II.
MATERIALS AND METHODS
Sampling sites.
Sampling for all analyses was carried out in the Blanes Bay Microbial Observatory, in the Mediterranean Sea, ca. ∼800 m offshore (41°39.90′N, 2°48.03′E). Surface water was directly withdrawn with a bucket, filtered through a 200-μm mesh net, dispensed into 25-liter polycarbonate carboys, and transported to the laboratory under dim light (within 1.5 h). Data for the Atlantic cruise were collected from on board the R/V Hespérides vessel in the northwest Atlantic during cruise COCA-2. Details about sampling, prokaryote activity, and abundance determinations can be found in Arístegui et al. (4) and Alonso-Sáez et al. (2) but do not significantly differ from the protocols used at Blanes Bay (see below).
Basic experimental design.
Natural bacterioplankton (we use this term to indicate all apparently heterotrophic planktonic prokaryotes, bacteria, or archaea) communities were exposed to factors that induce cell stress and death, including exposure to heat, UVC, and antibiotics. For heat, 10-ml tubes containing bacteria were immersed for 1, 5, or 15 min in a water bath maintained at 70°C. For UVC, 100-ml seawater samples were exposed to UV radiations in spherical quartz glass bottles for 10, 20, and 40 min at a 20-cm distance from a Philips UV G30T8 30-W lamp. For antibiotics, we added an antibiotic mixture at the following final concentrations: 0.3 mg ml−1 penicillin G (Sigma), 0.5 mg ml−1 streptomycin (Bristol-Myers Squibb), 0.2 mg ml−1 gentamicin (Essex, Italy), and 5 μg ml−1 amphotericin B (Sigma). Incubation was performed at room temperature in the dark.
Additionally, we established two replicated seawater microcosms prepared in 1.5-liter polycarbonate bottles to evaluate the effects of natural predation and starvation on bacterial viability as measured by the NADS protocol. One set of microcosms was constructed with seawater filtered through a 3-μm filter (polycarbonate membrane; Millipore), with the aim of freeing heterotrophic nanoflagellates (HNF) from predation and increasing their effect on bacteria. The other microcosms were filled with seawater filtered through a <0.8-μm filter (polycarbonate membrane; Millipore). We expected bacteria to develop in these microcosms until they ran out of some essential nutrient and entered a nutrient limitation status. The microcosms were started within 2 h after sampling and incubated at room temperature for 7 days. Subsamples were taken daily from the microcosms for bacterial and HNF analysis.
Flow cytometry.
Bacteria were analyzed with a FACSCalibur flow cytometer (Becton Dickinson) equipped with a blue (488-nm) argon laser set at 15 mV. All fluorescent filters and detectors used were the standard machine equipment, with green fluorescence collected in the FL1 channel, orange fluorescence collected in the FL2 channel, and red fluorescence collected in the FL3 channel (>670 nm). All parameters were collected as logarithmic signals. We generally acquired data at a low speed (ca. 15 μl min−1), and the sample concentration was adjusted to keep the count lower than 1,000 events s−1 with MilliQ water. A daily calibrated fluorescent microsphere (yellow-green 0.92-μm Polysciences latex beads [106 ml−1]) solution was added to each sample as an internal reference. Population concentration was estimated with CellQuest and PaintAGate software (Becton Dickinson, Palo Alto, CA).
NADS protocol.
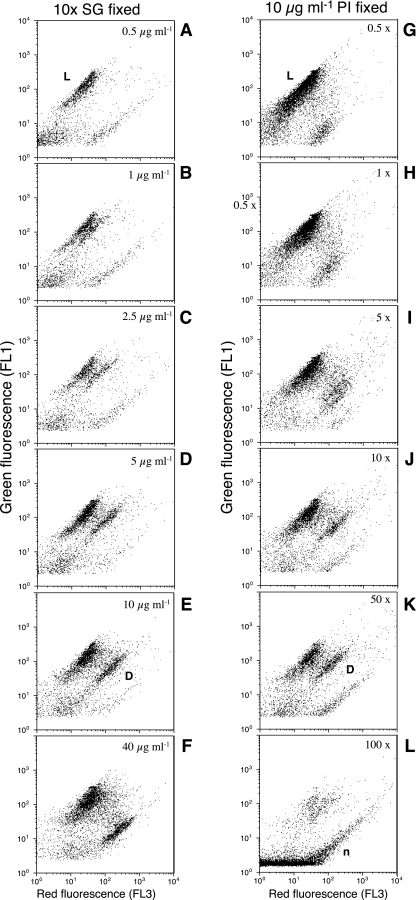
The NADS viability protocol is based on the combination of the cell-permanent nucleic acid stain Sybr green I (SG1; Molecular Probes, Eugene, OR) and the cell-impermeant PI (Sigma Chemical Co.) fluorescent probe. They both stain RNA and DNA (31). Fluorochrome concentrations were optimized by testing a range of PI concentrations (0.25, 0.5, 1, 2.5, 5, 10, 20, and 40 μg ml−1) with respect to a fixed SG1 concentration (10×, a 1,000-fold dilution of the commercial stock in dimethyl sulfoxide) and a range of SG1 concentrations (0.25×, 0.5×, 1×, 5×, 10×, 50×, 100×, and 200×) with respect to a fixed PI concentration (10 μg ml−1) on seawater samples that had received different treatments (1-min and 5-min 70°C treatments). With the experiments reported in Fig. 1 and 2, we determined the optimal concentrations, 1:10× SG1 and 10 μg ml−1 PI, which were thereafter used in all other experiments. Note that SG1 comes with a stock concentration of 10,000× in dimethyl sulfoxide. A 1,000-fold dilution of this stock corresponds to a concentration of 10×. After simultaneous addition of each stain, the samples were incubated for 20 min in the dark at room temperature and then analyzed flow cytometrically. Initial tests showed the population structure to be stable after 10 min of incubation and to remain for at least 30 min (details not shown). SG1 and PI fluorescence were detected in the green (FL1) and orange-red (FL3) cytometric channels, respectively. A dot plot of red versus green fluorescence allowed distinction of the “live” cell cluster (i.e., cells with intact membranes and DNA present) from the “dead” cell one (i.e., with compromised membranes) (Fig. 1). A plot of right-angle scatter versus green fluorescence is useful for distinguishing beads from cells and also helps in differentiating photosynthetic prokaryotes, which might appear in between the “live” and “dead” clusters of the FL1-FL3 plot. We used a threshold in the green channel and ran the samples at a low speed (ca. 15 μl min−1). Note that the samples cannot be stored, as fixation will alter membrane states.
FIG. 1.
Flow cytometric dot plots of red (FL3) versus green (FL1) fluorescence of seawater bacteria stained simultaneously with SG1 and PI at different dye concentrations. The samples correspond to Mediterranean seawater subjected at 70°C in a water bath for 1 min. Left panels (A to F) show results for an SG concentration fixed at 10× and various concentrations of PI. Right panels (G to L) show results for a PI concentration fixed at 10 μg ml−1 and various concentrations of SG. “Live” cells (not stained by PI) are labeled as L. Dead cells (stained by PI) are labeled as D. Flow cytometric noise is labeled as n.
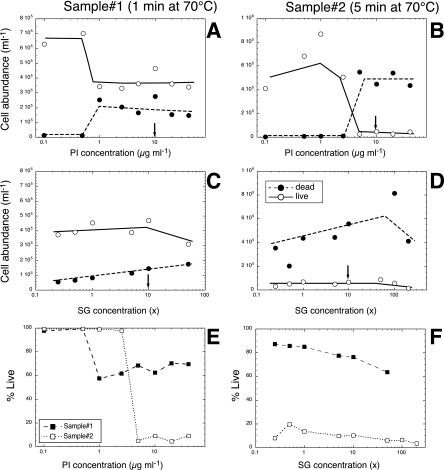
FIG. 2.
Effects of various concentrations of PI (A and B) and SG1 (C and D) on the enumeration of “live” (○) and “dead” (•) cells in a sample of Mediterranean seawater subjected at 70°C in a water bath for 1 min (A and C) and the same seawater sample subjected for 5 min at 70°C (B and D). Panels A and B show results for staining with various concentrations of PI and a fixed SG concentration of 10×. Panels C and D show results for staining with various concentrations of SG and a fixed concentration of 10 μg ml−1 of PI. (E) Evolution of the percentages of live cells at different concentrations of PI with a fixed SG concentration (10×). (F) Evolution of the percentages of live cells at different concentrations of SG with a fixed PI concentration (10 μg ml−1). Symbols: ▪, a sample maintained at 70° for 1 min; □, a sample maintained at 70° for 5 min. Arrows in panels A to D indicate the determined optimal concentrations of PI and SG.
Our study is not designed to test whether the red-labeled cells are “dead,” “inactive,” “membrane damaged,” or “membrane compromised” or whether the green-labeled cells are “active,” “live,” “viable,” or “intact.” However, for the sake of simplicity, we will use the terminology live/dead, following others (e.g., reference 29) and also following commercial kits based on the same principle (e.g., the Molecular Probes Live/Dead BacLight bacterial viability kit).
Bacterial abundance.
Samples (0.4 ml) for total prokaryote enumeration were stained with 4 μl of a 10× SG1 (Molecular Probes) solution (final dilution, 1:1,000 [vol/vol]) for 10 min and run through the FACSCalibur flow cytometer at a low speed, with beads as an internal standard. Bacteria were detected in a dot plot of side scatter versus green fluorescence (FL1) as reported elsewhere (27).
CTC.
The prokaryotes able to reduce 5-cyano-2,3-diotolyl tetrazolium chloride (CTC; Polysciences) are considered to be respiring bacteria, because CTC intercalates in the electron transport chain and is reduced instead of oxygen (60). CTC turns into a red fluorescent formazan that is detectable by epifluorescence and flow cytometry (60, 61). Sample aliquots (0.4 ml) received 5 mM CTC (from a fresh stock solution, 50 mM) immediately following collection and were incubated for 90 min in the dark at room temperature. CTC-positive (CTC+) cells were enumerated by flow cytometry using the FL2-versus-FL3 dot plot (see reference 28 for details). For these analyses, we used a high speed (ca. 100 μl min−1) and a threshold set in red fluorescence.
HNF enumeration by epifluorescence microscopy.
HNF were fixed with glutaraldehyde (10%). Fifty milliliters was filtered using 0.6-μm-pore-diameter polycarbonate filters, and HNF cell counts were performed after DAPI staining (0.5-mg-ml−1 final concentration) for 10 min. The filters were then frozen at −20°C until enumeration with an Olympus BS40 epifluorescence microscope.
Heterotrophic prokaryotic bulk activity and production.
Prokaryotic heterotrophic production was estimated from the uptake of [3H]leucine (protein biosynthesis; 40 nM final concentration) and thymidine (DNA biosynthesis; 20 nM final concentration). We used both tracers in the Atlantic and only leucine in the laboratory experiments. The incubation times were 1 to 2 h in the Mediterranean and 2 to 4 h in the Atlantic. For each sample, three or four aliquots (1.2 ml) plus one or two trichloroacetic acid-killed controls were incubated with the tracers at temperatures close to the in situ ones. Incubation was stopped with 50% trichloroacetic acid, and the samples were processed by the centrifugation method. Leucine-to-carbon empirical conversion factors were calculated with dilution cultures, following the cumulative method (see details in reference 2).
RESULTS
Protocol optimization.
Different protocol characteristics have been used in the past in the choices of green stain and, particularly, of stain concentration (Table 1). We carried out initial tests to determine the optimum concentrations of the SG1 and PI dyes. We considered “optimal” the concentrations that would clearly differentiate the two populations, clearly separate noise from the cells, and unambiguously allow enumeration of each type of cells.
The simultaneous staining with SG1 and PI produces two clusters of cells, one preferentially green and another that also has red fluorescence. The two clusters are clearly observed in a plot of red versus green fluorescence (Fig. 1) (SG1 concentrations of >5× and PI concentrations of >2.5 μg ml−1). We consider “intact” or “live” the cluster that has more green fluorescence than red fluorescence (population L in Fig. 1) and “damaged” or “dead” the population that has more red than green fluorescence (population labeled D in Fig. 1). With some brands of cytometers, electronic compensation can be applied to move the L population to the channel with no red fluorescence and the D population to the channel with no green fluorescence (see reference 29). However, this is not possible in the machine that we used, a FACSCalibur.
When planktonic bacteria were stained with variable concentrations of PI and a fixed SG1 concentration (10×), a very dim red fluorescent population was observed with concentrations from 0.5 μg ml−1 to 1 μg ml−1 of PI (Fig. 1). At concentrations of >1 μg ml−1, the cells stained by PI could clearly be observed (Fig. 1) and remained quite constant in number with increasing PI concentration (Fig. 2A). A complementary trend was observed in the range of SG concentrations studied, although the two populations were visible at all concentrations of SG (Fig. 1). The green and red clusters were better separated at concentrations between 10× and 50× of SG1 when the distance between the two clusters was higher (Fig. 1). At lower SG1 concentrations, the red fluorescence could be confounded with the debris signal, and at very high SG1 concentrations, there was too much electronic noise above the established fluorescence threshold. Interestingly, PI staining with an additional small concentration of SG1 already separated the two populations, even though the D population was very close to the threshold applied and to the noise (labeled n) (Fig. 1).
The ranges of SG1 and PI concentrations were applied to two samples that had different amounts of presumably intact and damaged cells. The cells with membranes compromised by heat exposure were detected after staining with >1 μg ml−1 of PI in one sample (1-min exposure in Fig. 2A) but at 10 μg ml−1 in the second sample (5-min exposure in Fig. 2B). Detection of both types of bacteria was less sensitive to changes in SG1 concentration (Fig. 2C and D), but more dead bacteria seemed to be detectable with increasing SG1 concentration. The percentage of live cells decreased to approximately 5% at a 5-μg-ml−1 PI concentration when the bacteria were exposed to 70°C for 5 min and to approximately 60% when the bacteria were exposed to 70°C for 1 min (Fig. 2E). These percentages did not change significantly with higher PI concentrations after the 1- or 10-μg-ml−1 threshold. The percentage of live cells decreased slightly with increasing SG concentrations in the two samples studied (Fig. 2F). Our results indicate an optimal PI staining concentration between 5 and 20 μg ml−1 and an optimal SG1 concentration of 10×.
Performance of the protocol in artificial death-causing treatments.
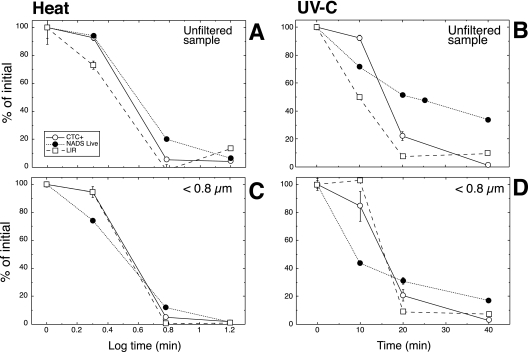
When bacteria were exposed to a high temperature (70°C), the number of NADS-determined live cells and the number of respiring bacteria (CTC+) declined concomitantly with the rates of leucine uptake, reaching values of between 0 and 20% after 5 min (Fig. 3A). A similar trend was observed when the bacteria were exposed to UVC radiations (Fig. 3B). Not all the samples in all death-causing treatments showed exactly the same correspondence in decrease of leucine incorporation, CTC, or number of NADS-determined live cells: the UVC treatment on the unfiltered sample maintained more live cells than CTC+ cells or leucine incorporation after 20 min of exposure (Fig. 3B). However, the decrease in viability measured by the NADS methods was significantly correlated with that measured by CTC (n = 13, Pearson's r = 0.90, P < 0.001) and by leucine incorporation (n = 13, Pearson's r = 0.79, P < 0.001).
FIG. 3.
Effects of exposure to UV radiations (UVC lamp, right panels) and heat (70°C, left panels) on viability of a marine bacterioplankton community, measured with the NADS protocol (•), with the electron chain respiratory activity probe (CTC+; ○), and with a radioactive tracer commonly used to measure bacterial production (leucine incorporation; □). Two samples are shown, an untouched marine sample and the same sample after having been filtered through a polycarbonate 0.8-μm filter. Results are expressed as percentages of the initial value and are the averages for two replicated experiments ± standard errors.
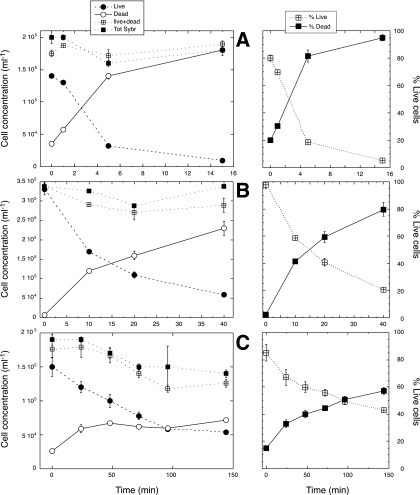
During all these treatments, the number of live cells declined in parallel to the increase in dead cells (Fig. 4) and there was an inverse relationship between the proportions of live and dead cells. The proportion of live cells declined from 80% to 5%, and the PI-stained dead cells increased from 20% to 95% in the heat treatment (Fig. 4A). During UVC exposure, live cells declined from 99% to 20%, and dead cells increased from 1% to 80% (Fig. 4B). When bacteria were incubated with the antibiotic cocktail, live cells gently declined, reaching values of 1 order of magnitude lower than the initial ones (Fig. 4C), while dead cells varied from 15% in the fresh seawater sample to 60% after approximately 144 h (Fig. 4C). The total number of cells did decrease after 2 h of incubation, indicating the possibility that some antibiotic-generated dead cells could be damaged in such a way that the nucleic acids would no longer be visible with the SG1 staining or with the combined SG1-PI staining. Indeed, the number of dead and live cells combined (as determined with NADS) was well correlated with the total for the values measured independently, with a slope not significantly different from 1 and an intercept not significantly different from 0 (n = 28, regression r2 = 0.81, all tests’ P values < 0.0001). The total for live and dead cells does not seem to miss any of the bacteria that could be detected with a total bacterial determination protocol.
FIG. 4.
NADS-determined “live” and “dead” marine bacteria after exposure to heat (70°C for 1, 5, and 15 min) (A), exposed to UV radiation (UVC) for 10, 20, and 40 min (B), and incubated with an antibiotic cocktail (gentamicin, penicillin G, streptomycin, and amphotericin B) for up to 144 h (C). Averages plus standard errors for two replicates are shown. The addition of the “live” and “dead” cells is also shown in the left-side panels (live+dead) as well as an independent determination of total bacterial abundance (Tot Sybr). Each experiment was done in a different day, with a different initial microbial community.
Natural sources of bacterial mortality.
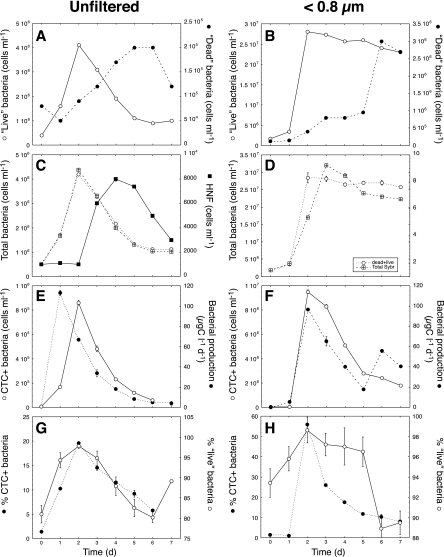
In the unfiltered-seawater microcosms, HNF cell numbers increased strongly (from ∼500 to 8,000 ml−1) in the first 4 days and then gradually decreased to 3,000 ml−1 in the last days of the experiment (Fig. 5C). An opposite trend was observed for the NADS-determined live bacteria (Fig. 5A), indicating that HNF had caused the decline in live bacteria and a corresponding increase in dead bacteria. Total bacterial abundance increased before the development of the HNF and could be measured either with NADS or with SG1 alone (Fig. 5C). The number of CTC+ cells and leucine incorporation peaked on days 1 and 2, respectively (Fig. 5E), and then decreased concomitantly with the increase in HNF abundance (Fig. 5E) and with the decrease in live bacteria (Fig. 5A). A good relationship was observed between the percentages of CTC+ bacteria and the percentages of NADS-determined live bacteria (Fig. 5G), even though the ranges were different (2 to 18% for the CTC cells and 79 to 99% for the live cells).
FIG. 5.
Microcosm experiments designed to generate HNF growth (unfiltered; left panels) and bacterial nutrient limitation (<0.8 μm; right panels). (A, B) Changes in “live” (○) and “dead” (•) bacteria (evaluated by the NADS protocol). (C, D) Changes in total bacterial abundance as determined by the addition of live and dead bacteria (dead+live; ○) and by an independent determination. Also in panel C, the concentrations of HNF (▪) are shown. (E, F) Numbers of respiring (CTC+) bacteria (○) and changes in bacterial production (•). (G, H) Percentages of CTC+ (•) and “live” (○) bacteria.
In the second set of microcosms (<0.8-μm-filtered seawater), we tested the effects of nutrient limitation on the detection of dead cells. In these microcosms, initial increases in numbers of “live” bacteria (Fig. 5B), total bacterial numbers (Fig. 5D), CTC+ cell numbers, and leucine incorporation (Fig. 5F) were observed in the first 2 days of the experiment. Once the bacterial population reached its maximum, the number of live cells started to slowly decrease, with a small increase in the number of dead cells (Fig. 5B), together with strong decreases in leucine incorporation and number of CTC+ cells (Fig. 5F). As in the other experiments (Fig. 5E), leucine incorporation and number of CTC+ cells were quite similar (Fig. 5F). The trends for percent CTC+ bacteria and percent live bacteria were less similar than those in the previous experiments (Fig. 5H), probably because the range for percent live bacteria (89 to 99%) was much smaller than the range for percent CTC+ bacteria (2 to 55%).
Application to the open ocean.
We tested the NADS protocol in a series of offshore stations in the northwest Atlantic Ocean (2). Through the epi- and mesopelagic zones of station 32, total prokaryotic abundance ranged from 1.8 × 105 to 6.8 × 105 cells ml−1 while the numbers of CTC+ cells ranged from 0.6 × 104 to 6.7 × 104 cells ml−1 and the numbers of live bacteria from 0.2 × 105 to 4.4 × 105 cells ml−1 (Table 2). The percentages of CTC+ cells ranged from 2 to 12%, the percentages of live cells from 21 to 70%, and the percentages of cells with high nucleic acid content (HNA) from 24 to 58%. The percentages of live and CTC+ cells decreased with depth, and there was a tendency for the percentage of cells with HNA to increase with depth. The number of CTC+ bacteria and that of live bacteria were significantly correlated (n = 9, Pearson's r = 0.89, P = 0.01). Interestingly, this station showed deep chlorophyll maxima at ca. 100 to 120 m of depth and an increase in bacterial production below these maxima, concomitant with increases in live and CTC+ cell abundance and the respective percentages at those depths (120 to 150 m). In the whole set of stations, total prokaryote abundance decreased with depth, with a slope of −0.60 ± 0.04 (n = 56, P < 0.001), while the number of CTC+ bacteria decreased with a slope of −0.44 ± 0.09 (n = 43, P < 0.001), the number of live bacteria decreased with a slope of −0.75 ± 0.05 (n = 55, P < 0.001), and that of dead bacteria decreased with a slope of −0.48 ± 0.05 (n = 55, P < 0.001). Bacterial production decreased with a slope of −0.70 ± 0.10 (n = 59). These differences in the rates of change of all these variables with depth indicate that the different methods are measuring different characteristics of the cells, which vary differentially with depth. Bacterial production decreases at a rate similar to that for live bacteria, while CTC+ and dead bacteria decreased at significantly lower rates.
TABLE 2.
Vertical profile of some bacterial subgroup abundances through the epi- and mesopelagic zones in the northwest Atlantic Ocean during cruise COCA-2, at station 32, 26.00°N, 26.00°Wa
| Depth (m) | Chl concn (mg m−3) | TPA (no. of cells [105] ml−1) | % HNA cells | PHP (μg carbon liter−1 day−1) | SGR (day−1) | No. of CTC+ cells (104) ml−1 | No. of “live” cells (105) ml−1 | % CTC+ cells | % “Live” cells |
|---|---|---|---|---|---|---|---|---|---|
| 5 | 0.04 | 6.68 | 38.6 | 6.32 ± 0.65 | 0.398 | 4.79 | 4.61 | 7.2 | 69.6 |
| 50 | 0.05 | 6.76 | 37.2 | 6.18 ± 0.73 | 0.383 | 2.32 | 3.4 | ||
| 100 | 0.15 | 4.14 | 26.6 | 4.09 ± 0.39 | 0.416 | 2.38 | 4.06 | 5.7 | 61.0 |
| 120 | 0.25 | 5.24 | 24.2 | 1.99 ± 0.16 | 0.181 | 6.22 | 3.94 | 11.9 | 63.4 |
| 130 | 0.65 | 6.79 | 34.8 | 10.99 ± 1.42 | 0.580 | 6.59 | 4.41 | 9.7 | 46.0 |
| 150 | 0.45 | 6.11 | 53.0 | 21.22 ± 1.11 | 0.954 | 2.58 | 3.93 | 4.2 | 46.5 |
| 200 | 0.04 | 2.85 | 51.4 | 0.94 ± 0.04 | 0.158 | 1.05 | 2.59 | 3.7 | 49.6 |
| 300 | <0.01 | 2.41 | 52.6 | 0.26 ± 0.05 | 0.053 | 1.04 | 1.49 | 4.3 | 44.0 |
| 400 | <0.01 | 2.20 | 54.1 | 0.15 ± 0.03 | 0.034 | 0.98 | 44.7 | ||
| 500 | <0.01 | 2.38 | 50.4 | 0.19 ± 0.04 | 0.040 | 0.68 | 28.5 | ||
| 600 | <0.01 | 2.36 | 57.9 | 0.21 ± 0.07 | 0.042 | 0.83 | 0.46 | 3.5 | 31.4 |
| 700 | <0.01 | 2.06 | 49.5 | 0.11 ± 0.04 | 0.026 | 0.69 | 42.4 | ||
| 800 | <0.01 | 1.86 | 54.9 | 0.06 ± 0.01 | 0.015 | 0.30 | 21.9 | ||
| 900 | <0.01 | 1.88 | 47.7 | 0.11 ± 0.01 | 0.030 | ||||
| 1,000 | <0.01 | 2.41 | 38.6 | 0.05 ± 0.02 | 0.011 | 0.57 | 0.24 | 2.4 | 21.1 |
Prokaryote heterotrophic production (PHP) is the average of leucine and thymidine incorporation values converted with empirically derived conversion factors. Chl, chlorophyll; TPA, total prokaryote abundance determined by SG1 staining; SGR, heterotrophic prokaryote specific growth rate.
DISCUSSION
The state of the membrane provides much information on the general cell physiological condition (13). It is usually assumed that bacterial permeability to exclusion stains, such as PI (with a molecular weight of 668.4) is associated with the presence of large and presumably irreparable gaps in the membrane (52). It is likely that membrane damage is also associated with the loss of the capsular envelope, which in turn has been linked to loss of intracellular integrity in bacterioplankton cells (32). The NADS protocol, in the way that we have tested and calibrated it here, is one approach to the differentiation between bacterioplankton cells with damaged membranes and those with intact mechanisms of isolation from the environment (29). The central component of the stain mixture is PI, which acts depending upon a number of factors: temperature, pH, ion composition, stain concentration, staining time, and cell storage conditions (67). In an unclear, little-investigated way, some of these factors affect the state of the membrane, and some others affect the binding of PI to the nucleic acids. It has been shown, e.g., that very low temperatures and low or high pH values produce higher numbers of PI-stained cells in a growing Escherichia coli culture than medium-range temperatures and pHs (67), but it is not known whether this is due to membrane breaches or to the kinetics of the nucleic acid-PI reaction.
Few researchers have used PI alone in studies of environmental samples (but see reference 42), maybe because of the problems that occur with the conditions of optimal PI staining when it is used alone. In the 1990s, Molecular Probes introduced the BacLight Live/Dead kit, which consists of a mixture of a generic DNA stain (which we now know is Syto9) and the membrane-impermeant stain PI. Both are added at the same time and interact so that live cells are stained green and dead cells are stained red. The interaction between the dyes means that the red dye (PI) quenches the green emission by the other dye (65). The methodology was first applied by Lloyd and Hayes (41) and to marine bacteria by Nagamura (49) and Choi et al. (15). Barbesti et al. (6) devised a similar protocol, but using SG combined with PI (so that the researchers did not need to pay for the kit). Other authors have used other combinations of stains (e.g., Hoechst and PI [51] and Syto13 and Sytox Orange [11]). Using flow cytometry, the interpretation of the green-versus-red-fluorescence cytograms could provide information not only on “live” (green) and “dead” (red) cells but also on “green-plus-red” particles, which have been identified as “damaged” cells (30).
One of the critical aspects in the use of PI is the concentration of the stain, as pointed out by Williams et al. (67). These authors recommended values between 1 and 5 μg ml−1 because with higher concentrations cells became leaky (44). Schumann et al. (59) used a 2.2-μg-ml−1 final PI concentration, as recommended in the instructions for the BacLight viability kit, and Pirker et al. (58) used a PI concentration of 0.6 μg ml−1. While high PI concentrations resulted in higher percentages of PI-positive cells in some cases (67), we found no changes in the range of 5 to 20 μg ml−1 (Fig. 2) but unclear results with PI concentrations below 5 μg ml−1. Note that it is possible that differences occur when PI is assayed alone compared to when PI is combined with the green nucleic acid stain. When PI is used in combination with SG1, the brightness of the PI red fluorescence is higher than that observed when PI is used alone (unpublished observations). The efficiency of the combined staining is amplified by the energy transfer (FRET) from SG1 to PI when both are bound to the nucleic acids at a very small distance (<6 nm) (6). This might be the reason why the double-staining protocols are particularly appropriate for working with natural samples, where some background noise in the red channel is always present.
While PI needs to be at a certain concentration to be effective, and at a sufficient quantity to stain all present DNA (cf. 65 for BacLight staining), this does not seem to be the case for SG1 (Fig. 2). Our data also suggest that the ideal set of concentrations might vary depending on the amount and severity of the mortality induced on the bacteria (compare Fig. 2A and B). This could be a disadvantage in working with commercial kits (such as the BacLight kit) where the dyes are present in fixed concentrations.
Since the FRET mechanism seems to be a mechanism that increases fluorescence from the individual particles, it is important to be sure to double stain simultaneously with the two dyes. Results for protocols in which the staining is not simultaneous should be interpreted with caution. In the work of Pirker et al. (58), for example, the SG dye was added after 30 min of incubation with PI and rinsing with MilliQ. Our experience in the use of the NADS protocol suggests that simultaneous addition of the two dyes is essential for interaction between them. Preliminary results obtained using confocal laser scanning microscopy (details not shown) suggest that nonsimultaneous addition produces significantly different results.
Previous studies have used different concentrations of the two components of the NADS protocol (Table 1): 10 μg ml−1 of PI and a 10× concentration of SG1 for use on bacterial pure cultures (6), 1 μg ml−1 of PI and a final dilution of 1:100,000 for SG1 for staining of freshwater samples with fewer than 0.5 × 106 bacteria ml−1, and 10 μg ml−1 of PI and 1:10,000 for SG1 for more-concentrated samples (up to 2 × 107 bacteria ml−1) (29). During our staining calibration, we observed optimal working concentrations for marine planktonic bacterial staining and detection by flow cytometry in the range of 5 to 20 μg ml−1 of PI and a 10× concentration of SG1. In this range, the green and red fluorescent subgroups were well separated and also separated from the noise (Fig. 1). Outside this range, at high PI and low SG1 concentrations, there was overlap of the dead population with the noise signal (Fig. 2). The exact positions of the bacterial clusters depend on the type of flow cytometer instrument. The dot plot distribution obtained with the flow cytometer used in the work of Grégori et al. (29) (Bryte-HS; Bio-Rad), e.g., is different from that obtained with our flow cytometer. In the work of Grégori et al. (29), the red (dead) population is located at an angle of 90° with respect to the green (live) one, because true fluorescence compensation is possible. The FACSCalibur has optical system differences that do not allow a full compensation of these fluorescences.
Note that for these double-staining protocols to identify a cell as “dead,” the cell must contain enough nucleic acids to allow staining by PI. This, in theory, would allow the methodology to distinguish at least an extra group of cells if PI were combined with a universal dye not based on nucleic acids (e.g., Sypro, which stains proteins [71], or DAPI, which also stains membranes [72]). In fact, the difference that we observe in Fig. 4 (particularly panel C) between the initial and final total numbers of cells is in cells that died and lost the nucleic acids. Heissenberger et al. (32) used transmission electron microscopy to analyze the structure of the cellular envelope as well as those of the internal parts of bacterioplankton cells and could tell apart three types: intact bacteria, having all cell elements; damaged bacteria, usually lacking an intact membrane but having internal content; and empty cells, lacking any recognizable plasm and often a membrane. One-third of the cells analyzed were intact, 43% were damaged, and 24% were empty. This allows us to hypothesize that the last group would not be counted by any nucleic acid stain (SG1, Syto9, or PI) and that the damaged subgroup would be the one counted as “dead” by the NADS protocol.
In any case, we should expect to find the number of dead and live cells measured by the NADS protocol equal to the total number of cells measured independently by a DNA stain. Some studies have found the same number of total cells by adding the red and green particles (49, 15, 19), while some others have found smaller amounts of particles (26, 59, 69). This difference can be due to differences in the standard chosen: DAPI stains particles even if they do not have nucleic acids (72), while green stains such as SG1 and Syto13 might not do so. It also might depend on the type of “mortality agent” that could affect the membrane or could destroy all macromolecules. Here, we found mostly the same number of cells (Fig. 4).
These additional physiological categories could, in theory, be combined with an additional one, as observed by Grégori et al. (30) in marine bacterioplankton and Berney et al. (9) in cultures and in freshwater bacterioplankton, in which cells having ambiguous scores as “red” or “green” are interpreted as “damaged” cells. These accounted for 2 to 18% of the total cells in the Bay of Marseille. We chose not to consider this additional category, because it was very difficult to delimit and enumerate in most of the cases (see Fig. 1 for situations in which this category is impossible to outline).
The NADS protocol targets loss of membrane integrity, but this does not necessarily imply cell death, because there is evidence that cells can recuperate and resume growth under the appropriate conditions (8). Recently, Manini and Danovaro (44) used a protocol including SG1 destaining, and they observed cells that responded positively to PI but also maintained a cell nucleoid. These cells, which would be labeled “dead” by the NADS protocol, could be competent cells prepared for DNA exchange and thus not at all dead.
Pirker et al. (58) combined microautoradiography and PI to simultaneously assess the metabolic activity and viability of individual bacterioplankton cells in the coastal and open North Sea. In this case, the overwhelming majority (97%) of cells taking up glucose and leucine were also PI positive. Apparently, the uptake of radiolabeled substrate was related to PI accumulation in the cells, generating doubts as to its utility as a dead-cell marker. Our results in which we compared the NADS protocol to leucine incorporation (Fig. 3) are incompatible with the results presented by Pirker et al. (58), and we postulate that the fact that these authors did not double stain the samples simultaneously might account for this disagreement.
While membrane potential and membrane integrity are two aspects of cellular function that should a priori be strongly linked, some studies have shown that this expectation is not always held, because loss of membrane integrity is not always accompanied by loss of potential and vice versa (52). For example, in a study along a freshwater-saltwater estuarine gradient, del Giorgio and Bouvier (21) showed peaks in depolarized [measured with bis-(1,2-dibutylbarbituric acid) trimethine oxonol dye] and injured (PI-positive) cells within the salinity transition zone but not always coincident in the same station, which suggests that loss of membrane potential is not always synonymous with cell injury and death.
As an additional methodological problem, if the samples are dominated by Prochlorococcus organisms, which have very low red fluorescence in surface oceanic waters, they often appear in between the red and green cell clusters, thus making sample analysis more complicated. An additional sample, without addition of the stains, is necessary for enumerating these cells, which are later on discounted from the NADS cell counts.
To validate the NADS methodology, Grégori et al. (29) compared its performance after heat and ozone treatments of seawater samples. That the method worked correctly was shown by some cell-sorting experiments in which river water was NADS stained, red and green cells were sorted onto plates with rich culture medium, and only the green cells produced colonies. But neither of these sources of mortality are likely to be experienced by marine bacteria, nor are CFU counts a valid estimator of marine bacterial viability (i.e., reference 3). We instead compared the performance of the method to that of two accepted estimators of bacterial bulk (leucine incorporation [37]) and single-cell (CTC [60]) activity, with the assumption that if a treatment induces loss of leucine incorporation and CTC+ cells, this should be reflected in the NADS response, and vice versa. Furthermore, we combined classical sources of mortality with natural ones. When bacterioplankton was exposed to biotic factors that induced bacterial death, the proportions of “live” and respiring bacteria and the rates of leucine uptake concomitantly declined with incubation time at similar rates (Fig. 3). In the case of the heat-treated samples, no rates of leucine uptake were measurable after 5 min of incubation (Fig. 3). Other authors have reported a similar trend in lake water samples that were heat exposed (45).
We also observed that the types of mortality caused by UVC, heat, and antibiotics were different. Heat (70°C) produced membrane damage in ca. 5 min and in all cells (Fig. 4A), while UVC produced damage in 80% of the cells after 40 min (Fig. 4B). The antibiotic cocktail produced 60% damaged cells after 150 min of incubation. This indicates that each process that causes bacterial mortality does so in a different manner, and a given methodology (like NADS) will reflect only one cellular characteristic. It is advisable to use a mixture of protocols to describe cell viability in the ocean (19, 21, 22). The NADS protocol correctly identified mortality caused by flagellates (Fig. 5), most probably through sloppy feeding or production of picopellets (50). When we inspected the detectability of cell damage caused by starvation, we first detected an increase in live cells that used the carbon stored in the microcosm and then, slowly, there was a steady increase in damaged bacteria, something that was less well correlated with the changes in the number of respiring bacteria and in total leucine incorporation (Fig. 5). Again, the mechanisms causing bacterial death in the ocean, which include programmed cell death (53) and viruses, cause different kinds of damage in the different constituents of the bacterial cell and these might not necessarily all be detected by the NADS protocol. But it seemed to correctly detect viability changes caused by predators and by starvation.
We were able to measure the variability in percent live bacteria in different oceanic situations. Our average results are 47% live cells in the northwest Atlantic and 74% in the northwest Mediterranean Sea (Table 3), which are within the range of other studies that have used the live/dead or the NADS protocol in oceanic environments. In the vertical profile of the Atlantic Ocean, the percentage of live cells decreased from ca. 65% at the surface to ca. 22% at 1,000 m. The rate of decrease of live cells with depth was similar to that of bacterial production and higher than the rates of decrease of total and CTC+ cells. It is possible to hypothesize that death-generating mechanisms are more important in the mesopelagic zone than in the epipelagic zone (i.e., reference 14), but it could equally be possible that the rates of decomposition of dead cells in the epipelagic zone are higher than those in the mesopelagic zone because total bacterial production is higher there. Thus, the rate of accumulation of dead cells would be higher at depth than at the surface. The fact that the number of respiring cells decreases significantly less than the number of intact-membrane cells indicates that it is probable that different mechanisms act on each type of cell. In any case, our data show that it is possible to use the NADS protocol in the mesopelagic zone and that this produces data which might help understand the mechanisms controlling bacterial activity and distribution in the ocean.
TABLE 3.
Published data on planktonic bacterial viability measured with the BacLight live/dead method or the NADS protocol
| Site | Method | TBAa (no. of cells [106] ml−1) | % “Live” cells | Source or reference |
|---|---|---|---|---|
| Kitahashi River | BacLight | 4.03 | 61.9 | 68 |
| Warnow River | BacLight | 12.02 | 27.1 | 25 |
| Lake Frederiksborg Slotssø | BacLight | 8.46 | 18.5 | 64 |
| Freshwater | BacLight | 8.58 | 64.9 | 59 |
| Estuary | BacLight | 14.60 | 74.0 | 59 |
| Choptank River Estuary | BacLight | 5.60 | 90.8 | 21 |
| Seto Inland Sea | BacLight | 1.16 | 54.2 | 49 |
| Hiroshima Bay | BacLight | 1.90 | 66.0 | 20 |
| Coastal Mediterranean (Masnou) | BacLight | 1.43 | 72.4 | 26 |
| Canadian Arctic | BacLight | 2.95 | 54.0 | 66 |
| Baltic Sea | BacLight | 2.48 | 67.3 | 59 |
| Oregon coastal seawater | BacLight | 0.90 | 14.0 | 15 |
| Tasmania coastal seas | BacLight | 0.54 | 29.3 | 19 |
| Mediterranean Sea | BacLight | 0.42 | 15.2 | 69 |
| Eastern English Channel | BacLight | 1.40 | 56.7 | 38 |
| Coastal Antarctica | BacLight | 0.54 | 14.2 | 54 |
| Mediterranean coastal sites | NADS | 0.70 | 54.9 | 29 |
| Bay of Marseille | NADS | 0.59 | 63.4 | 30 |
| North Sea | Modified NADS | 0.82 | 21.3 | 58 |
| Blanes Bay Microbial Observatory | NADS | 0.90 | 65.0 | 1 |
| Northeast Atlantic | NADS | 0.65 | 47.0 | This work |
| Blanes Bay Microbial Observatory | NADS | 1.09 | 74.3 | This work |
Total bacterial abundance.
In the search for the mechanisms determining the growth and losses of different types of bacteria, the NADS protocol combined with flow cytometry is a fast and simple alternative to other techniques. We foresee also a combined NADS-FISH method being developed to gain insight into the roles of the different death mechanisms controlling different bacterial subgroups in the ocean.
Acknowledgments
This work was funded by EU project BASICS (EVK3-CT-2002-00078), Spanish MEC project MODIVUS (CTM2005-04795/MAR), and CIPE research project 17-2003 of the Italian Marche Region.
We thank C. Cardelús and V. Balagué for help with the Blanes Bay Microbial Observatory sampling program, L. Alonso-Sáez for sharing the Atlantic data, scientists and technicians on R/V Hesperides for the basic oceanographic data of cruise COCA, and T. Lefort for comments on the manuscript.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Alonso-Sáez, L., J. M. Gasol, T. Lefort, J. Hofer, and R. Sommaruga. 2006. Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 72:5806-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Sáez, L., J. M. Gasol, J. Arístegui, J. C. Vilas, D. Vaqué, C. M. Duarte, and S. Agustí. 2007. Large-scale variability in surface bacterial carbon demand and growth efficiency in the subtropical North East Atlantic Ocean. Limnol. Oceanogr. 52:533-546. [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arístegui, J., C. M. Duarte, J. M. Gasol, and L. Alonso-Sáez. 2005. Active mesopelagic prokaryotes support high respiration in the subtropical northeast Atlantic Ocean. Geophys. Res. Lett. 32:L03608-L03612. [Google Scholar]
- 5.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Reil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 6.Barbesti, S., S. Citterio, M. Labra, M. D. Baroni, M. G. Neri, and S. Sgorbati. 2000. Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry 40:214-218. [PubMed] [Google Scholar]
- 7.Barcina, I., P. Lebaron, and J. Vives-Rego. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23:1-9. [Google Scholar]
- 8.Barer, M. R., and C. R. Harwood. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:93-137. [DOI] [PubMed] [Google Scholar]
- 9.Berney, M., F. Hammes, F. Bosshard, H. U. Weilenmann, and T. Egli. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidle, K. D., and G. Falkowski. 2004. Cell death in planktonic, photosynthetic microorganisms. Nat. Rev. Microbiol. 2:643-655. [DOI] [PubMed] [Google Scholar]
- 11.Biggerstaff, J. P., M. Le Puil, B. L. Weidow, J. Prater, K. Glass, M. Radosevich, and D. C. White. 2006. New methodology for viability testing in environmental samples. Mol. Cell. Probes 20:141-146. [DOI] [PubMed] [Google Scholar]
- 12.Boulos, L., M. Prévost, B. Barbeau, J. Coallier, and R. Desjardins. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37:77-86. [DOI] [PubMed] [Google Scholar]
- 13.Brehm-Stecher, B. F., and E. A. Johnson. 2004. Single-cell microbiology: tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 68:538-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho, B. C., S. C. Na, and D. H. Choi. 2000. Active ingestion of fluorescently labeled bacteria by mesopelagic heterotrophic nanoflagellates in the East Sea, Korea. Mar. Ecol. Prog. Ser. 206:23-32. [Google Scholar]
- 15.Choi, J. W., E. B. Sherr, and B. F. Sherr. 1996. Relation between presence-absence of a visible nucleoid and metabolic activity in bacterioplankton cells. Limnol. Oceanogr. 41:1161-1168. [Google Scholar]
- 16.Choi, J. W., E. B. Sherr, and B. F. Sherr. 1999. Dead or alive? A large fraction of ETS-inactive marine bacterioplankton cells, as assessed by reduction of CTC, can become ETS active cells with incubation and substrate addition. Aquat. Microb. Ecol. 18:105-115. [Google Scholar]
- 17.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 19.Davidson, A. T., P. G. Thompson, K. Westwood, and R. van den Enden. 2004. Estimation of bacterioplankton activity in Tasmanian coastal waters and between Tasmania and Antarctica using stains. Aquat. Microb. Ecol. 37:33-45. [Google Scholar]
- 20.Decamp, O., and N. Rajendran. 1998. Assessment of bacterioplankton viability by membrane integrity. Mar. Pollut. Bull. 36:739-741. [Google Scholar]
- 21.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic succession in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 22.del Giorgio, P. A., and J. M. Gasol. Physiological structure and single-cell activity in marine bacterioplankton. In D. L. Kirchman (ed.), Microbial ecology of the ocean, 2nd ed., in press.
- 23.Ericsson, M., D. Hanstorp, P. Hagberg, J. Enger, and T. Nyström. 2000. Sorting out bacterial viability with optical tweezers. J. Bacteriol. 182:5551-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falcioni, T., A. Manti, P. Boi, B. Canonico, M. Balsamo, and S. Papa. 2006. Comparison of disruption procedures for enumeration of activated sludge floc bacteria by flow cytometry. Cytometry B 70:149-153. [DOI] [PubMed] [Google Scholar]
- 25.Freese, H. M., U. Karsten, and R. Schumann. 2006. Bacterial abundance, activity, and viability in the eutrophic river Warnow, Northeast Germany. Microb. Ecol. 51:117-127. [DOI] [PubMed] [Google Scholar]
- 26.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Å. Hagström. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasol, J. M., and P. A. del. Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 28.Gasol, J. M., and J. Arístegui. 2007. Cytometric evidence reconciling the toxicity and usefulness of CTC as a marker of bacterial activity. Aquat. Microb. Ecol. 46:71-83. [Google Scholar]
- 29.Grégori, G., S. Citterio, A. Ghiani, M. Labra, S. Sgorbati, S. Brown, and M. Denis. 2001. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl. Environ. Microbiol. 67:4662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grégori, G., M. Denis, D. Lefevre, and J. C. Romano. 2003. Viability of heterotrophic bacteria in the Bay of Marseille. C. R. Biol. 326:739-750. [DOI] [PubMed] [Google Scholar]
- 31.Haugland, R. P. 1998. Handbook of fluorescent probes and research chemicals. Molecular Probes, Eugene, OR.
- 32.Heissenberger, A., G. G. Leppard, and G. J. Herndl. 1996. Relationship between the intracellular integrity and the morphology of the capsular envelope in attached and free-living marine bacteria. Appl. Environ. Microbiol. 62:4521-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoefel, D., W. L. Grooby, P. T. Monis, S. Andrew, and C. P. Saint. 2003. Enumeration of water-borne bacteria using viability assays and flow cytometry: a comparison to culture-based techniques. J. Microbiol. Methods 55:585-597. [DOI] [PubMed] [Google Scholar]
- 34.Jones, K. H., and J. A. Senf. 1985. An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide. J. Histochem. Cytochem. 33:77-79. [DOI] [PubMed] [Google Scholar]
- 35.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 36.Karner, M., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchman, D., E. Knees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein-synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamy, D., L. F. Artigas, C. Jauzein, F. Lizon, and V. Cornille. 2006. Coastal bacterial viability and production in the eastern English channel: a case study during a Phaeocystis globosa bloom. J. Sea Res. 56:227-238. [Google Scholar]
- 39.Lebaron, P., N. Parthuisot, and P. Catala. 1998. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl. Environ. Microbiol. 64:1725-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloyd, D., and A. J. Hayes. 1995. Vigor, vitality and viability of microorganisms. FEMS Microbiol. Lett. 133:1-7. [Google Scholar]
- 42.López-Amorós, R., D. J. Comas, and J. Vives-Rego. 1995. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl. Environ. Microbiol. 61:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovejoy, C., L. Legendre, B. Klein, J. E. Tremblay, R. G. Ingram, and J. C. Therriault. 1996. Bacterial activity during early winter mixing (Gulf of St. Lawrence, Canada). Aquat. Microb. Ecol. 10:1-13. [Google Scholar]
- 44.Manini, E., and R. Danovaro. 2006. Synoptic determination of living/dead and active/dormant bacterial fractions in marine sediments. FEMS Microbiol. Ecol. 55:416-423. [DOI] [PubMed] [Google Scholar]
- 45.Maranger R., P. A. del Giorgio, and D. F. Bird. 2002. Accumulation of damaged bacteria and viruses in lake water exposed to solar radiation. Aquat. Microb. Ecol. 28:213-227. [Google Scholar]
- 46.Marie, D., F. Partensky, S. Jacquet, and D. Valout. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mason, C. A., G. Hamer, and J. D. Bryers. 1986. The death and lysis of microorganisms in environmental processes. FEMS Microbiol. Rev. 39:373-401. [Google Scholar]
- 48.Mason, D. J., R. López-Amorós, R. Allman, J. M. Stark, and D. Lloyd. 1995. The ability of membrane potential dyes and calcafluor white to distinguish between viable and non-viable bacteria. J. Appl. Bacteriol. 78:309-315. [DOI] [PubMed] [Google Scholar]
- 49.Nagamura, T. 1986. Differential enumeration of intact and damaged planktonic bacteria based on cell membrane integrity. J. Aquat. Ecosyst. Health 5:217-222. [Google Scholar]
- 50.Nagata, T. 2000. “Picopellets” produced by phagotrophic nanoflagellates: role in material cycling within marine environments, p. 241-256. In N. Handa, E. Tanoue, and T. Hama (ed.), Dynamics and characterization of marine organic matter. Terrapub/Kluwer, Tokyo, Japan.
- 51.Nebe-von-Caron, G., and R. A. Badley. 1995. Viability assessment of bacteria in mixed populations using flow cytometry. J. Microsc. 179:55-66. [Google Scholar]
- 52.Novo, D. J., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyström, T. 2003. Nonculturable bacteria: programmed survival forms or cells at death's door? Bioessays 25:204-211. [DOI] [PubMed] [Google Scholar]
- 54.Pearce, I., A. T. Davidson, E. M. Bell, and S. Wright. 2007. Seasonal changes in the concentration and metabolic activity of bacteria and viruses at an Antarctic coastal site. Aquat. Microb. Ecol. 47:11-23. [Google Scholar]
- 55.Pernthaler, A., and J. Pernthaler. 2005. Diurnal variation of cell proliferation in three bacterial taxa from coastal North Sea waters. Appl. Environ. Microbiol. 71:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phe, M. H., M. Dossot, H. Guilloteau, and J. C. Block. 2005. Nucleic acid fluorochromes and flow cytometry prove useful in assessing the effect of chlorination on drinking water bacteria. Water Res. 39:3618-3628. [DOI] [PubMed] [Google Scholar]
- 57.Pianetti, A., T. Falcioni, F. Bruscolini, L. Sabatini, E. Sisti, and S. Papa. 2005. Viability of Aeromonas hydrophila in different types of water by flow cytometry and comparison with classical methods. Appl. Environ. Microbiol. 71:7948-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pirker, H., C. Pausz, K. E. Stoderegger, and G. J. Herndl. 2005. Simultaneous measurement of metabolic activity and membrane integrity in marine bacterioplankton determined by confocal laser-scanning microscopy. Aquat. Microb. Ecol. 39:225-233. [Google Scholar]
- 59.Schumann, R., U. Schiewer, U. Karsten, and T. Rieling. 2003. Viability of bacteria from different aquatic habitats. II. Cellular fluorescent markers for membrane integrity and metabolic activity. Aquat. Microb. Ecol. 32:137-150. [Google Scholar]
- 60.Sherr, B. F., P. A. del Giorgio, and E. B. Sherr. 1999. Estimating abundance and single-cell characteristics of actively respiring bacteria via the redox dye CTC. Aquat. Microb. Ecol. 18:117-131. [Google Scholar]
- 61.Sieracki, M. E., T. L. Cucci, and J. Nicinski. 1999. Flow cytometric analysis of 5-cyano-2,3-ditolyl tetrazolium chloride activity of marine bacterioplankton in dilution cultures. Appl. Environ. Microbiol. 65:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, E. M., and P. A. del Giorgio. 2003. Low fractions of active bacteria in natural aquatic communities? Aquat. Microb. Ecol. 31:203-208. [Google Scholar]
- 63.Reference deleted.
- 64.Søndergaard, M. 2000. Bacterioplankton: how many and how active? Verh. Int. Ver. Theor. Agnew. Limnol. 27:859-865. [Google Scholar]
- 65.Stocks, S. M. 2004. Mechanism and use of the commercially available viability stain, BacLight. Cytometry A 61:189-195. [DOI] [PubMed] [Google Scholar]
- 66.Tam, L., P. G. Kevan, and J. T. Trevors. 2003. Viable bacterial biomass and functional diversity in fresh and marine waters in the Canadian Arctic. Polar Biol. 26:287-294. [Google Scholar]
- 67.Williams, S. C., Y. Hong, D. C. A. Danavall, M. H. Howard-Jones, D. Gibson, M. E. Frischer, and P. G. Verity. 1998. Distinguishing between living and nonliving bacteria: evaluation of the vital stain propidium iodide and its combined use with molecular probes in aquatic samples. J. Microbiol. Methods 32:225-236. [Google Scholar]
- 68.Yokomaku, D., N. Yamaguchi, and M. Nasu. 2000. Improved direct viable count procedure for quantitative estimation of bacterial viability in freshwater environments. Appl. Environ. Microbiol. 66:5544-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zampino, D., R. Zaccone, and R. La Ferla. 2004. Determination of living and active bacterioplankton: a comparison of methods. Chem. Ecol. 20:411-422. [Google Scholar]
- 70.Ziglio, G., G. Andreottola, S. Barbesti, G. Boschetti, L. Bruni, P. Foladori, and R. Villa. 2002. Assessment of activated sludge viability with flow cytometry. Water Res. 36:460-468. [DOI] [PubMed] [Google Scholar]
- 71.Zubkov, M. V., B. M. Fuchs, H. Eilers, P. H. Burkill, and R. Amann. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl. Environ. Microbiol. 65:3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zweifel, U. L., and Å. Hagström. 1995. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts). Appl. Environ. Microbiol. 61:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]