Abstract
Microfluidic devices permit direct observation of microbial behavior in defined microstructured settings. Here, the swimming speed and dispersal of individual marine ciliates in straight and bent microfluidic channels were quantified. The dispersal rate and swimming speed increased with channel width, decreased with protozoan size, and was significantly impacted by the channel turning angle.
Micron-scale structure is ubiquitous in virtually all microbial habitats, including suspended aggregates (11, 18) and biofilms on solid surfaces (10, 16, 23). In terrestrial and benthic systems, interstitial pore spaces may be connected to form tortuous pore networks (3). In virtually all microbial habitats, physical structure impacts water flow (17, 21), gas diffusion (2, 20), and transport of substrates and products to and from microbial cells (24). Physical structure offers bacteria a refuge from predation (14, 19) by impeding the mobility of protozoans and other bacteriovores (5, 15). However, the impact of microscale habitat structure on the behavior of individual microbes is generally not well understood.
Our approach is to use custom-made microfluidic devices to better understand microbe-habitat interactions. The advantages of the microfluidic approach include direct observation and reproducible quantification of the behaviors and interactions of individual microbes in researcher-defined microstructured habitats. Previous work focused on the impact of microchannel constrictions on protozoan mobility (22). Here, we observed and quantified the impacts of microchannel dimensions and channel turning angles on protozoan swimming speed and dispersal. More broadly, this work demonstrated how certain features of a natural system may be systematically examined using a well-defined microfluidic test system.
Protozoan cultures.
Two species of marine ciliates were chosen for this work. Both species are widely distributed and have a significant impact on bacterial populations in natural systems (8), but they are quite different in terms of size, swimming speed, and typical habitat. Euplotes vannus (CCAP 1624/12 from the Culture Collection of Algae and Protozoa, Cumbria, United Kingdom) is a medium-size hypotrich ciliate (length, 82 ± 11 μm; width, 47 ± 7 μm; height, 26 ± 5 μm [means ± standard deviations; n = 20]) normally found associated with solid surfaces (9), including suspended aggregates (4); E. vannus has also been isolated from interstitial environments (7).
Uronema sp. (clone BBcil, provided by David Caron, University of Southern California) is a small, ovoid scuticociliate (length, 28 ± 6 μm; width, 9 ± 3 μm; height, 9 ± 3 μm [means ± standard deviations; n = 20]) that is usually found swimming freely in the water column but also is associated with natural aggregates (6). Protozoans were cultured and prepared for use in experiments as described elsewhere (22).
Experiment design.
The experiments reported here tested the mobility of both ciliates in microfluidic devices with four different channel turning angles and two different channel widths (2 by 4 by 2). At least five replicates of each treatment were conducted.
Microfluidic device design and fabrication.
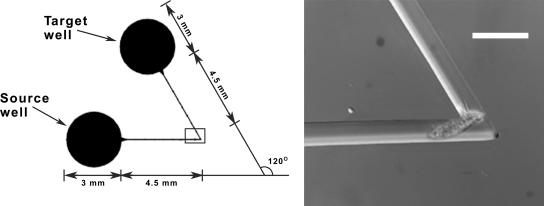
The microfluidic devices consisted of sets of two 3-mm-diameter, 6-mm-high cylindrical wells connected by a single 8.5-mm-long microchannel including a single channel bend with a turning angle of 0, 60, 90, or 120°. The devices were designed so that multiple sets fit simultaneously on a single 75- by 50-mm slide to facilitate high-throughput analysis with automated microscopy. Similar devices with channel widths of 30, 20, and 10 μm and a uniform channel height of 40 μm were created using standard soft lithography methods as described previously (22). The devices were constructed so that three sides (the “walls” and the “ceiling” of each square channel) were polydimethyl siloxane and all bottom surfaces of the wells and channels were the top surface of a glass microscope slide. Prior to use, the devices were filled with filtered, sterile artificial seawater for protozoans via application of a vacuum. A device with a microfluidic turning angle of 120° is shown in Fig. 1.
FIG. 1.
Schematic diagram of a microchannel with a 120° bend connecting a source well and a target well and micrograph of E. vannus trying to pass a 40-μm-high, 20-μm-wide 120° channel bend (scale bar = 40 μm).
Statistical analysis.
Swimming speeds were analyzed by using two-factor analysis of variance (ANOVA) with channel width and channel angle factors. Three-factor ANOVA were performed using channel width, channel angle, and time as factors for each species with square root-transformed dispersal data. Tukey's honestly significant difference test was used for a posteriori comparison of means when a significant effect of channel angle was found by ANOVA. An alpha level of 0.05 was used as the criterion for rejecting the null hypothesis of equal means. All statistical tests were performed using JMP IN software (version 6.0.0; SAS Institute, Cary, NC).
Protozoan observations.
In all experiments, protozoans were initially introduced into one of two identical wells (referred to as the “source well” below) and were free to locate, enter, and migrate along the microchannel to the target well. No bacterial food or other enticement was given to promote migration. Protozoans were observed and their movements and interactions were quantified using an automated inverted microscope system described elsewhere (22). Micrographs of protozoans in microfluidic devices are shown in Fig. 1 and elsewhere (22). In agreement with the observations of Holyoak and Lawler (12), both species of protozoans quickly emigrated from the source well, explored new spaces, and dispersed toward the target well, although there was no food in the target well. We found it quite interesting that a planktonic species like Uronema sp. would so readily locate and enter narrow channels from a larger reservoir. While the physical structure of our microfluidic devices clearly differs from structures found in nature, our observations suggest that protozoans may interact more with microstructured habitat features than is generally believed.
By tracking individual protozoans moving within the device, we also observed several interesting behaviors. Both species altered their physical dimensions to some extent to pass the bends in the bent channels. Uronema, in particular, showed great flexibility and an ability to gain access to restricted bent crevices by elongating its dimensions. Interactive behavior was also observed for E. vannus, especially in 20-μm-wide channels, where the movement of individuals was most restricted. When several E. vannus individuals were in the observation area at the same time, individuals pushed other ciliates in front of them forward, possibly to move the obstruction past the channel bend, and also moved backwards frequently, possibly to expel individuals behind them from the channel.
Swimming speed.
Swimming speeds were measured in straight channel segments before and after channel bends for five Uronema and E. vannus individuals for each microfluidic design (Table 1). The speed per individual was computed from the average of 20 separate measurements for the distance traveled in 0.5 s.
TABLE 1.
Average measured swimming speeds of protozoans in channel angle devicesa
| Protozoan | Channel width (μm) | Ratio of channel cross-sectional area to protozoan cross-sectional areab | Swimming speed (μm/s) with the following channel bend anglec:
|
|||
|---|---|---|---|---|---|---|
| 0° | 60° | 90° | 120° | |||
| Uronema | 20 | 12.52 | 427 ± 64 | 404 ± 47 | 389 ± 58 | 411 ± 39 |
| 10 | 6.26 | 167 ± 33 | 73 ± 31 | 87 ± 20 | 147 ± 28 | |
| E. vannus | 30 | 1.23 | 70 ± 16 | 58 ± 9.3 | 59 ± 9.2 | 65 ± 28 |
| 20 | 0.82 | 43 ± 11 | 19 ± 8.7 | 21 ± 9.5 | 27 ± 11 | |
| 10 | 0.41 | NAd | 6.2 ± 1.3 | NA | NA | |
Swimming speeds were measured 20 times for five randomly picked individuals in straight channels before and after the channel bends. The values are means ± standard deviations (n = 100).
Ratios may be <1 for two reasons: (i) an individual may have had a smaller unconstrained cross-sectional area than the average for the population and (ii) some individuals were observed to elongate in narrow channels.
The turn angle is expressed from the perspective of the protozoan, where a 0o turn is a straight channel with no bend.
NA, not applicable (the protozoan stayed in the source well and did not enter the channels within 2 days).
Consistent with a previous study (22), the small protozoan (Uronema) moved at a significantly higher velocity than the large protozoan (E. vannus) (Table 1) (P < 0.0001). For example, in 20-μm-wide channels bent at a 90° angle, the swimming speed of Uronema was nearly 20 times greater than that of E. vannus. For both Uronema and E. vannus, the swimming speeds were greater in wider channels (P < 0.0001). In the case of E. vannus, the average swimming speeds before and after a 60° bend were 58 μm s−1 in a 30-μm-wide channel, 19 μm s−1 in a 10-μm-wide channel, and 6.2 μm s−1 in a 20-μm-wide channel. Both protozoans had slower swimming speeds when the channel dimensions were smaller (Table 1). A key parameter impacting swimming speed was the ratio of the cross-sectional area of an individual protozoan to the cross-sectional area of the microchannel (P < 0.0001).
Surprisingly, the influence of channel angle on protozoan swimming speed was also significant (P < 0.0001). The swimming speeds of both protozoan species were greater in devices without channel bends than in straight segments that were the same size before and after a channel bend. We speculate that the channel bend structure may interact hydrostatically or hydrodynamically with individual protozoans, affecting the swimming speed of the protozoans in the straight channels before and after the bend. Another possible explanation is that stretching and squeezing through the twisted channel bends may damage protozoan cirri or cilia and therefore decrease the swimming speed. Finally, it is possible that the subset of individuals in a population which have traversed a narrow bend may have a different average swimming speed than the parent population. Additional studies are needed to investigate any underlying tradeoff in swimming speed versus the efficiency of passage through a constriction.
The swimming speeds reported here are several orders of magnitude less than the speeds reported in other studies without microscale physical structure. Fenchel (9) found that the speeds of starved E. vannus and feeding E. vannus were 430 and 220 μm s−1, respectively. Jonsson and Johansson (13) reported that the average speed of Euplotes sp. in still water was 1.62 mm s−1 without food and 1.39 mm s−1 with food. In natural habitats, however, microstructures are ubiquitous and have been shown to restrict protozoan movements. The swimming speed parameters reported here may be more appropriate for modeling the dispersal of protozoans in habitats with microstructures.
Dispersal experiments.
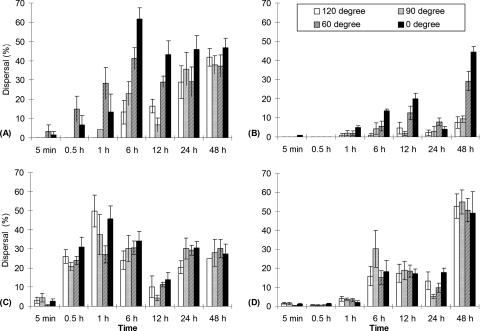
To investigate how populations of protozoans dispersed in the microfluidic devices over time, approximately 15 protozoans from the same monoxenic culture were added to the source wells of at least five replicates for each channel angle-channel width design combination. At different times following introduction (5 and 30 min and 1, 6, 12, 24, and 48 h), all protozoans in the devices were fixed with formaldehyde (final concentration, 3.7%), and the dispersal of protozoans, expressed as a percentage, was quantified by determining the proportions of all the protozoans that were in the target well and in the portion of the channel beyond the midpoint bend. Since the protozoans were free to travel in both directions, both from the source well to the target well and back from the target well to the source well, an oscillating pattern for the percent dispersal versus time was obtained (Fig. 2). The dispersal percentage increased from 0% at the start of the experiment and approached 50% (i.e., even dispersal) over time.
FIG. 2.
Dispersal of (A) E. vannus in 30-μm-wide microchannels, (B) E. vannus in 20-μm-wide microchannels, (C) Uronema in 20-μm-wide microchannels, and (D) Uronema in 10-μm-wide microchannels. The error bars indicate one standard deviation (n = 5).
For a given device, the percent dispersal of Uronema was higher than that of E. vannus at all sampling times. Protozoan species was a significant parameter impacting the percent dispersal (P < 0.0001). For instance, in 20-μm-wide channels with a 60° angle, no E. vannus had passed the channel bends at 30 min after introduction, while 24% of Uronema individuals had passed. The dispersal of protozoans varied and was positively correlated with channel width (P < 0.0001 for both species). In the case of E. vannus in channels with 90° bends, 23% of the individuals had passed the 30-μm-wide bends at 6 h. When the channel width was decreased to 20 μm, the percent dispersal dropped to 4.3%. No E. vannus individuals could enter 10-μm-wide channels.
Estimating the dispersal rate and dispersal time.
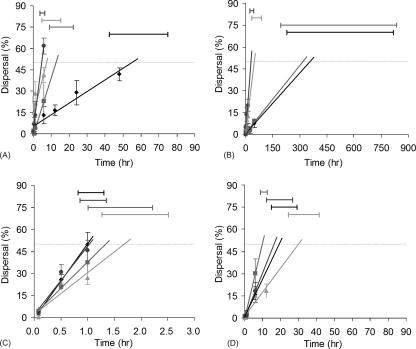
To estimate the dispersal rate for a given experimental treatment, we focused on the time when the percent dispersal was strictly increasing for all turning angles in each channel width treatment. (For example, the selected time intervals for the straight channel [0° bend] were the first 6 h for E. vannus in 30-μm-wide channels, the first 12 h for E. vannus in 20-μm-wide channels, the first 1 h for Uronema in 20-μm-wide channels, and the first 6 h for Uronema in-10 μm-wide channels.) The percent dispersal within each interval was regressed against time (with the y intercept fixed at 0). The resulting slope of the regression line was defined as the relative dispersal rate (in h−1), and the intersection of the regression with 50% dispersal was operationally defined as the dispersal time (in h) (Fig. 3).
FIG. 3.
Regressions of percent dispersal versus time showing the dispersal rate (slope) and dispersal time (intercept with 50% dispersal) for (A) E. vannus in 30-μm-wide microchannels, (B) E. vannus in 20-μm-wide microchannels, (C) Uronema in 20-μm-wide microchannels, and (D) Uronema in 10-μm-wide microchannels. Symbols correspond to bend angle: 0°, circles; 60°, triangles; 90°, squares; and 120°, diamonds. The horizontal bars indicate the 95% confidence intervals for calculation of the intercepts.
Impact of channel properties on the dispersal rate.
The ratio of channel cross-sectional area to protozoan cross-sectional area had a statistically significant effect on the log of dispersal time for both species (E. vannus, P = 0.008; Uronema, P = 0.001) (Table 2). This result confirms that pore structure impacts protozoan transport at the population level. Previous work of Adl (1) suggested that the average migration rate of protozoans varies with pore size in soil columns. The smallest pore size used in that study, however, was 500 μm. Our work extended the results by investigating protozoan dispersal in much smaller channels.
TABLE 2.
Calculated average times required for dispersal beyond the channel bend (the time required to locate and enter the channel, to move along the channel, and to pass the bend) and times required for passing the bend alone
| Protozoan | Channel width (μm) | Dispersal time (h) with the following channel bend anglea:
|
Bend time (h) with the following channel bend angleb:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0° | 60° | 90° | 120° | 60° | 90° | 120° | ||
| Uronema | 20 | 1.0 | 1.7 | 1.3 | 1.0 | |||
| 10 | 17 | 30 | 11 | 20 | ||||
| E. vannus | 30 | 4.8 | 7.4 | 13 | 55 | 2.6 | 7.9 | 50 |
| 20 | 30 | 48 | 320 | 360 | 19 | 290 | 330 | |
The dispersal times were predicted from a linear regression of protozoan dispersal versus elapsed time for each experimental treatment.
The bend time is the difference between the dispersal time in the bent channel and the dispersal time in the corresponding straight channel.
The channel angle had a different effect on the dispersal rate of E. vannus than it had on the dispersal rate of Uronema. In both 30-μm-wide channels and 20-μm-wide channels, the relationship of the dispersal rates for E. vannus in channels with different bend angles was 0° ≈ 60° > 90° ≈ 120°, where differences between the 0° and 60° values and between the 90° and 120° values were not statistically significant. In contrast, channel angle was not a significant factor affecting the dispersal rate of Uronema (P = 0.102). This may have been the result of the difference in relative channel dimensions. In all devices, Uronema individuals were able to pass one another in narrow channels, while Euplotes individuals were not; thus, the observed migration of E. vannus was akin to a multistage process occurring in sequence, while the migration of Uronema was akin to a multistage process occurring in parallel. Future work should compare the dispersal properties of different populations in systems with similar channel/protozoan cross-sectional area ratios to see if there are differences in migration between species or if what we observed was most likely the influence of simultaneous versus sequential migration ability.
Computing the bend time.
The average time required for an individual to disperse beyond the bend (dispersal time) can be thought of as the sum of the time needed to find and enter the channel entrance from the source well (scouting time), the time swimming in the straight channel segments before and after the bend (channel time), and the time required to pass the channel bend (bend time). Since straight channels and bent channels had identical entrance areas and straight channel segment dimensions, the scouting time and channel time were the same for straight channels and bent channels for a given species and given microfluidic channel dimensions. Therefore, the bend time in a given treatment could be estimated by subtracting the dispersal time in the straight channel from the corresponding time for a bent channel (Table 2).
Comparing bend times.
The difference in the dispersal time of Uronema between bent channels and straight channels was not significant, resulting in negligible time delays due to channel bends. E. vannus, on the other hand, took a long time to pass the channel bends (Table 2). For instance, the bend time was 330 h in 20-μm-wide channels with a 120° bend, which was more than 90% of the dispersal time. Based on microscopic observations, the bend time in bent channels was due to the protozoans swimming back and forth before the bend. Since the swimming speed for E. vannus in 20-μm-wide channels was 19 ± 8.7 μm/s, we estimated that, on average, individuals traveled the equivalent of 6.2 cm before passing the bend. In our devices, on average, this corresponds to 14 trips of the entire distance between the source well and the bend. However, we observed that protozoans of both species in all devices changed direction after approximately 350 μm (data not shown), so on average E. vannus individuals circulated before a channel bend nearly 200 times. This result is an elegant illustration of how microstructured settings provide refuge and directly impose foraging costs on predators.
Conclusions.
Our results suggest that both the number and the characteristics (i.e., acute versus obtuse turning angles) of bends influence the dispersal of protozoans in model microstructured systems. Our approach allows us to work in a well-defined setting to directly observe protozoan behaviors and to collect controlled, reproducible data. We provide evidence that channel structure strongly affects the dispersal rate of populations and the swimming speed of individuals. The empirical results and qualitative observations obtained in this study provide useful insights into the behavior of protozoans in physically structured habitats and also provide a foundation for understanding migration behavior in more complex systems.
Acknowledgments
This study was supported by grants 0120453 and 0649883 from the National Science Foundation and in part by the Vanderbilt Institute for Integrative Biosystems Research and Education through a grant from the Vanderbilt University Academic Venture Capital Fund.
We thank David Gruber for providing protozoan samples and technical assistance with culturing protozoans. We also thank Ronald Reiserer and Philip Samson for assistance with the microfabrication process.
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Adl, S. M. 2007. Motility and migration rate of protozoa in soil columns. Soil Biol. Biochem. 39:700-703. [Google Scholar]
- 2.Arah, J. R. M., and A. J. A. Vinten. 1995. Simplified models of anoxia and denitrification in aggregated and simple-structured soils. Eur. J. Soil Sci. 46:507-517. [Google Scholar]
- 3.Babel, U., H. Vogel, M. Krebs, G. Leithold, and C. Hemmann. 1995. Morphological investigations on genesis and functions of soil fabric—places, pathways, boundaries, p. 11-30. In K. H. Hartge and B. A. Stewart (ed.), Soil structure: its development and function. CRC Press, Inc., Boca Raton, FL.
- 4.Caron, D. A., P. G. Davis, L. P. Madin, and J. M. Sieburth. 1982. Heterotrophic bacteria and bacterivorous protozoa in oceanic macroaggregates. Science 218:795-797. [DOI] [PubMed] [Google Scholar]
- 5.Darbyshire, J. F. 2005. The use of soil biofilms for observing protozoan movement and feeding. FEMS Microbiol. Lett. 244:329-333. [DOI] [PubMed] [Google Scholar]
- 6.Davoll, P. J., and M. W. Silver. 1986. Marine snow aggregates: life history sequence and microbial community of abandoned larvacean houses from Monterey Bay, California. Mar. Ecol. Prog. Ser. 33:111-120. [Google Scholar]
- 7.Dini, F., and D. Nyberg. 1999. Growth rates of marine ciliates on diverse organisms reveal ecological specializations within morphospecies. Microb. Ecol. 37:13-22. [DOI] [PubMed] [Google Scholar]
- 8.Fenchel, T. 1987. Ecology of protozoa: the biology of free-living phagotrophic protists. Science Tech Publishers, Madison, WI.
- 9.Fenchel, T. 2004. Orientation in two dimensions: chemosensory motile behaviour of Euplotes vannus. Eur. J. Protistol. 20:49-55. [Google Scholar]
- 10.Ghannoum, M. A., and G. A. O'Toole (ed.). 2004. Microbial biofilms. ASM Press, Washington, DC.
- 11.Heissenberger, A., G. G. Leppard, and G. J. Herndl. 1996. Ultrastructure of marine snow. 11. Microbiological considerations. Mar. Ecol. Prog. Ser. 135:299-308. [Google Scholar]
- 12.Holyoak, M., and S. P. Lawler. 1996. Persistence of an extinction-prone predator-prey interaction through metapopulation dynamics. Ecology 77:1867-1879. [Google Scholar]
- 13.Jonsson, P. R., and M. Johansson. 1997. Swimming behaviour, patch exploitation and dispersal capacity of a marine benthic ciliate in flume flow. J. Exp. Mar. Biol. Ecol. 215:135-153. [Google Scholar]
- 14.Killham, K., M. Amato, and J. N. Ladd. 1993. Effect of substrate location in soil and soil pore-water regime on carbon turnover. Soil Biol. Biochem. 25:57-62. [Google Scholar]
- 15.Krembs, C., R. Gradinger, and M. Spindler. 2000. Implications of brine channel geometry and surface area for the interaction of sympagic organisms in Arctic sea ice. J. Exp. Mar. Biol. Ecol. 243:55-80. [Google Scholar]
- 16.Kuiper, M. W., B. A. Wullings, A. D. L. Akkermans, R. R. Beumer, and D. van der Kooij. 2004. Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl. Environ. Microbiol. 70:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y. S., E. J. LeBoeuf, P. K. Basu, and S. Mahadevan. 2005. Stochastic modeling of the permeability of randomly generated porous media. Adv. Water Resour. 28:835-844. [Google Scholar]
- 18.Meyer, R. L., R. J. Zeng, V. Giugliano, and L. L. Blackall. 2005. Challenges for simultaneous nitrification, denitrification, and phosphorus removal in microbial aggregates: mass transfer limitation and nitrous oxide production FEMS Microbiol. Ecol. 52:329-338. [DOI] [PubMed] [Google Scholar]
- 19.Postma, J., C. H. Hok-A-Hin, and J. A. van Veen. 1990. Role of microniches in protecting introduced Rhizobium leguminosarum biovar trifolii against competition and predation in soil. Appl. Environ. Microbiol. 56:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sexstone, A. J., N. P. Revsbech, T. B. Parkin, and J. M. Tiedje. 1985. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci. Soc. Am. J. 49:645-651. [Google Scholar]
- 21.Vogel, H. J. 2000. A numerical experiment on pore size, pore connectivity, water retention, permeability, and solute transport using network models. Eur. J. Soil Sci. 61:99-105. [Google Scholar]
- 22.Wang, W., L. M. Shor, E. J. LeBoeuf, J. P. Wikswo, and D. S. Kosson. 2005. Mobility of protozoa through narrow channels. Appl. Environ. Microbiol. 71:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf, G., J. G. Crespo, and M. A. M. Reis. 2002. Optical and spectroscopic methods for biofilm examination and monitoring. Rev. Environ. Sci. Biotechnol. 1:227-251. [Google Scholar]
- 24.Wright, D. A., K. Killham, L. A. Glover, and J. Prosser. 1995. Role of pore size location in determining bacterial activity during predation by protozoa in soil. Appl. Environ. Microbiol. 61:3537-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]